Abstract
Mitochondrial dysfunction has been implicated in the pathogenesis of Huntington disease (HD), a primarily neurodegenerative disorder that results from an expansion in the polymorphic trinucleotide CAG tract in the HD gene. In order to evaluate whether mitochondrial DNA (mtDNA) variation contributes to HD phenotype we genotyped 13 single nucleotide polymorphisms (SNPs) that define the major European mtDNA haplogroups in 404 HD patients. Genotype-dependent functional effects on intracellular ATP concentrations were assessed in peripheral leukocytes. In patients carrying the most common haplogroup H (48.3%), we demonstrate a significantly lower age at onset (AO). In combination with PGC-1alpha genotypes, 3.8% additional residual variance in HD AO can be explained. Intracellular ATP concentrations in HD patients carrying the cytochrome c oxidase subunit I (CO1) 7028C allele defining haplogroup H were significantly higher in comparison to non-H individuals (mean ± SEM, 599 ± 51.8 ng/ml, n = 14 vs. 457.5 ± 40.4 ng/ml, p = 0.03, n = 9). In contrast, ATP concentrations in cells of HD patients independent from mtDNA haplogroup showed no significant differences in comparison to matched healthy controls. Our data suggest that an evolutionarily advantageous mitochondrial haplogroup is associated with functional mitochondrial alterations and may modify disease phenotype in the context of neurodegenerative conditions such as HD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Huntington disease (HD) is an autosomal dominantly transmitted, progressive neurodegenerative disease associated with expansions in a stretch of CAG repeats in the 5' part of the HD gene encoding the protein huntingtin (htt) [1]. The length of the polyglutamine tract is the most critical factor that determines age at onset (AO), accounting for more than half of the overall variance [2–4]. Yet, there remains considerable variation up to more than 40 years in AO of neurological symptoms in individuals with identical repeat lengths. Several candidate modifier genes of HD have already been described in independent studies [5–9]. Recently, variations in the PPARGC1A gene that codes for the peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1α), a master regulator of mitochondrial genes, were shown to exert modifying effects on the AO in HD [10, 11].
Further evidence for a pathogenetically relevant role of mitochondrial dysfunction with biochemical, morphological and functional mitochondrial abnormalities has been provided in cell culture models, animal models and in HD patients [12, 13]. NMR-spectroscopy demonstrated increased lactate levels in cerebral cortex and basal ganglia and impaired ATP production in muscle of symptomatic HD patients and pre-symptomatic mutation carriers [14–16]. Additionally, patients show unintended weight loss associated with CAG repeat length, thus suggesting hypermetabolic state and/or reduced efficiency of mitochondria [17].
Whereas mostly nuclear genes have been intensively investigated as modifiers thus far, the role of the mitochondrial genome has been neglected. Yet, particular combinations of certain mt variations define mtDNA haplogroups, which tend to be associated with subtle differences in oxidative phosphorylation (OXPHOS) capacity and the generation of reactive oxygen species (ROS) [18]. Specific European and Asian mtDNA haplogroups were found to be associated with longevity as well as with other age-related complex traits such as Alzheimer and Parkinson diseases (AD, PD) [19–22]. Therefore, the goal of the present study was to assess the contribution of mtDNA SNPs or haplogroups to the AO of HD.
Materials and methods
Patient recruitment and diagnosis
The study population consisted of 404 unrelated patients with the clinical and genetic diagnosis of HD, recruited from the Huntington Center NRW, Bochum (Germany). Clinical assessment and determination of the motor AO was performed exclusively by two experienced neurologists of the Center. The expanded CAG repeats ranged from 40 to 66 trinucleotide units and AO ranged from 16 to 76 years of age, with a mean of 45 years. The normal CAG repeats ranged from 10 to 32 trinucleotide units. HD CAG repeat sizes were determined by polymerase chain reaction using an assay counting the perfectly repeated (CAG)n units. Informed consent was obtained from all patients or if necessary, their legal guardian, respectively, and healthy controls (HC). The studies were performed in a manner that fully complies with the Code of Ethics of the World Medical Association (Declaration of Helsinki) and was approved by the Ruhr-University Bochum ethics review board.
Genotyping
In order to analyse mitochondrial haplotypes, we used 13 core mtSNPs that define the major European haplogroups: SNPs 1719, 3010, 4580, 6776, 7028, 8251, 9055, 10398, 11719, 12308, 13368, 13708 and 16391. PCR and restriction enzyme digestions on the mtDNA were performed to ascertain the mitochondrial haplogroup of each patient. Additionally, nuclear polymorphisms were tested. The common UCP2–866G/A polymorphism [23] (rs659366) was tested for the whole group and rs7665116 in PGC-1alpha [11], rs1969060 in GRIN2A, rs1806201 in GRIN2B [24], rs5880308 in ASK1 and rs2521354 in MAP2K6 [7] were caught up for parts of the study population. Primers, PCR conditions and restriction enzymes used for RFLP analysis are available upon request.
Assessment of intracellular ATP level
Sodium heparin blood was obtained from a subgroup of 23 HD patients and 48 age-/sex-matched healthy controls devoid of acute infections (clinically, white blood cell count, C-reactive protein). After mitogen stimulation with phytohemagglutinin (PHA, 2.5 μg/ml) for 15 h (37°C, 5% CO2), CD4+ cells were immuno-selected with anti-human CD4-coated magnetic beads (Dynal, Oslo, Norway). After cell lysis ATP concentrations were measured in a luciferase-activity based assay according to the manufacturer’s specifications in a FDA-approved assay (ImmunoKnow™, Cylex, Columbia, USA). CD4/CD8 cell counts were determined by flow cytometry (FACS-Calibur, Multiset IMK Kit, Becton Dickinson, Heidelberg, Germany).
Statistical analysis
Statistical analyses were performed using SPSS software, release 15 (SPSS, Chicago, IL, USA). P values less than 0.05 were considered significant. Variability in AO attributable to the CAG repeat number was controlled by linear regression using the logarithmically transformed AO as the dependent variable and each haplogroup with all other haplogroups pooled into a single group as independent variables. Parametric t test was performed for ATP production in CD4+ cells (GraphPad Prism, CA, USA), p < 0.05 (*) was considered significant. Data are given as mean ± standard error.
Results
Association of mt haplogroup H with earlier age at onset
We analysed 13 core mtSNPs that define the common European haplotypes. Overall, frequencies did not differ substantially from those reported in previous studies by other authors who analysed different European populations [25, 26] (48.3% for haplogroup H, 14.4% for U, 8.2% for T, 5.9% for J1b, 1.7% for J, 4.5% for K, 4% for V, 2.7% for W/I, 2.5% for preH/HV, 1% for X and 6.9% for others).
The dependence of AO on CAG repeat number in the HD gene was assessed by linear regression. The best fit estimated by the R 2 value was obtained after log transformation of AO. The CAG repeat explained ∼73% of the variance in AO. Inclusion of the interaction term between mutant and normal CAG repeat sizes did not raise the adjusted R 2 value. Multiple regression models were used to test the effect of the different haplogroups. Addition of haplogroup H (cytochrome c oxidase subunit I (CO1) 7028C, n = 195) vs. all other haplogroups to the effect of CAG repeat lengths resulted in significant increase of the R 2 value (0.736 to 0.740, p = 0.006, Table 1). Therefore, in all subjects belonging to haplogroup H, SNPs corresponding to the frequent sub-haplogroups H1 (G3010A) and H3 (C6776T) were genotyped. But addition of H1 (n = 76) and H3 (n = 15) to the model resulted in no increase of the R 2 value (Table 1).
The same applied for the addition of other frequent haplogroups. Yet, lack of statistical power for analysis of the infrequent haplogroups and sub-haplogroups may account for the lack of significance observed. Addition of haplogroup H* (excluding H1 and H3) resulted in significant increase of the R 2 value (0.736 to 0.743, p = 0.003, Table 1). In contrast to the CO1 7028 genotypes, which can distinguish between haplogroup H and non-H individuals, most of the other mtDNA polymorphisms are found in several haplogroups. In order to test for their isolated influence, the genotype for each SNP was coded as 0 or 1, representing a two-level categorical variable. Yet, beside the effect of CO1 C7028T, no other mt genotype explained a significant part of the variability in AO. Additionally, interaction analyses were performed by using multiple regression models based on the combinations of described HD modifier SNPs with the C7028T variation. No significant interaction was detected with the SNPs in GRIN2A, GRIN2B, ASK1 and MAP2K6 and UCP2 genes. Yet, a significant interaction was found concerning SNP rs7665116 in the PGC-1alpha gene. In combination, PGC-1alpha and haplogroup H or H* explain more variability in AO than isolated analyses (H*: 0.736 to 0.746, p = <0.0005;Table 1). Together, these genotype variations explain 3.8% additional variance in HD AO.
Mt haplogroup H is associated with high ATP concentrations in HD patients
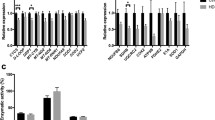
No significant differences were seen when comparing the mean intracellular ATP concentrations in unstimulated and PHA-stimulated CD4+ cells of all HD patients (543.7 ± 23.8 ng/ml) with age- and sex-matched healthy controls (489.8 ± 30 ng/ml). There was no significant correlation between ATP concentration, CAG triplet repeat length, CAG index, years of age, years of disease or clinical severity as defined by UHDRS (Unified Huntington’s Disease Rating Scale). One homozygous HD patient with 45/45 CAG repeats revealed ATP concentrations within the lowest range observed in these populations (321.9 ng/ml). ATP concentrations remained stable over an interval of 1–67 days (eight HD patients, five healthy controls). When correlating the ATP concentrations with different mitochondrial genotypes, HD patients carrying the CO1 7028C allele defining haplogroup H, showed significantly higher ATP concentrations (599 ± 51.8 ng, n = 14) than non-H individuals (457.5 ± 40.4 ng/ml, p = 0.03; Fig. 1). These genotype specific differences were not observed in healthy donors (H: 478.7 ± 30.1 ng/ml, n = 18 vs. non-H: 496.4 ± 30.4 ng/ml, n = 30). Neither in HD patients nor in controls was a significant association between ATP concentrations and the other genotypes obvious.
Discussion
The present study shows that mtDNA haplogroup H (7028C) is significantly associated with an earlier AO in HD patients, relative to all other mt haplogroups (7028 T). Furthermore, when comparing the mean ATP concentrations in peripheral leukocytes, significant differences were observed in HD patients belonging to the H haplogroup compared to patients with other (non-H) haplogroups, who showed significantly lower ATP levels.
Since imaging studies implicated bioenergetic defects in HD pathogenesis, a number of studies have examined whether alterations in mitochondrial respiration contribute to disease mechanisms. Yet, bioenergetic mitochondrial dysfunction is not restricted to the CNS, but has also been demonstrated in lymphoblastoid cells from HD patients and in early asymptomatic mutation carriers by 31P-MRS of skeletal muscle with significant reduction of ATP production [15, 16, 27]. Furthermore, HD mitochondria from lymphoblasts show increased vulnerability to mitochondrial toxins, including 3-nitropropionic acid targeting complex II and sodium cyanide, targeting complex IV, in a polyglutamine length-dependent manner [28, 29]. Since in lymphoblasts bioenergetic defects are not secondary to tissue damage, they likely result from direct influence of mutant htt expression on mitochondrial function.
Interestingly, by correlating the ATP values with the different mitochondrial genotypes, HD patients carrying the CO1 7028C allele defining haplogroup H showed significantly higher ATP concentrations than non-H individuals. This effect of the 7028 variation, a synonymous SNP located in the CO1 gene with unknown consequence on protein function, was not obvious in healthy controls. Therefore, against the background of a neurodegenerative condition, potential haplogroup differences in the OXPHOS metabolism become evident. Given the relatively small sample sizes, the association between haplogroups, AO and ATP production should be regarded as preliminary. Nevertheless, these data combined with published evidence may trigger cautious speculations: Considering the hypothesis that climatic adaptation has influenced the geographic distribution of mtDNA diversity, some of the mutations characterising European mtDNA haplogroups may, therefore, influence the coupling efficiency of OXPHOS and heat generation [18]. Assuming that haplogroup H mtDNA variations result in tightly coupled OXPHOS, they would generate more ATP, thereby increasing ROS production and apoptosis, which could be reflected in earlier AO. We therefore suggest that under conditions of impaired OXPHOS, cells harbouring the mitochondrial haplogroup H have better potential to compensate energetic bottlenecks. Maybe selection during epidemics or longer periods of exposure to infectious diseases has contributed to the rapid expansion of the relatively young haplogroup H in European populations rather than climatic adaptation. This scenario could be beneficial in conditions of temporary limited OXPHOS (e.g., septic shock and stroke) [26, 30, 31], but it would be disadvantageous in the case of chronic (neuro-) degenerative processes with increased ROS-mediated cell death via augmented ATP generation [32].
Contrary to our report, a previous study did not support any association between mtDNA haplogroups and HD [33]. Yet, this apparent inconsistency may be the result of lack of statistical power due to smaller sample size in that study.
Consistent with our results, haplogroup H is reported to be negatively associated with age-related, neurodegenerative complex traits (e.g., AD, PD) as compared to haplogroups J, T, K and U [34–37]. In the case of haplogroups J, T, K and U, polymorphisms partially uncoupling OXPHOS would cause the mitochondria to “burn” metabolites excessively in order to generate heat, thus leaving fewer excess electrons for ROS generation. This could explain their association with longevity and protection against neurodegenerative diseases. On the other hand, the same partial uncoupling polymorphisms would reduce the efficiency of ATP production, thereby explaining reduced sperm mobility [38] and the predisposition to diseases caused by ATP deficiency (e.g., Leber’s hereditary optic neuropathy).
Moreover, it has been shown that cytoplasmic hybrids repopulated with mtDNA belonging to haplogroup H or J differentially express IL-1β, TNFR2, and IL-6, not only at basal but also under oxidative stress conditions [39]. These findings provide initial experimental evidence that genetic mitochondrial background appears to modulate the expression of nuclear genes related to stress response. Our findings also suggest a more pervasive effect like interplay between mitochondrial and nuclear genome. Variations in the nuclear gene PGC-1alpha implicated in mitochondrial biogenesis and in the polymorphism that defines haplogroup H may contribute interactively to systemic metabolic disturbances relevant to the pathogenesis of HD, thus highlighting the complex interrelation of mitochondrial dysfunction in HD. Our findings warrant more detailed assessment also in other age-related and neurodegenerative diseases in order to quantify the effect specific to mtDNA haplogroups.
References
The Huntington’s Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72:971–983
Snell RG, MacMillan JC, Cheadle JP, Fenton I, Lazarou LP, Davies P, MacDonald ME, Gusella JF, Harper PS, Shaw DJ (1993) Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington’s disease. Nat Genet 4:393–397
Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman MA et al (1993) The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nat Genet 4:398–403
Duyao M, Ambrose C, Myers R, Novelletto A, Persichetti F, Frontali M, Folstein S, Ross C, Franz M, Abbott M et al (1993) Trinucleotide repeat length instability and age of onset in Huntington’s disease. Nat Genet 4:387–392
Andresen JM, Gayán J, Cherny SS, Brocklebank D, Alkorta-Aranburu G, Addis EA; US-Venezuela Collaborative Research Group, Cardon LR, Housman DE, Wexler NS (2007) Replication of twelve association studies for Huntington’s disease residual age of onset in large Venezuelan kindreds. J Med Genet 44:44–50
Arning L, Saft C, Wieczorek S, Andrich J, Kraus PH, Epplen JT (2007) NR2A and NR2B receptor gene variations modify age at onset in Huntington disease in a sex-specific manner. Hum Genet 122:175–182
Arning L, Monté D, Hansen W, Wieczorek S, Jagiello P, Akkad DA, Andrich J, Kraus PH, Saft C, Epplen JT (2008) ASK1 and MAP2K6 as modifiers of age at onset in Huntington’s disease. J Mol Med 86:485–490
Metzger S, Rong J, Nguyen HP, Cape A, Tomiuk J, Soehn AS, Propping P, Freudenberg-Hua Y, Freudenberg J, Tong L et al (2008) Huntingtin-associated protein-1 is a modifier of the age-at-onset of Huntington’s disease. Hum Mol Genet 17:1137–1146
Gusella JF, Macdonald ME (2009) Huntington’s disease: the case for genetic modifiers. Genome Med 8:80
Weydt P, Soyal SM, Gellera C, Didonato S, Weidinger C, Oberkofler H, Landwehrmeyer GB, Patsch W (2009) The gene coding for PGC-1alpha modifies age at onset in Huntington’s disease. Mol Neurodegener 4:3
Taherzadeh-Fard E, Saft C, Andrich J, Wieczorek S, Arning L (2009) PGC-1 alpha as modifier of onset age in Huntington disease. Mol Neurodegener 4:10
Lin MT, Beal MF (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443:787–795
Bossy-Wetzel E, Petrilli A, Knott AB (2008) Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci 31:609–616
Jenkins BG, Koroshetz WJ, Beal MF, Rosen BR (1993) Evidence for impairment of energy metabolism in vivo in Huntington’s disease using localized 1H NMR spectroscopy. Neurology 43:2689–2695
Lodi R, Schapira AH, Manners D, Styles P, Wood NW, Taylor DJ, Warner TT (2000) Abnormal in vivo skeletal muscle energy metabolism in Huntington’s disease and dentatorubropallidoluysian atrophy. Ann Neurol 48:72–76
Saft C, Zange J, Andrich J, Müller K, Lindenberg K, Landwehrmeyer B, Vorgerd M, Kraus PH, Przuntek H, Schöls L (2005) Mitochondrial impairment in patients and asymptomatic mutation carriers of Huntington’s disease. Mov Disord 20:674–679
Aziz NA, van der Burg JM, Landwehrmeyer GB, Brundin P, Stijnen T; EHDI Study Group, Roos RA (2008) Weight loss in Huntington disease increases with higher CAG repeat number. Neurology 71:1506–1513
Wallace DC, Brown MD, Lott MT (1999) Mitochondrial DNA variation in human evolution and disease. Gene 238:211–230
Niemi AK, Moilanen JS, Tanaka M, Hervonen A, Hurme M, Lehtimäki T, Arai Y, Hirose N, Majamaa K (2005) A combination of three common inherited mitochondrial DNA polymorphisms promotes longevity in Finnish and Japanese subjects. Eur J Hum Genet 13:166–170
van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Haines JL, Koller WC, Lyons K et al (2003) Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet 72:804–811
van der Walt JM, Dementieva YA, Martin ER et al (2004) Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett 365:28–32
Takasaki S (2008) Mitochondrial SNPs associated with Japanese centenarians, Alzheimer’s patients, and Parkinson’s patients. Comput Biol Chem 32:332–337
Vogler S, Goedde R, Miterski B, Gold R, Kroner A, Koczan D, Zettl UK, Rieckmann P, Epplen JT, Ibrahim SM (2005) Association of a common polymorphism in the promoter of UCP2 with susceptibility to multiple sclerosis. J Mol Med 83:806–811
Arning L, Kraus PH, Valentin S, Saft C, Andrich J, Epplen JT (2005) NR2A and NR2B receptor gene variations modify age at onset in Huntington disease. Neurogenetics 6:25–28
Wiesbauer M, Meierhofer D, Mayr JA, Sperl W, Paulweber B, Kofler B (2006) Multiplex primer extension analysis for rapid detection of major European mitochondrial haplogroups. Electrophoresis 27:3864–3868
Rosa A, Fonseca BV, Krug T, Manso H, Gouveia L, Albergaria I, Gaspar G, Correia M, Viana-Baptista M, Simões RM et al (2008) Mitochondrial haplogroup H1 is protective for ischemic stroke in Portuguese patients. BMC Med Genet 9:57
Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT (2002) Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci 5:731–736
Sawa A, Wiegand GW, Cooper J, Margolis RL, Sharp AH, Lawler JF Jr, Greenamyre JT, Snyder SH, Ross CA (1999) Increased apoptosis of Huntington disease lymphoblasts associated with repeat length-dependent mitochondrial depolarization. Nat Med 5:1194–1198
Seong IS, Ivanova E, Lee JM, Choo YS, Fossale E, Anderson M, Gusella JF, Laramie JM, Myers RH, Lesort M et al (2005) HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum Mol Genet 14:2871–2880
Baudouin SV, Saunders D, Tiangyou W, Elson JL, Poynter J, Pyle A, Keers S, Turnbull DM, Howell N, Chinnery PF (2005) Mitochondrial DNA and survival after sepsis: a prospective study. Lancet 366:2118–2121
Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, Poole JC, Lautenberger J, Sezgin E, Kingsley L, Goedert JJ, Vlahov D, Donfield S et al (2008) Mitochondrial DNA haplogroups influence AIDS progression. AIDS 22:2429–2439
Brand MD (2000) Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Exp Gerontol 35:811–820
Mancuso M, Kiferle L, Petrozzi L, Nesti C, Rocchi A, Ceravolo R, Orsucci D, Maluccio MR, Bonuccelli U, Filosto M et al (2008) Mitochondrial DNA haplogroups do not influence the Huntington’s disease phenotype. Neurosci Lett 444:83–86
De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafe M, Monti D, Baggio G, Bertolini S et al (1999) Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J 13:1532–1536
Rose G, Passarino G, Carrieri G, Altomare K, Greco V, Bertolini S, Bonafè M, Franceschi C, De Benedictis G (2001) Paradoxes in longevity: sequence analysis of mtDNA haplogroup J in centenarians. Eur J Hum Genet 9:701–707
Niemi AK, Hervonen A, Hurme M, Karhunen PJ, Jylhä M, Majamaa K (2003) Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum Genet 112:29–33
Ghezzi D, Marelli C, Achilli A, Goldwurm S, Pezzoli G, Barone P, Pellecchia MT, Stanzione P, Brusa L, Bentivoglio AR et al (2005) Mitochondrial DNA haplogroup K is associated with a lower risk of Parkinson’s disease in Italians. Eur J Hum Genet 13:748–752
Ruiz-Pesini E, Lapeña AC, Díez-Sánchez C, Pérez-Martos A, Montoya J, Alvarez E, Díaz M, Urriés A, Montoro L, López-Pérez MJ et al (2000) Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am J Hum Genet 67:682–696
Bellizzi D, Cavalcante P, Taverna D, Rose G, Passarino G, Salvioli S, Franceschi C, De Benedictis G (2006) Gene expression of cytokines and cytokine receptors is modulated by the common variability of the mitochondrial DNA in ybrid cell lines. Genes Cells 11:883–891
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Larissa Arning and Aiden Haghikia contributed equally to this work.
Rights and permissions
About this article
Cite this article
Arning, L., Haghikia, A., Taherzadeh-Fard, E. et al. Mitochondrial haplogroup H correlates with ATP levels and age at onset in Huntington disease. J Mol Med 88, 431–436 (2010). https://doi.org/10.1007/s00109-010-0589-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-010-0589-2