Abstract
Colorectal cancer (CRC) is the third most common form of cancer and the second cause of cancer-related death in the Western world, leading to 655,000 deaths worldwide per year (Jemal et al. in CA Cancer J Clin 56:106–130, 2006). Despite the emergence of new targeted agents and the use of various therapeutic combinations, none of the treatment options available is curative in patients with advanced cancer. A growing body of evidence is increasingly supporting the idea that human cancers can be considered as a stem cell disease. According to the cancer stem cell model, malignancies originate from a small fraction of cancer cells that show self-renewal and pluripotency and are capable of initiating and sustaining tumor growth (Boman and Wicha in J Clin Oncol 26:2795–2799, 2008). The cancer-initiating cells or “cancer stem cells” were first identified in hematologic malignancies and most recently in several solid tumors, including CRC. The hypothesis of stem cell-driven tumorigenesis in colon cancer raises questions as to whether current treatments are able to efficiently target the tumorigenic cell population that is responsible for tumor growth and maintenance. This review will focus on the different aspects of stem cell biology in the context of CRC, which might help to understand the mechanisms that give rise to tumor development and resistance to therapy. First, we will briefly revise the knowledge available on normal intestinal stem cells and recent advances in understanding crypt biology, which have led to new theory on the origins of colon adenomas and cancers. Then, we will summarize the evidence and current status on colon cancer stem cells, focusing on their relevance and promises for the treatment of colorectal carcinoma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Colonic crypt organization
The colon is organized into four histologically distinct layers. The epithelial layer, at the luminal surface, consists of a single sheet of columnar epithelial cells folded into finger-like invaginations that are supported by the lamina propria to form the functional unit of the intestine called crypts of Lieberkühn (Fig. 1a). Approximately 14,000 crypts per square centimeter are located in the adult human colon. Given a rate of 5 days for colonic epithelium renewal, it has been estimated that over 6 × 1014 colonocytes are produced during the individual lifetime [3, 4]. The terminally differentiated cells, which are found in the top third of the crypt, are continually extruded into the lumen. They are derived from multipotent stem cells located at the bottom of the crypt. During asymmetric division, these cells undergo self-renewal and generate a population of transit-amplifying cells that, upon migration upward the crypt, proliferate and differentiate into one of the epithelial cell types of the intestinal wall. There are three major terminally differentiated epithelial lineages in the colon: the colonocytes, also termed absorptive enterocytes; the mucus-secreting goblet cells; and the less abundant enteroendocrine cells (Fig. 1b). Finally, Paneth cells, functionally similar to neutrophils, are scattered at the bottom of the crypt only in the small intestine epithelium and do not follow the downward migratory pathway. Transit-amplifying cells and stem cells occupy the lower two thirds of the crypt [5]. The maintenance of the stem cell compartment as well as the transition from proliferation to differentiation are finely regulated by Wnt signaling ligands that are thought to be produced by mesenchymal cells of the myofibroblast lineage, closely applied to the basal lamina that surrounds the crypt [6]. Other factors, such as the bone morphogenetic protein (BMP) antagonists gremlin 1 (GREM1) and gremlin 2 (GREM2) [7], Notch signaling pathways, ephrin-B1 (Eph-B1) and its receptors Eph-B2 and Eph-B3, contribute to stem cell behavior, migration, and differentiation [8–10].
Colonic crypt organization. a In the epithelial lining of normal colonic mucosa, stem cells (red) are located at the bottom of the crypts. Upon asymmetrical divisions, the daughter cells undergoing differentiation migrate upward to give rise in turns to transit-amplifying (TA) precursors (light blue) and terminally differentiated cells (pink). b Cell types in the colon epithelium. Intestinal stem cells generate three epithelial cell types: the absorptive columnar cells, the hormone-producing enteroendocrine cells, and the mucous-producing goblet cells
Intestinal stem cell identification
Stem cells (SCs) are defined by two functional properties: the ability to perpetuate themselves throughout an extended period of time (self-renewal) and the potential to generate all the differentiated cells of the tissue of origin (multipotency). Despite the significant progress made in recent years in the field of stem cell biology, the identification, isolation, and characterization of SCs of the colonic crypt remain elusive. The concept that all terminally differentiated epithelial cell types within the intestine can be derived from a single multipotent stem cell was first formulated by Chen and Leblond in 1974 and it is well documented in mouse small intestine. Many obstacles have hindered the identification of intestinal SCs among which the lack of clonogenic and reconstitution assays and the complexity of the crypt structure that limits the retrieval of putative SCs from their niche where they are interspersed among more differentiated daughter cells. The exact identity of the intestinal stem cells has proven to be controversial over the last 30 years. Several studies have attempted to identify intestinal SCs within colonic crypts by using indirect techniques based on biological features restricted to the stem cell compartment. Long-term retention of label DNA has been exploited as surrogate marker of stemness based on the observation that SCs in adult tissues usually divide at a slow rate when compared to the progenitor population [11]. This functional difference is highlighted by labeling the genetic material of proliferating cells in mouse intestinal crypts with tritiated thymidine [12] or by the administration of the DNA-labeling dye bromodeoxyuridine to rats in drinking water [13]. These approaches have allowed the identification of low mitotic index cells that undergo only limited dilution of label over time and are located at the bottom of the crypts. The “immortal strand hypothesis” formulated by Cairns in 1975 proposed an alternative mechanism behind DNA label retention to identify putative intestinal SCs. This hypothesis is based on the assumption that SCs selectively retain their original DNA strands, while donating the newly synthesized DNA strand to their daughter cells. However, this theory has recently proven controversial due to the demonstration that in the well-characterized hematopoietic stem cell compartment, the asymmetric segregation of genetic material is not observed [14]. The studies performed in the last three decades to identify the intestinal SCs have lead to the formulation of two different models, known respectively as the “+4 position” and the “stem cell zone” models. In the “+4 position” model, proposed in the late 1950s, it was assumed that the SCs are located at the +4 position just above the Paneth cells which occupy the small intestine crypt base. The multipotent SCs give rise to the transit-amplifying population that undergoes vigorous division and differentiation into enterocytes, goblet cells, and enteroendocrine cells as they migrate out of the crypt onto the villi. In contrast, Paneth cells differentiate while migrating down from the +4 position toward the crypt base. A more recent model, “the stem cell zone,” is based on the identification of small undifferentiated cycling cells interspersed within the Paneth cells, termed crypt base columnar cells (CBC), that are believed to be the true intestinal SCs (Fig. 2). Definitive proof for either model has proven elusive due to the lack of specific markers for these cells. The recognition of specific cell surface markers has allowed the identification of SCs in many tissues including the hematopoietic system [15] and mammary gland [16]. Several molecules have been proposed as markers of SCs in the intestine including the RNA-binding protein musashi-1 (Msi-1) that is thought to be involved in asymmetric division during neuronal development in Drosophila melanogaster [17]. Immunohistochemical analysis performed in normal human colon crypts revealed that the majority of cells expressing Msi-1 reside in the lower region of the crypt, which corresponds to the expected position of the colonic SCs [18]. However, immunoreactivity was also observed above the bottom of the crypt, suggesting that Msi-1 is still expressed by early transient-amplifying progenitor cells. Similarly, the expression of Hes1, a transcriptional repressor transactivated by Msi-1, has been evaluated in the mouse small intestine epithelium [19]. Hes1 and Msi-1 were coexpressed by the putative SCs at the crypt base, although Hes1 was expressed by a broader population of cells. Other putative biomarkers have been evaluated to distinguish the SC population within the colon, such as members of the integrin superfamily of transmembrane glycoproteins including α2 and β1 subunits [20]. More recently, Eph-B receptors have been described as important regulators of migration and proliferation in the intestinal epithelium. The expression of both Eph-B2 and Eph-B3 tyrosine kinase receptors has been reported at the bottom of the crypt in the mouse colon [21]. Inhibition of Eph-B2/Eph-B3 signaling has shown to reduce the number of proliferating cells without altering the stem cell number, suggesting that Eph-B receptors is unlikely to be an independent biomarker of colonic SCs. Conversely, a more promising intestinal SC marker might be Bmi-1, a factor involved in the self-renewal of hematopoietic and neural stem cells, as it was recently reported to be expressed within the bottom crypts in the small intestine predominantly by the cells at the +4 position [22].
The Wnt signaling pathway in stem cell compartment
The homeostatic self-renewal of the intestine depends on a complex network of interplay involving many cellular processes, including proliferation, differentiation, migration, and cell death. All these phenomena are finely regulated by a relative small number of evolutionary highly conserved signaling pathways, among which the Wnt signaling cascade plays a major role. In the so-called canonical Wnt pathway, the central player is the cytoplasmic protein β-catenin. In the absence of the Wnt ligand, free cytoplasmic β-catenin is targeted to degradation by recruitment to a multiprotein degradation complex containing the scaffold protein Axin, the tumor suppressor gene product adenomatous polyposis coli (APC), as well as casein kinase I (CKI) and glycogen synthase kinase 3β (GSK3β). Upon sequential phosphorylation of highly conserved serine and threonine residues at the N terminus domain, β-catenin is ubiquitinated and subsequently degraded by the proteasome machinery. In the presence of secreted Wnt ligand, signal occurs via interaction with a serpentine receptor of the frizzled (Fz) family and a member of the low-density lipid receptor family (Lpr5 or Lpr6). Following binding, Axin is recruited to the plasma membrane through direct interaction with Lpr5/6 or indirectly through an interaction with Fz via dishevelled (Dsh). Membrane docking results in Axin degradation and/or dissociation of the “destruction complex.” GSK3β also might be displaced from this complex through Dsh action. Consequently, β-catenin accumulates in the cytoplasm in a stabilized nonphosphorylated form, enters the nucleus, and binds to a member of the Tcf/Lef family converting the Tcf/Lef transcription factors from transcriptional repressors to activators [23, 24]. Several genetic studies have indicated the dominant role of the Wnt signaling pathway in the pathophysiology of the intestine. Tcf4 or β-catenin knockout mice fail to develop colonic crypts [25, 26], as do transgenic mice for the expression of Wnt-specific secreted inhibitor Dkk-1 in the intestine [27]. The Wnt signaling gradient controls the expression of the Eph-B receptors and ligands allowing the correct positioning of epithelial cells along the crypt–villus axis as well as the positioning of Paneth cells at the bottom of the crypt [28]. Recently, during a study aimed at determining the genetic program that is deregulated in APC-mutant human colon cancer cells, several Wnt target genes were selected for its restricted crypt expression. One of these, the leucine-rich repeat containing G protein-coupled receptor 5 (Lgr5), also known as Gpr49, has been proposed as a biomarker of colonic SCs [29]. Lgr5 is predicted to encode a seven-transmembrane protein with a large extracellular domain for ligand binding and a short cytoplasmic tail for coupling to G proteins. In the mouse colon, the Lgr5 expression is restricted to cycling columnar cells at the crypt base and it has been demonstrated that Lgr5-expressing cells differentiate into the expected functional lineages of the colonic epithelium [29]. More recently, it has been described that single sorted Lgr5 positive stem cells can also initiate long-term culture by generating crypt–villus organoid in which all differentiated cell lines are present. These self-organizing structures can be established by exposing a single cell to a uniform set of growth signals including R-spondin 1, Noggin, and epidermal growth factor (EGF), without any requirement for mesenchymal niche [30]. Table 1 (in italics) summarizes the markers that have been proposed to characterize normal intestinal SCs.
Cancer stem cell theory: the case of colorectal cancer
The concept that tumors are composed by a heterogeneous population of cells different for morphology, marker expression, proliferation ability, and tumorigenic potential has long been recognized. Traditional models of carcinogenesis assumed that this heterogeneity can be explained by stochastic genetic events and microenvironmental influence leading to clonal selection. By contrast, the cancer stem cell model proposes that only a small fraction of cells within a tumor possesses cancer-initiating potential and that these cancer stem cells (CSCs) are able to initiate and sustain tumor growth. Like normal tissue SCs that support the cellular hierarchy of the tissue over the lifespan of an individual, CSCs are defined by the ability to self-renew while maintaining their ability to generate a progeny of both tumorigenic and nontumorigenic cancer cells through asymmetric division. Although the cancer stem cell model is an old idea, it was not until 1997 that malignant stem cells were isolated from patients with acute myeloid leukemia [31]. Subsequently, CSCs have been isolated from breast and brain cancers and several other solid tumors, including colorectal cancer (CRC). To date, the practical translation of CSCs definition is the ability to generate a phenocopy of the original tumor upon transplantation into immunodeficient mice.
Colorectal carcinogenesis
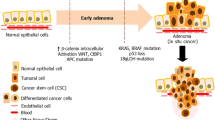
By a molecular point of view, colorectal carcinoma is one of the best-characterized cancer, mainly due to the studies performed on hereditary cases, which account for about 15% of colorectal carcinomas, that have significantly contributed to the understanding of many biological aspects of this neoplasm. The observation that the accumulation of mutations involving oncogenes and tumor suppressor genes follows the progression of the disease along the adenoma–carcinoma sequence-induced Fearon and Vogelstein to formulate in the 1990s the “adenoma–carcinoma model” in which the neoplastic process, initiated by APC or β-catenin mutations and tumor progression, results from the sequential mutation of other genes, such as K-Ras and p53, in the context of a growing genomic instability. This model has been further refined and the studies performed on relatively rare inherited cases led to the identification of genetic alterations that play a major role in the development of sporadic CRC. According to the cancer stem cell hypothesis, it can be assumed that the first mutational hit occurs in a colonic SC located at the crypt bottom that, being long-lived, can accumulate oncogenic mutations over years or decades. Once transformed, mutated SCs can divide symmetrically and asymmetrically giving rise to other CSCs and progenitors, which in turn generate other cancer cells devoid of self-renewal ability. Eventually, the entire niche will be colonized by mutant stem cells, and the crypt will be filled with their progeny, an event termed monoclonal conversion (Fig. 3). The proliferating cancer cells will be subjected to further changes that may result in the progression of cancer. This scenario has been suggested by early studies performed on patients heterozygous for the O-acetyltransferase, a gene responsible for O-acetylation of sialomucins in the goblet cell lineage [32]. Subsequently, monoclonal conversion has been further supported by the observation that the more frequent lesions in patient affected by the familial adenomatous polyposis coli (FAP), an inherited condition characterized by a germline mutation of the APC gene, is the unicryptal or monoclonal adenoma [33]. In situ hybridization studies of Y chromosome, performed on XO/XY mosaic patients who underwent a prophylactic colectomy for FAP, revealed that colonic crypts were composed exclusively of either X0 or XY cells [34]. More recently, it has been shown that two-color enzyme histochemistry can be used to simultaneously detect the mitochondrial-encoded cytochrome c oxidase (COX), relatively frequently mutated in colonic SCs, and the nuclear DNA-encoded succinate dehydrogenase [35]. The identification of wholly COX-deficient crypts in which all lineages are mutated confirms the ability of a single mutated stem cell to repopulate a crypt. All these experiments definitively show that human colonic crypts are indeed a clonal population derived from a multipotent SC.
Models for stem cells location in intestinal crypts. a The “+4 position” model suggests that intestinal stem cells are located just above the Paneth cells at position +4 relative to the crypt bottom (green). The most important marker that identifies these cells is Bmi-1. b The “stem cell zone” model assumes that small, undifferentiated, cycling cells, so-called crypt base columnar (CBC) cells (red), interspersed with Paneth cells, are the true intestinal stem cells. The most important marker to identify these cells is Lgr5
Colorectal cancer stem cell identification
Although the concept that colorectal carcinoma develops as a result of a sequence of mutations occurring during the clonal expansion of a single SC has been widely accepted, the existence of colonic CSCs had not been experimentally demonstrated until recently. A number of studies have been conducted that provide evidence for the existence of colon CSCs and demonstrate that the CRC tumorigenic cell population can be isolated by means of the expression of specific cell surface biomarkers. In the first two of these studies, CD133 was employed to identify a colon cancer-initiating cell (CC-IC) population in human tumors [36, 37]. CD133, also known as prominin-1, is a five-transmembrane domain molecule that has been shown to be located in the apical plasma membrane protrusions of embryonal epithelial structures [38]. Due to its location, a functional role was ascribed to CD133 as an “organizer” of the plasma membrane topology [39]. However, to date, its function remains unknown. The tumorigenic potential of CD133-positive CC-IC was evaluated in both studies by sorting freshly dissociated tumor cells and injecting them into immunocompromised mice. CD133+ cells, which account for approximately 2.5% of the bulk tumor cells, were shown to be devoid of the intestinal epithelial differentiation marker cytokeratin 20 (CK20), while expressing the epithelial adhesion molecule BerEp4 (also known as EpCAM) [36]. In the other study, O'Brien et al. isolated CD133+ cells from seven primary CRC and ten CRC metastases and injected the sorted cells in the renal capsule of nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice. Using limiting dilution analysis, the frequency of CC-ICs was calculated in an unfractionated population of cancer cells as to be one out of 5.7 × 104. However, CC-IC fraction is enriched in CD133+ cells, resulting in a frequency of one out of 262 cells. CD133+ cells readily gave rise to tumors in mice, whereas the CD133− cell population was unable to generate tumors even after serial transplantation in mice. More importantly, tumor xenografts generated by CD133+ CC-ICs displayed the same morphologic features of the parental tumor and were reproducibly maintained upon serial transplantation, suggesting that the molecular heterogeneity of the original tumor was recapitulated, as demonstrated by the presence of CD133+ and CD133− cells at similar ratios to the original tumor. Both studies demonstrated the expression of CD133 also in normal colon tissue, although at lower frequency, suggesting that CD133+ CC-ICs in cancer samples might result from oncogenic transformation of normal colonic SCs. Colonic cancer cells, obtained from dissociation of cancer specimens, can be perpetuated in vitro as floating aggregates called “tumor spheres,” which can be maintained in culture in serial passages and are able to generate tumors when injected in SCID mice. Upon growth factor deprivation, CD133+ spheres gradually differentiate, acquiring adherent cell morphology and the expression of differentiation markers such as CK20. Conversely, the expression of CD133 is progressively lost during differentiation, as well as the ability to transfer the tumor into immunocompromised mice. An alternative protocol for the isolation of CC-ICs employed CD44 and the epithelial surface antigen EpCAM [40]. Subcutaneous injection of purified CD44+/EpCAMHIGH cells into NOD/SCID mice resulted in high-frequency generation of tumor xenograft, whereas CD44−/EpCAMLOW cells lack tumor-initiating activity. Further subfractionation of the CD44+/EpCAMHIGH cell population by using the mesenchymal stem cell marker CD166 increased the success of the tumor xenograft. However, immunohistochemical analysis of normal colonic epithelium shows that CD44 expression occurs not only in the stem cell compartment at the crypt bottom but also in cells within the proliferative compartment, thus the specificity of CD44 for colonic SC remain to be determined.
Controversy exists concerning the markers that are more robust in identifying CC-ICs. Shmelkov et al. questioned CD133 as a CRC-SCs marker using a knockin LacZ reporter mouse in which the expression of LacZ is driven by the endogenous CD133 promoter to demonstrate that CD133 expression at the mRNA level in the mouse colon is not restricted to SCs [41]. The authors concluded that CD133 is widely expressed in human primary colon cancer epithelial cells, whereas the CD133− population is composed mostly of stromal and inflammatory cells. Such hypothesis is in evident contrast with our data, which show that the majority of EpCAM/BerEp4+ epithelial cells in colon cancer are of tumor origin [36]. Moreover, Horst et al. have recently shown that CD133 expression correlates with poor prognosis and is an independent prognostic marker for low survival in CRC [42]. Using three different antibodies against the CD133 antigen, the authors show that CD133+ cells lack CK20 expression, but they are positive for EpCAM and conclude that evaluation of CD133 and nuclear β-catenin can identify colon cancer cases with dramatic reduced survival [43]. More recently, aldehyde dehydrogenase 1 (ALDH) has been proposed as a promising new marker for normal and malignant human colonic SCs [44]. Flow cytometric isolation of ALDH1+ cancer cells and implantation of as few as 25 cells in NOD/SCID mice generate tumor xenografts. Further isolation of cancer cells using a second marker (CD44 or CD133 serially) only modestly increased enrichment based on tumor-initiating ability.
Identification of biomarkers for CRC-SCs will improve the understanding of the mechanism underlying tumor growth and progression. Once again, studies performed on mouse model could offer helpful suggestion. Recently, Clevers and coworkers provide a very convincing demonstration of the origin of intestinal cancer from Lgr5+ CBC cells. The authors show that deletion of APC in these SCs leads to their transformation within days. Transformed SCs remain located at the crypt bottom while feeding a growing microadenoma that develops in a macroscopic adenoma within 3–5 weeks. At the same time, Zhu et al. have published another paper showing that Prom1+ cells are located at the base of crypts in the small intestine, coexpress Lgr5, generate the entire intestinal epithelium, and are susceptible to neoplastic transformation [45]. Table 1 (in bold) summarizes the markers that have been used to isolate CRC-ICs.
Clinical implications of colorectal cancer stem cells
The identification of the cell type capable of initiating and sustaining tumor growth has major implications for cancer prognosis and therapy. At present, anticancer therapies for CRC include surgery, radiation, chemotherapy, and anti-VEGF or EGFR monoclonal antibodies. Whatever the therapeutic approach, none of these treatment modalities is curative in most of advanced cancer cases. One of the major concerns on the use of cytotoxic agents is that they are designed to kill actively proliferating cells, which represent the bulk of the tumor cell population, thus reducing tumor mass but potentially leaving behind CSCs. In most cases, such therapy exclusively targets the pool of differentiated cells while sparing the CSC compartment. Similarly, CRC-SCs have been found to be enriched in colon tumors following classical chemotherapeutic regimens intended to shrink tumors [46]. Failure is thought to be often due to the acquisition of resistance in subsets of cells through genetic mutation and natural selection. In support of this hypothesis, it has been recently demonstrated that resistance to radiation of CD133+ glioblastoma stem cells can result from elevated expression of DNA damage response gene [47]. Todaro et al. recently demonstrated that CD133+ CC-ICs produce interleukin-4 (IL-4) as an autocrine growth factor that promotes tumor resistance to the chemotherapeutic agents 5-fluorouracil and oxaliplatin. On the basis of this finding, they devised a strategy to sensitize the CRC-SCs to chemotherapy through the targeting of IL-4 [48]. CSCs share with normal SCs not only the capacity for self-renewal, but they also have other properties that might explain the failure of current therapies. Similarly to normal SCs, CSCs could also express active transmembrane ABC transporter family members, such as multidrug resistance transporter 1 (MDR1) and ABCG2 that can facilitate the efflux of anticancer drugs. Moreover, signaling pathways that regulate self-renewal of normal colon SC population are altered in colon cancer.
An ideal interventional approach should target molecular pathways preferentially active in CSCs without affecting normal tissues. Such goal would require new technical advances that allow stringent purification and extensive characterization of CSCs. Different groups are testing potential therapeutic agents aiming at either inhibiting CSC survival pathways or forcing CSC differentiation. Such attempts should take into account the different microenvironments surrounding the metastatic CRC-SCs, as clinical evidence suggests that the site of metastatic formation may have a considerable impact on the therapeutic response.
References
Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ (2006) Cancer statistics, 2006. CA Cancer J Clin 56:106–130
Boman BM, Wicha MS (2008) Cancer stem cells: a step toward the cure. J Clin Oncol 26:2795–2799
Potten CS, Kellett M, Rew DA, Roberts SA (1992) Proliferation in human gastrointestinal epithelium using bromodeoxyuridine in vivo: data for different sites, proximity to a tumour, and polyposis coli. Gut 33:524–529
Cheng H, Bjerknes M, Amar J (1984) Methods for the determination of epithelial cell kinetic parameters of human colonic epithelium isolated from surgical and biopsy specimens. Gastroenterology 86:78–85
Radtke F, Clevers H (2005) Self-renewal and cancer of the gut: two sides of a coin. Science 307:1904–1909
Fevr T, Robine S, Louvard D, Huelsken J (2007) Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol 27:7551–7559
Kosinski C, Li VS, Chan AS, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW, Yuen ST, Leung SY, Chen X (2007) Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci USA 104:15418–15423
Crosnier C, Stamataki D, Lewis J (2006) Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat Rev Genet 7:349–359
van Es JH, Clevers H (2005) Notch and Wnt inhibitors as potential new drugs for intestinal neoplastic disease. Trends Mol Med 11:496–502
Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, Clevers H (2002) Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 111:251–263
Cotsarelis G, Sun TT, Lavker RM (1990) Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61:1329–1337
Potten CS, Owen G, Booth D (2002) Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci 115:2381–2388
Kim SJ, Cheung S, Hellerstein MK (2004) Isolation of nuclei from label-retaining cells and measurement of their turnover rates in rat colon. Am J Physiol Cell Physiol 286:C1464–C1473
Kiel MJ, He S, Ashkenazi R, Gentry SN, Teta M, Kushner JA, Jackson TL, Morrison SJ (2007) Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature 449:238–242
Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ (2005) SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121:1109–1121
Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE (2006) Generation of a functional mammary gland from a single stem cell. Nature 439:84–88
Nakamura M, Okano H, Blendy JA, Montell C (1994) Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron 13:67–81
Nishimura S, Wakabayashi N, Toyoda K, Kashima K, Mitsufuji S (2003) Expression of Musashi-1 in human normal colon crypt cells: a possible stem cell marker of human colon epithelium. Dig Dis Sci 48:1523–1529
Kayahara T, Sawada M, Takaishi S, Fukui H, Seno H, Fukuzawa H, Suzuki K, Hiai H, Kageyama R, Okano H, Chiba T (2003) Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett 535:131–135
Fujimoto K, Beauchamp RD, Whitehead RH (2002) Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology 123:1941–1948
Holmberg J, Genander M, Halford MM, Anneren C, Sondell M, Chumley MJ, Silvany RE, Henkemeyer M, Frisen J (2006) EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell 125:1151–1163
Sangiorgi E, Capecchi MR (2008) Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40:915–920
Gregorieff A, Clevers H (2005) Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev 19:877–890
Katoh M (2007) WNT signaling pathway and stem cell signaling network. Clin Cancer Res 13:4042–4045
Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H (1998) Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet 19:379–383
Pinto D, Gregorieff A, Begthel H, Clevers H (2003) Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev 17:1709–1713
Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J, Nusse R, Kuo CJ (2004) Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci USA 101:266–271
Batlle E, Bacani J, Begthel H, Jonkheer S, Gregorieff A, van de Born M, Malats N, Sancho E, Boon E, Pawson T, Gallinger S, Pals S, Clevers H (2005) EphB receptor activity suppresses colorectal cancer progression. Nature 435:1126–1130
Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449:1003–1007
Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459:262–265
Bonnet D, Dick JE (1997) Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3:730–737
Campbell F, Williams GT, Appleton MA, Dixon MF, Harris M, Williams ED (1996) Post-irradiation somatic mutation and clonal stabilisation time in the human colon. Gut 39:569–573
Nakamura S, Kino I (1984) Morphogenesis of minute adenomas in familial polyposis coli. J Natl Cancer Inst 73:41–49
Preston SL, Wong WM, Chan AO, Poulsom R, Jeffery R, Goodlad RA, Mandir N, Elia G, Novelli M, Bodmer WF, Tomlinson IP, Wright NA (2003) Bottom-up histogenesis of colorectal adenomas: origin in the monocryptal adenoma and initial expansion by crypt fission. Cancer Res 63:3819–3825
Taylor RW, Barron MJ, Borthwick GM, Gospel A, Chinnery PF, Samuels DC, Taylor GA, Plusa SM, Needham SJ, Greaves LC, Kirkwood TB, Turnbull DM (2003) Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest 112:1351–1360
Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R (2007) Identification and expansion of human colon-cancer-initiating cells. Nature 445:111–115
O'Brien CA, Pollett A, Gallinger S, Dick JE (2007) A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445:106–110
Corbeil D, Roper K, Hellwig A, Tavian M, Miraglia S, Watt SM, Simmons PJ, Peault B, Buck DW, Huttner WB (2000) The human AC133 hematopoietic stem cell antigen is also expressed in epithelial cells and targeted to plasma membrane protrusions. J Biol Chem 275:5512–5520
Corbeil D, Roper K, Fargeas CA, Joester A, Huttner WB (2001) Prominin: a story of cholesterol, plasma membrane protrusions and human pathology. Traffic 2:82–91
Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, Shelton AA, Parmiani G, Castelli C, Clarke MF (2007) Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA 104:10158–10163
Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, St Clair R, Baljevic M, White I, Jin DK, Chadburn A, Murphy AJ, Valenzuela DM, Gale NW, Thurston G, Yancopoulos GD, D'Angelica M, Kemeny N, Lyden D, Rafii S (2008) CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J Clin Invest 118:2111–2120
Horst D, Kriegl L, Engel J, Kirchner T, Jung A (2008) CD133 expression is an independent prognostic marker for low survival in colorectal cancer. Br J Cancer 99:1285–1289
Horst D, Kriegl L, Engel J, Jung A, Kirchner T (2009) CD133 and nuclear beta-catenin: the marker combination to detect high risk cases of low stage colorectal cancer. Eur J Cancer 45:2034–2040
Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM (2009) Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res 69:3382–3389
Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, Poppleton H, Zakharenko S, Ellison DW, Gilbertson RJ (2009) Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 457:603–607
Dylla SJ, Beviglia L, Park IK, Chartier C, Raval J, Ngan L, Pickell K, Aguilar J, Lazetic S, Smith-Berdan S, Clarke MF, Hoey T, Lewicki J, Gurney AL (2008) Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS ONE 3:e2428
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN (2006) Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444:756–760
Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, Tripodo C, Russo A, Gulotta G, Medema JP, Stassi G (2007) Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 1:389–402
Acknowledgements
We thank the Italian Ministry of Health, the Italian Ministry for University and Research (FIRB_RBIP06ZJ78), and the Italian Association for Cancer Research (AIRC) for supporting the colon cancer stem cell research.
Disclosure of potential conflict of interests
The authors declare that they have no conflicting interests related to this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ricci-Vitiani, L., Fabrizi, E., Palio, E. et al. Colon cancer stem cells. J Mol Med 87, 1097–1104 (2009). https://doi.org/10.1007/s00109-009-0518-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-009-0518-4