Abstract
Background
Magnetic resonance (MR) relaxometry is of increasing scientific relevance in neurodegenerative disorders but is still not established in clinical routine. Several studies have investigated relaxation time alterations in disease-specific areas in Parkinson’s disease (PD), all using manually drawn regions of interest (ROI). Implementing MR relaxometry into the clinical setting involves the reduction of time needed for postprocessing using an investigator-independent and reliable approach. The aim of this study was to evaluate an automated, atlas-based ROI method for evaluating T2* relaxation times in patients with PD.
Method
Automated atlas-based ROI analysis of quantitative T2* maps were generated from 20 PD patients and 20 controls. To test for the accuracy of the atlas-based ROI segmentation, we evaluated the spatial overlap in comparison with manually segmented ROIs using the Dice similarity coefficient (DSC). Additionally, we tested for group differences using our automated atlas-based ROIs of the putamen, globus pallidus, and substantia nigra.
Results
A good spatial overlap accuracy was shown for the automated segmented putamen (mean DSC, 0.64 ± 0.04) and was inferior but still acceptable for the substantia nigra (mean DSC, 0.50 ± 0.17). Based on our automated defined ROI selection, a significant decrease of T2* relaxation time was found in the putamen as well as in the internal and external globus pallidus in PD patients compared with healthy controls.
Conclusion
Automated digital brain atlas-based approaches are reliable, more objective and time-efficient, and therefore have the potential to replace the time-consuming manual drawing of ROIs.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disease, estimated to occur in approximately 1 % of individuals over 60 years of age, with 4.1–4.6 million people affected worldwide [1]. Although loss of dopaminergic neurons due to accumulation of aggregated α‑synuclein in the substantia nigra (SN) is a prerequisite for the pathological confirmation of the disease, the pathological lesions are much more extensive and involve a number of ascending projection pathways in the brainstem and specific areas of the neocortex [2].
Despite rigorous research efforts, patient management and clinical research are still impeded by suboptimal methods for diagnosis, i. e., the lack of reliable and applicable in vivo biomarkers [3]. Since magnetic resonance imaging (MRI) is a noninvasive and widely available imaging tool, MRI-based techniques are increasingly used in the ongoing search for sensitive and reliable biomarkers in PD. As altered levels of iron concentration were found in the basal ganglia and in the SN in patients with PD [4], MRI mapping techniques using T2* relaxation times are postulated to be promising imaging tools for diagnosis and especially for the assessment of disease progression in PD. During a 3-year evolution of PD, a recent study showed increased R2* (=1/T2*) in the SN and in the putamen, without significant changes in controls [5].
All studies investigating relaxation times to quantify in vivo the signal alterations of disease-specific areas in PD used manually drawn regions of interest (ROIs) [5–10]. An exact manual definition of a specific anatomical structure is time consuming. As was shown in the previously mentioned studies, more and more frequently small spheres are placed, resulting in arbitrary region coverage.
Therefore, the aim of the present study was to assess an automated, investigator-independent, atlas-based ROI approach for evaluating T2* relaxation changes in disease-specific brain regions in patients with PD.
Methods
The study included 20 PD patients and 20 age- and gender-matched healthy controls. All study patients fulfilled the standard UK Brain Bank criteria for PD (see Table 1 for demographic and clinical data of the subjects). Additionally, all study participants had to fulfill the common criteria for performing MRI. Subjects with concomitant brain diseases such as ischemic stroke, tumor, or severe cerebral microangiopathy were excluded from this study.
Patients were individually matched to healthy controls using a multidimensional matching approach [11]. Compatible controls were drawn from a larger pool of healthy subjects while simultaneously minimizing pair-wise differences in age and sex. The resulting groups did not differ in their mean age (p = 0.970) or in the underlying age distribution (Levene test, p = 0.448). Furthermore, all pairs comprised individuals of the same sex (Table 1).
All subjects were interviewed regarding their previous medical history. In the PD group, disease duration, levodopa equivalent dose (LED) [12], Hoehn and Yahr (HY) [13] stages, and the motor part of Unified Parkinson’s Disease Rating Scale (UPDRS III) [14] were recorded, the latter two being assessed without dopaminergic medication (OFF condition) in 17 of 20 patients (Table 1). For the remaining three patients, it was not possible to obtain these parameters.
The study was approved by the local research ethics committee and all subjects gave written informed consent prior to participation.
Image Acquisition
Image acquisition was performed on a 3-T MRI scanner (Magnetom TRIO, Siemens Medical Solutions, Erlangen, Germany) using a 12-channel head coil for reception. In addition to the standard fixing device of the head coil, foam pads were used to reduce involuntary head movements.
For mapping the T2* relaxation time we used a 2D gradient echo sequence with 25 slices, which were acquired at five different echo times (6.82, 12.04, 18.00, 25.00, and 33.00 ms) with TR = 422 ms, FOV = 220 × 220 mm2, matrix = 132 × 132, slice thickness = 4 mm, and a total scan duration of 7 min. For spatial transformation, additional MP-Rage images (T1-weighted 3D dataset) were acquired with a voxel size of 1.0 × 1.0 × 1.0 mm, TE = 2.14 ms, TR = 2,200 ms, TI = 1,100 ms, and a flip angle of 12°.
Image Postprocessing
Postprocessing of imaging data was performed using SPM8 (http://www.fil.ion.ucl.ac.uk/spm/software/spm8/). T1-weighted anatomical images were segmented using the VBM8 toolbox (http://dbm.neuro.uni-jena.de/vbm/download/). Deformation field parameters for nonlinear transformation into the stereotactic Montreal Neurological Institute (MNI) space were derived from the DARTEL approach [15] implemented in VBM8 (using the provided MNI template of the IXI-550 cohort).
The T2* signal amplitudes were fitted to a monoexponential decay with Matlab functionality. The resulting spatial maps of T2* relaxation times in individual space were transformed into stereotactic MNI space using the DARTEL deformation fields (see previous paragraph). Spatially normalized images were resampled to a resolution of 1.5 × 1.5 × 1.5 mm3. For group analyses, T2* relaxation times were extracted and median-aggregated based on ROIs for the left and right substantia nigra (SN), putamen, as well as internal and external globus pallidus (GPi and GPe), as predefined in the WFU PickAtlas (http://fmri.wfubmc.edu/software/PickAtlas; Fig. 1a). For these analyses, left and right regions were merged.
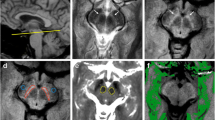
Illustration of hand-drawn region of interest (ROIs; red) and automated generated ROIs (green) of the putamen (a) and the substantia nigra (b) on six different axial slices of an individual T1-weighted image in MNI space (position on z‑axis in mm = −13.5, −10.5, −7.5, −4.5, −1.5, 0) of a study participant with a representative DSC (0.67 for the putamen bilaterally, 0.50 for the left SN, and 0.52 for the right SN). c T2* values in milliseconds (ms) in each ROI (mean ± standard deviation) in patients with Parkinson’s disease (PD) and healthy controls (HC). For group analyses, left and right regions were merged. SN substantia nigra, PUT putamen, GPe external globus pallidus, GPi pallidus. **p < 0.01, *p < 0.05, °p = 0.066
To test for the spatial overlap accuracy, we additionally performed manual delineation of individual ROIs for the SN and the putamen bilaterally, based on the T2*- and T1-weighted images of each participant. The spatial overlap of atlas-based and manually drawn ROIs was analyzed using the Dice Similarity Coefficient (DSC) [16]. Finally, we performed group analyses with the manually extracted T2* relaxation times of the putamen and the SN.
Results
The spatial overlap accuracy of atlas-based ROIs with manually drawn ROIs analyzed with the DSC showed good results regarding the putamen (mean DSC, 0.64 ± 0.04) and inferior but still acceptable DSC results for the SN (mean DSC, 0.50 ± 0.17).
The atlas-based derived T2* values presented in this study (putamen, 34.2 ± 4.4; GPe, 23.5 ± 4.3; GPi, 23.4 ± 4.1; SN mean, 25.9 ± 6.5) were within the range of previously reported T2* values [5–10]. A multivariate ANOVA on the T2* relaxation times in the four specified anatomical regions as dependent variables did not show a significant main effect but a trend for the between-subject factor group (p = 0.071). Subsequent univariate testing revealed that patients with PD showed, in comparison with the individually matched healthy controls, significantly reduced T2* relaxation times in the putamen (p = 0.048), GPi (p = 0.006), GPe (p = 0.004), and a trend thereof in the SN (p = 0.066; Fig. 1b). These findings were confirmed using the manually segmented T2* relaxation times within the putamen and SN (p = 0.037 and p = 0.113, respectively).
Discussion
In the present study we evaluated an investigator-independent atlas-based ROI method to obtain individual T2* relaxation times within PD-specific brain regions.
First, atlas-based putaminal ROIs showed good spatial alignment with the manually defined ROIs in the same individuals (mean DSC, 0.64 ± 0.04), whereas the DSC analyzes of the SN was inferior, but still acceptable (mean DSC 0.50 ± 0.17) (Fig. 1). Additionally, T2* relaxation values of the atlas-based ROIs were within the range of previously reported values [5–10].
When comparing healthy controls versus patients with PD, T2* relaxation rates were significantly different in the putamen as well as in the globus pallidus (GPi and GPe), while the result within the SN just missed the significance threshold, and is therefore reported as a trend only. The following group comparison of the manually derived T2* relaxation rates confirmed the significant finding within the putamen, as well as the nonsignificant finding within the SN, although the patients of the present study were more strongly affected than in other studies. Therefore, we hypothesize that this lack of significant SN differences might primarily be due to the inferior spatial resolution of the T2*-weighted sequence used with a rather high slice thickness of 4 mm. An additional reason might be the fact that in this study the whole SN was segmented, which is different to previous studies, in which spheres were placed in the middle of the ROI and/or subregions of the SN were segmented [5, 8–10].
The inferior coverage accuracy of the atlas-based SN ROI shown with the manually delineated SN could be explained by the fact that the SN is a rather small, anatomically heterogeneous structure, located within the brainstem, again a structure with a relatively small cross-sectional dimension, and with a relatively poor local magnetic field homogeneity causing image distortion and lower signal-to-noise ratios (SNR) [17]. This, together with the fact that there is still no reliable automated method available to exactly delineate substructures of the SN, indicates that hand-drawn ROIs might remain the most reliable segmenting method for the SN.
Interestingly, a very recent relaxometry study showed that quantitative R2* (=1/T2*) relaxometry within the basal ganglia is able to serve as a reliable biomarker of disease progression in patients with PD. Longitudinal variation of relaxation rates observed in the putamen of patients with PD correlated significantly with clinical biomarkers of disease worsening, while results within the SN failed [5].
Regarding this, the automated ROI approach, as presented in our study, could be a valuable tool especially in longitudinal PD studies. Determination of clinically interesting ROIs using an automated atlas-based approach provides a reliable individual anatomical region coverage, which is essential for longitudinal studies, is investigator independent, and leads to a significant reduction in time needed for postprocessing. That the reduction of time needed for postprocessing is of major interest becomes apparent when considering that the most recent ROI-based studies use small spheres in the center of the region instead of delineating the whole structure of interest. The total time needed for individual atlas-based postprocessing was less than 2 min, regardless of how many ROIs are included. A limitation regarding time effectiveness is the indispensable need for a visual inspection to exclude possible misregistration.
As already mentioned, a limitation of this study is the T2* multi-echo sequence used with a rather high slice thickness. Especially in areas heavily affected by field inhomogeneities (e. g., the brainstem) additional improvement might be achieved by applying a magnetic field inhomogeneity correction as described previously [18, 19]. Another option for measuring the iron content within disease-specific brain regions could be quantitative SWI using a field estimation method [20, 21]. However, this was not in the scope of this work.
In conclusion, the present study confirmed the potential of a fully automated, investigator-independent, atlas-based ROI approach, as presented in this study, especially when evaluating supratentorial brain structures.
References
Guttmacher AE, Collins FS. Welcome to the genomic era. N Engl J Med. 2003;349:996–8.
Halliday G, Lees A, Stern M. Milestones in Parkinson’s disease – clinical and pathologic features. Mov Disord. 2011;26:1015–21.
Shtilbans A, Henchcliffe C. Biomarkers in Parkinson’s disease: an update. Curr Opin Neurol. 2012;25:460–5.
Dexter DT, Carayon A, Javoy-Agid F, Agid Y, Wells FR, Daniel SE, Lees AJ, Jenner P, Marsden CD. Alterations in the levels of iron, ferritin and other trace metals in Parkinson’s disease and other neurodegenerative diseases affecting the basal ganglia. Brain. 1991;114(4):1953–75.
Ulla M, Bonny JM, Ouchchane L, Rieu I, Claise B, Durif F. Is R2* a new MRI biomarker for the progression of Parkinson’s disease? A longitudinal follow-up. PLoS ONE. 2013;8:e57904.
Baudrexel S, Nürnberger L, Rüb U, Seifried C, Klein JC, Deller T, Steinmetz H, Deichmann R, Hilker R. Quantitative mapping of T1 and T2* discloses nigral and brainstem pathology in early Parkinson’s disease. Neuroimage. 2010;51:512–20.
Ordidge RJ, Gorell JM, Deniau JC, Knight RA, Helpern JA. Assessment of relative brain iron concentrations using T2-weighted and T2*-weighted MRI at 3 Tesla. Magn Reson Med. 1994;32:335–41.
Du G, Lewis MM, Sen S, Wang J, Shaffer ML, Styner M, Yang QX, Huang X. Imaging nigral pathology and clinical progression in Parkinson’s disease. Mov Disord. 2012;27:1636–43.
Rossi M, Ruottinen H, Soimakallio S, Elovaara I, Dastidar P. Clinical MRI for iron detection in Parkinson’s disease. Clin Imaging. 2013;37:631–6.
Martin, Wayne WR, Wieler M, Gee M. Midbrain iron content in early Parkinson disease: a potential biomarker of disease status. Neurology. 2008;70:1411–7.
Kaller CP, Reisert M, Katzev M, Umarova R, Mader I, Hennig J, Weiller C, Köstering L. Predicting planning performance from structural connectivity between left and right mid-dorsolateral prefrontal cortex: moderating effects of age during postadolescence and midadulthood. Cereb Cortex. 2015;25:869–83.
Thobois S. Proposed dose equivalence for rapid switch between dopamine receptor agonists in Parkinson’s disease: a review of the literature. Clin Ther. 2006;28:1–12.
Hoehn MM, Yahr MD. Parkinsonism: onset, progression and mortality. Neurology. 1967;17:427–42.
Fahn S, Bressman SB, Marsden CD. Classification of dystonia. Adv Neurol. 1998;78:1–10.
Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113.
Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26:297.
Stroman PW, Figley CR, Cahill CM. Spatial normalization, bulk motion correction and coregistration for functional magnetic resonance imaging of the human cervical spinal cord and brainstem. Magn Reson Imaging. 2008;26:809–14.
Baudrexel S, Volz S, Preibisch C, Klein JC, Steinmetz H, Hilker R, Deichmann R. Rapid single-scan T2*-mapping using exponential excitation pulses and image-based correction for linear background gradients. Magn Reson Med. 2009;62:263–8.
Preibisch C, Volz S, Anti S, Deichmann R. Exponential excitation pulses for improved water content mapping in the presence of background gradients. Magn Reson Med. 2008;60:908–16.
Zheng W, Nichol H, Liu S, Cheng YN, Haacke EM. Measuring iron in the brain using quantitative susceptibility mapping and X‑ray fluorescence imaging. Neuroimage. 2013;78:68–74.
Acosta-Cabronero J, Williams GB, Cardenas-Blanco A, Arnold RJ, Lupson V, Nestor PJ. In vivo quantitative susceptibility mapping (QSM) in Alzheimer’s disease. PLoS ONE. 2013;8:e81093.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
K. Egger, F. Amtage, S. Yang, M. Obmann, R. Schwarzwald, L. Köstering, I. Mader, J. Koenigsdorf, C. Weiller, C.P. Kaller, and H. Urbach state that there are no conflicts of interest.
Additional information
K. Egger and F. Amtage contributed equally to this study.
Rights and permissions
About this article
Cite this article
Egger, K., Amtage, F., Yang, S. et al. T2* Relaxometry in Patients with Parkinson’s Disease. Clin Neuroradiol 28, 63–67 (2018). https://doi.org/10.1007/s00062-016-0523-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00062-016-0523-2