Abstract
The novel series of piperidine conjugated benzophenone analogs with amide link 11a–l were synthesized in a multistep process. The structures of these compounds were confirmed by IR, 1H, 13C, NMR, and mass spectra and also by elemental analyses. The newly synthesized molecules were screened for selectivity against cancers of different origin through cell based assay system using B16F10, A375, A549, HepG2, ACHN, and MCF7 cells. The results postulated that compound 11f with two bromo groups at the para position in rings A and E and two methyl groups at ortho position in rings B and D evokes target specific action against melanoma highlighting the importance of substituted groups. Down the line studies further inferred compound 11f evokes the apoptotic cellular event leading to cell death. Investigation of eventual mechanism revealed that compound 11f turned out to be a dual inhibitor of B-cell lymphoma-2 and X-linked inhibitors of apoptosis causing the up regulation of Bax and Bad. Further, the antiproliferative effects were mimicked in murine melanoma with similar mechanisms. Molecular docking experiments further confirmed that compound 11f possessed a superior affinity for of B-cell lymphoma-2 and X-linked inhibitors of apoptosis through strong hydrogen bonds. The study implies the identification of compound 11f with selective target against melanoma by inducing apoptogenic effect, which could be the future hope for the treatment of skin cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Skin cancers are by far the most common malignancy of humans and millions of cases detected each year (Gupta et al. 2016). The growing incidence of cutaneous malignancies has heralded the need for multiple treatment options. Although many therapeutic strategies available in the modern era, new research and fresh innovation are still required to reduce morbidity and mortality and urgent approaches for skin cancer may pass through new compounds. One of the most effective ways to combat skin cancers is through early diagnosis and administration of effective treatment with target specific drugs. Further, follow up and efficient monitoring will allow physicians to detect relapsing diseased condition.
Apoptosis, a normal physiological form of cell death, is critically involved in the regulation of cellular homeostasis. In normal physiological condition, the fate of the cells depends on the cellular microenvironment. Any mutation or damage will be detected and repair or elimination event takes place depending upon the damage to the cells. This event is regulated by different proteins which will sense and act. Whereas, in the pathogenesis of melanoma, the neoplastic cells are evolved by developing escaping strategy leading to programmed cell death, which is necessary for prognosis of the disease (Elmore 2007). These cancer cells regularly started to over express array of anti-apoptotic proteins that play an imperative roles in abiding the revival of apoptotic signaling cascade (Frenzel et al. 2009). Many members of the B-cell lymphoma 2 (Bcl-2) family of apoptosis-related genes have been found to be differentially expressed in various skin malignancies (Eberle and Hossini 2008). Most of the Bcl-2 family genes have been found to play a central regulatory role in apoptosis induction, which includes both pro and anti-apoptotic proteins. Bcl-2 is one of the prime anti-apoptotic proteins which primarily interact and neutralize the pro-apoptotic protein expression and their by halting the release of apoptogenic molecules from mitochondria (Frenzel et al. 2009). Another group of anti-apoptotic proteins expressed by the cancerous cells are inhibitors of apoptosis (IAPs), which interacts and antagonize the executioner caspases, such as caspase-3 and caspase-8. Modulating these proteins is an effective treatment strategy, which provides the basis for early diagnosis, understanding the prognosis, and establishment of therapeutic intervention that targets these apoptosis pathways (Parrish et al. 2013).
Targeting and mitigating the expression these anti-apoptotic proteins by novel molecules and thereby restoring the apoptotic event is considered to be an effective mode of treating cancer.
Current cancer therapy suffers from the major limitation of side effect and drug resistance, so continued search for newer and safer anticancer drugs remains critically important with target specificity. Nitrogen containing moieties occurs widely in synthetic and natural products. Besides, when one or more nitrogen atoms exist in the chemical structure, intercalating chromophore possesses a polarized character and optimal interaction occur (Ozkay et al. 2010). The nitrogen atom containing heterocycles occurs in pharmaceuticals, natural products, dyes, organic materials and in particular, in biologically active compounds (Yang et al. 2015). These heterocycles produces anticancer effects in different types of cancer through inhibiting cell growth and induction of cell differentiation and apoptosis. However, despite their wide range of biological activities along with their anticancer activity still there is a need for the development of novel, practical, and efficient methods for the synthesis of nitrogen containing heterocyclic compounds which nowadays becomes an important goal in modern organic synthesis.
Moreover, benzophenones are a class of compounds obtained from natural sources (Henry et al. 1999) or by synthetic methods (Khanum et al. 2005; Mohammed et al. 2018; Ranganatha et al. 2013). They are of enormous significance fundamentally due to their diverse biological properties. Further, benzophenones display significant antitumor activity both in vitro and in vivo (Al-Ghorbani et al. 2017; Ranganatha et al. 2013; Zabiulla et al. 2016). Moreover, synthetic benzophenones, such as benzophenone appended with oxadiazoles (Puttaswamy et al. 2018), benzimidazole benzophenones (Thirusangu et al. 2017), and pyridine conjugated benzophenones (Al-Ghorbani et al. 2016) have proven to be anticancer agents. Further, para-methoxy substituted benzophenones were evaluated as p38a inhibitors with high selectivity and efficacy (Revesz et al. 2004). Whereas, methoxy and amino substituted benzophenones have been reported to be potent cytotoxic agents against a panel of human cancer cell lines including multi-drug-resistant cell lines (Schlitzer et al. 2002). Also, analogs of benzophenone show a selective toxicity for proliferating endothelial cells by induction of apoptosis (Iyer et al. 1998), and polyprenylated benzophenone derivatives are also able to induce caspase mediated apoptosis (Balasubramanyam et al. 2004). In addition, piperidine derivatives have been found to display a wide range of biological and therapeutic activities in view of their wide range of occurrence and also due to low toxicity (Khanum et al. 2009; Hu et al. 2014; Wang et al. 2015; Xin-Hua et al. 2012). Besides, chemical modification and combination of two or more bioactive compounds is one of the most efficient approach in drug development. On this basis, an approach was made to the synthesis of potential apoptogenic molecules by combining two bioactive molecules like benzophenone and piperidine collectively through amide linkage 11a–l to attain pharmacologically efficacious single molecule as anticancer agent for both in vitro and in vivo studies. The study was also supported by molecular docking investigation which illustrated the interaction of the lead compound as dual inhibitor of both XIAP and Bcl-2 which is very promising.
Materials and methods
Experimental section
The chemicals were purchased from Sigma Aldrich, analytical thin layer chromatography (TLC) was performed on 0.25 mm silica gel plates (Merck 60 F 254) by using solvent system ethyl acetate: hexane (2:3). Melting point was determined on a Chemi Line Micro Controller based melting point apparatus with a digital thermometer. The IR spectrum was recorded by the potassium bromide pellet method on Cary 630 FTIR Agilent spectrophotometer, NMR spectrum was recorded on a VNMRS-400 MHz Agilent-NMR spectrophotometer in CDCl3 or DMSO. Mass spectrum was obtained with a VG70-70H spectrometer. Elemental analysis results are within 0.4% of the theoretical calculated value.
The human cancer lines, A549 (Human lung adenocarcinoma), MCF-7 (Human breast cancer), A375 (Human melanoma cancer), HepG2 (Human liver cancer), ACHN (Human renal cancer), B16F10 (Mouse melanoma cancer) were procured from National Center for Cell Science (NCCS), Pune, India. The protease inhibitor cocktail, anti mouse/rabbit IgG antibodies from Sigma Aldrich, USA. Dulbecco’s Modified Eagle Media (DMEM), Antibiotic-antimycotic solution, trypsin- ethylene diamine tetraacetic acid (EDTA) solution, fetal bovine serum (FBS) from In-vitrogen (Gibco), USA. Annexin V–FITC staining kit is from BD Bioscience, USA. A click-it TUNEL vision kit, Alexa fluor 488-anti mouse/rabbit IgG from Thermo Fisher Scientific, Watman, MA, USA. All other chemicals used in the current study are of molecular and analytical grade. Cell culture plastic wares were from corning Sigma and Eppendorf, Germany. All bright field and fluorescence image were taken in EVOS FL cell imaging, Thermo Scientific, USA and the results were assessed and analyzed by Image J software. All the gel and blotting images were documented using Bio-rad Gel Documentation TM XR + Imaging system and quantified using densitometric analysis. Experiments were performed at minimum three independent times and analyzed.
Chemistry
General procedure for the synthesis of phenyl benzoates 3a–f
The starting substituted benzoates 3a–f were synthesized by benzoylation of substituted phenols 1a–c with substituted benzoyl chlorides (2a–d, 1:1) in the presence of 10% sodium hydroxide solution. The reaction mixture was stirred for 3–4 h at 0–5 °C. The reaction was monitored by TLC using 4:1 n-hexane: ethyl acetate solvent mixture. After completion of the reaction, the oily product was extracted with ether layer (3 × 25 mL). Ether layer was washed with 10% sodium hydroxide solution (3 × 30 mL) followed by water (3 × 25 mL) and then dried over anhydrous sodium sulfate and the solvent was evaporated under reduced pressure to afford compounds 3a–f.
2-Methyl phenylbenzoate 3a
Yield: 93%. M.P.: 62–64 °C. IR (KBr) νmax (cm−1): 1715 (ester, C=O). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.45 (s, 3H, CH3), 7.11–8.25 (m, 9H,). LC-MS m/z 213 (M + 1). Anal. Cal. for C14H12O2 (212): C, 79.22; H, 5.70. Found: C, 79.20; H, 5.76%.
2,5-Dimethyl phenylbenzoate 3b
Yield: 82%. M.P.: 66–68 °C. IR (KBr) νmax (cm−1): 1725 (ester, C=O). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.42 (s, 6H, 2CH3), 7.20–8.25 (m, 8H, Ar–H). LC-MS m/z 227 (M + 1). Anal. Cal. for C15H14O2 (227): C, C, 79.62; H, 6.24. Found: C, 79.58; H, 6.13%.
2-Methylphenyl 4-bromobenzoate 3c
Yield: 90%. M.P.: 61–63 °C. IR (KBr) νmax (cm−1): 1715 (ester, C=O). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.46 (s, 3H, CH3), 7.09–8.18 (m, 8H, Ar-H). LC-MS m/z 291 (M+), 293 (M + 2). Anal. Cal. for C14H11BrO2 (291): C, 57.76; H, 3.81. Found: C, 57.70; H, 3.69%.
2-Chloro-6-fluorophenyl 4-chlorobenzoate 3d
Yield: 82%. M.P.: 50–52 °C. IR (KBr) νmax (cm−1): 1720 (ester, C=O). 1H NMR (400 MHz) (CDCl3) δ (ppm): 7.03–8.15 (m, 7H, Ar-H). LC-MS m/z 285 (M+), 287 (M + 2), 289 (M + 4). Anal. Cal. for C13H7Cl2FO2 (285): C, 54.77; H, 2.47. Found: C, 54.68; H, 2.40%.
2-Methylphenyl 4-methylbenzoate 3e
Yield: 93%. M.P.: 59–61 °C. IR (KBr) νmax(cm−1): 1710 (ester, C=O). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.37 (s, 6H, 2CH3), 7.11–8.28 (m, 8H, Ar–H). LC-MS m/z 227 (M + 1). Anal. Cal. for C15H14O2 (226): C, 79.62; H, 6.24. Found: C, 79.51; H, 6.13%.
2-Methylphenyl 2-methylbenzoate 3f
Yield: 83%. M.P.: 56–58 °C. IR (KBr) νmax (cm−1): 1715 (ester, C=O). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.44 (s, 6H, 2CH3), 7.12–8.24 (m, 8H, Ar–H). LC-MS m/z 227 (M + 1). Anal. Cal. for C15H14O2 (226): C, 79.62; H, 6.24. Found: C, 79.53; H, 6.19%.
General procedure for the synthesis of (4-hydroxy phenyl) phenyl methanones 4a–f
Substituted (4-hydroxy phenyl) phenyl methanone commonly known as hydroxy benzophenones 4a–f were synthesized by Fries rearrangement. Compounds 3a–f (0.002 mol) were treated with anhydrous aluminum chloride (0.005 mol) as a catalyst at 150–170 °C without using solvent condition for about 3–4 h. The reaction mixture was cooled to room temperature and quenched with 6N hydrochloric acid in the presence of ice cold water. The reaction mixture was stirred for about 2–3 h, filtered the solid then and recrystallized with methanol to obtain compounds 4a–f.
(4-Hydroxy-3-methyl-phenyl)-phenyl-methanone 4a
Yield: 76%. M.P.: 109–111 °C. IR (KBr) νmax (cm−1): 1630 (C=O), 3530–3630 (OH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.35 (s, 3H, CH3), 6.73–7.68 (m, 8H, Ar–H), 12.01 (bs, 1H, OH). LC-MS m/z 213 (M + 1). Anal. Cal. data for C14H12O2 (212): C, 79.22; H, 5.70. Found: C, 79.17; H, 5.68%.
(4-Hydroxy-2,5-dimethyl-phenyl)-phenyl-methanone 4b
Yield: 84%. M.P.: 121–123 °C. IR (KBr) νmax (cm−1): 1650 (C=O), 3505–3590 (OH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.35 (s, 6H, 2CH3), 6.61–7.70 (m, 7H, Ar–H), 11.80 (bs, 1H, OH). LC-MS m/z 227 (M + 1). Anal. Cal. data for C15H14O2 (226): C, 79.62; H, 6.24. Found: C, 79.54; H, 6.22%.
(4-Hydroxy-3-methyl-phenyl)-(4-Bromo-phenyl)-methanone 4c
Yield: 85%. M.P.: 153–156 °C. IR (KBr) νmax (cm−1): 1655 (C=O), 3520–3610 (OH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.35 (s, 3H, CH3), 6.51–7.60 (m, 7H, Ar–H), 12.02 (bs, 1H, OH). LC-MS m/z 291 (M+), 293 (M + 2). Anal. Cal. data for C14H11BrO2 (291): C, 57.76; H, 3.81. Found: C, 57.73; H, 3.75%.
(3-Chloro-5-fluoro-4-hydroxy-phenyl)-(4-chloro-phenyl)-methanone 4d
Yield: 79%. M.P.: 146–149 °C. IR (KBr) νmax (cm−1): 1660 (C=O), 3530–3625 (OH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 6.73–7.71 (m, 6H, Ar–H), 11.90 (bs, 1H, OH). LC-MS m/z 285 (M + 2), 287 (M + 2), 289 (M + 4). Anal. Cal. data for C13H7Cl2FO2 (285): C, 54.77; H, 2.47. Found: C, 54.68; H, 2.39%.
(4-Hydroxy-3-methyl-phenyl)-4-methylphenyl-methanone 4e
Yield: 82%. M.P.: 151–154 °C. IR (KBr) νmax (cm−1): 1640 (C=O), 3510–3620 (OH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.35 (s, 6H, 2CH3), 7.10–7.70 (m, 7H, Ar–H), 12.20 (bs, 1H, OH). LC-MS m/z 227 (M + 1). Anal. Cal. data for C15H14O2 (226): C, 79.62; H, 6.24. Found: C, 79.59; H, 6.20%.
(4-Hydroxy-3-methyl-phenyl)-2-methylphenyl-methanone 4f
Yield: 72%; M.P.: 124–127 °C: IR (KBr) νmax (cm−1): 1650 (C=O), 3500–3590 (OH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.34 (s, 6H, 2CH3), 6.74–7.61 (m, 7H, Ar–H), 11.60 (bs, 1H, OH). LC-MS m/z 227 (M + 1). Anal. Cal. data for C15H14O2 (226): C, 79.62; H, 6.24. Found: C, 79.60; H, 6.20%.
General procedure for the synthesis of ethyl 2-(4-benzoylphenoxy) acetates 5a–f
To a solution of compounds 4a–f (0.013 mol) in dry acetone (50 mL), anhydrous potassium carbonate (0.019 mol) and ethyl chloroacetate (0.026 mol) were added and the reaction mass was heated to 60 °C for 5–6 h. After completion of the reaction was monitored by TLC, the reaction mixture was cooled and the solvent was removed by distillation. The residual mass was triturated with ice water to remove potassium carbonate and then extracted with ether (3 × 40 mL). The ether layer was washed with 10% sodium hydroxide solution (3 × 25 mL) followed by water (3 × 25 mL) and then dried over anhydrous sodium sulfate and evaporated to dryness to obtain crude solid, which when recrystallized with ethanol, afforded compounds 5a–f.
Ethyl 2-(4-benzoyl-2-methylphenoxy)acetate 5a
Yield: 93%. M.P.: 50–52 °C. IR (KBr) νmax (cm−1): 1665 (C=O), 1755 (ester, C=O). 1H NMR (400 MHz) (CDCl3) δ (ppm): 1.20 (t, 3H, CH3 of ester), 2.31 (s, 3H, CH3), 4.13 (q, 2H, CH2 of ester), 4.54 (s, 2H, OCH2), 7.20–7.83 (m, 8H, Ar-H). LC-MS m/z 299 (M + 1). Anal. Cal. for C18H18O4 (298): C, 72.48; H, 6.04. Found: C, 72.38; H, 6.06%.
Ethyl 2-(4-benzoyl-2,5-dimethylphenoxy)acetate 5b
Yield: 92%. M.P.: 52–55 °C. IR (KBr) νmax (cm−1): 1650 (C=O), 1745 (ester, C=O). 1H NMR (400 MHz) (CDCl3) δ (ppm): 1.20 (t, 3H, CH3 of ester), 2.34 (s, 6H, 2CH3), 4.31 (q, 2H, CH2 of ester), 4.52 (s, 2H, OCH2), 6.93–7.82 (m, 7H, Ar-H). LC-MS m/z 313 (M + 1). Anal. Cal. for C19H20O4 (312): C, 73.06; H, 6.45. Found: C, 72.97; H, 6.36%.
Ethyl 2-(4-(4-bromobenzoyl)-2-methylphenoxy)acetate 5c
Yield: 89%. M.P.: 46–48 °C. IR (KBr) νmax (cm−1): 1650 (C=O), 1760 (ester, C=O). 1H NMR (400 MHz) (CDCl3) δ (ppm): 1.26 (t, 3H, CH3 of ester), 2.32 (s, 3H, CH3), 4.53 (q, 2H, CH2 of ester), 4.64 (s, 2H, OCH2), 6.92–7.83 (m, 7H, Ar-H). LC-MS m/z 377 (M+), 379 (M + 2). Anal. Cal. for C18H17BrO4 (377): C, 57.31; H, 4.54. Found: C, 57.22; H, 4.41%.
Ethyl 2-(2-chloro-4-(4-chlorobenzoyl)-6-fluorophenoxy)acetate 5d
Yield: 91%. M.P.: 55–57 °C. IR (KBr) νmax (cm−1): 1660 (C=O), 1755 (ester, C=O). 1H NMR (400 MHz) (CDCl3) δ (ppm): 1.35 (t, 3H, CH3 of ester), 4.16 (q, 2H, CH2 of ester), 4.83 (s, 2H, OCH2), 7.14–7.75 (m, 6H, Ar-H). LC-MS m/z 371 (M+), 373 (M + 2), 375 (M + 4). Anal. Cal. for C17H13Cl2FO4 (371): C, 55.01; H, 3.53. Found: C, 54.92; H, 3.46%.
Ethyl 2-(2-methyl-4-(4-methylbenzoyl)phenoxy)acetate 5e
Yield: 92%. M.P.: 48–50 °C. IR (KBr) νmax (cm−1): 1665 (C=O), 1750 (ester, C=O). 1H NMR (400 MHz) (CDCl3) δ (ppm): 1.22 (t, 3H, CH3 of ester), 2.46 (s, 6H, 2CH3), 4.13 (q, 2H, CH2 of ester), 4.55 (s, 2H, OCH2), 7.12–7.74 (m, 7H, Ar-H). LC-MS m/z 313 (M + 1). Anal. Cal. for C19H20O4 (312): C, 73.06; H, 6.45. Found: C, 73.04; H, 6.41%.
Ethyl 2-(2-methyl-4-(2-methylbenzoyl)phenoxy)acetate 5f
Yield: 90%. M.P.: 50–52 °C. IR (KBr) νmax (cm−1): 1660 (C=O), 1760 (ester, C=O). 1H NMR (400 MHz) (CDCl3) δ (ppm): 1.32 (t, 3H, CH3 of ester), 2.33 (s, 6H, 2CH3), 4.25 (q, 2H, CH2 of ester), 4.90 (s, 2H, OCH2), 6.84–7.63 (m, 7H, Ar-H). LC-MS m/z 313 (M + 1). Anal. Cal. for C19H20O4 (312): C, 73.06; H, 6.45. Found: C, 73.04; H, 6.55%.
General procedure for the synthesis of (4-benzoyl phenoxy)-acetic acid 6a–f
A mixture of compounds 5a–f (6.0 mmol) dissolved in ethanol (15 mL) and an aqueous solution of sodium hydroxide (15 mmol) was refluxed for 4–5 h. The reaction mass was cooled and acidified with 3N hydrochloric acid and the aqueous layer The precipitate was filtered, washed with ice water and recrystallized with ethanol to afford compounds 6a–f in a good yield.
(4-Benzoyl-2-methylphenoxy)-acetic acid 6a
Yield: 86%. M.P.: 131–133 °C. IR (KBr) νmax (cm−1): 1660 (C=O), 1720 (acid, C=O), 3420–3520 (acid OH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.44 (s, 3H, CH3), 4.81 (s, 2H, OCH2), 6.90–7.83 (m, 8H, Ar-H), 13.13 (s, 1H, COOH). LC-MS m/z 271 (M + 1). Anal. Cal. for C16H14O4 (270): C, 71.10; H, 5.22. Found: C, 71.06; H, 5.14%.
(4-Benzoyl-2,5-dimethylphenoxy)-acetic acid 6b
Yield: 87%. M.P.: 152–154 °C. IR (KBr) νmax (cm−1): 1655 (C=O), 1730 (acid, C=O), 3400–3530(acid OH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.44 (s, 6H, 2CH3), 4.53 (s, 2H, OCH2), 7.15–7.63 (m, 7H, Ar-H), 12.52 (s, 1H, COOH). LC-MS m/z 285 (M + 1). Anal. Cal. for C17H16O4 (284): C, 71.82; H, 5.67. Found: C, 71.70; H, 5.58%.
[4-(4-Bromo-benzoyl)-2-methylphenoxy]-acetic acid 6c
Yield: 92%. M.P.: 141–143 °C. IR (KBr) νmax (cm−1): 1610 (C=O), 1750 (acid, C=O), 3455–3560(acid OH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.21 (s, 3H, CH3), 4.41 (s, 2H, OCH2), 7.14–7.85 (m, 7H, Ar-H), 12.71 (s, 1H, COOH). LC-MS m/z 349 (M+), 351 (M + 2). Anal. Cal. for C16H13BrO4 (349): C, 55.04; H, 3.75. Found: C, 55.05; H, 3.78%.
[2-Chloro-4-(4-chloro-benzoyl)-6-fluorophenoxy]-acetic acid 6d
Yield: 94%. M.P.: 139–141 °C. IR (KBr) νmax (cm−1): 16440 (C=O), 1770 (acid, C=O), 3495–3600 (acid OH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 4.45 (s, 2H, OCH2), 6.84–7.86 (m, 6H, Ar-H), 12.93 (s, 1H, COOH). LC-MS m/z 343 (M+), 345 (M + 2), 347 (M + 4). Anal. Cal. for C15H9Cl2FO4 (343): C, 52.50; H, 2.64. Found: C, 52.50; H, 2.65%.
[2-Methyl-4-(4-methyl-benzoyl)-phenoxy]-acetic acid 6e
Yield: 93%. M.P.: 118–120 °C. IR (KBr) νmax (cm−1): 1680 (C=O), 1750 (acid, C=O), 3455–3570(acid OH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.32 (s, 6H, 2CH3), 4.39 (s, 2H, OCH2), 6.85–7.57 (m, 7H, Ar-H), 13.04 (s, 1H, COOH). LC-MS m/z 285 (M + 1). Anal. Cal. for C17H16O4 (284): C, 71.82; H, 5.67. Found: C, 71.74; H, 5.63%.
[2-Methyl-4-(2-methyl-benzoyl)-phenoxy]-acetic acid 6f
Yield: 84%. M.P.: 126–128 °C. IR (KBr) νmax (cm−1): 1620 (C=O), 1765 (acid, C=O), 3475–3570 (acid OH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.55 (s, 6H, 2CH3), 4.43 (s, 2H, OCH2), 6.95–7.82 (m, 7H, Ar–H), 12.76 (s, 1H, COOH). LC-MS m/z 285 (M + 1). Anal. Cal. for C17H16O4 (284): C, 71.82; H, 5.67. Found: C, 71.73; H, 5.60%.
General procedure for the synthesis of N-(2-amino-phenyl)-2-(4-benzoyl-phenoxy)-acetamide 8a–f
To a solution of compounds 6a–f (0.0037 mol) in dry dichloromethane (DCM), lutidine (0.0074 mol) was added at 25–30 °C, followed by the addition of o-phenylenediamine (7, 0.0037 mol). The reaction mixture was stirred for 30 min at room temperature, then the reaction was cooled to 0–5 °C, O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluroniumtetrafluoroborate (TBTU) (0.0037 mol) was added over a period of 30 min while maintaining the temperature below 5 °C. The reaction was allowed to stirred overnight and monitored by TLC using hexane and ethyl acetate (4:1). The reaction mixture was diluted with 25 mL of DCM and treated with 1.5 N HCl solution (30 mL). The organic layer was washed with water (3 × 30 mL) and brine (3 × 30 mL). Finally, the organic layer was dried over anhydrous sodium sulfate and concentrated to yield compounds 8a–f.
N-(2-Amino-phenyl)-2-(4-benzoyl-2-methyl-phenoxy)-acetamide 8a
Yield: 92%. M.P.: 191–193 °C. IR (KBr) νmax (cm−1): 1665 (C=O), 1735 (amide, C=O), 3110–3230 (NH), 3315–3410 (NH2). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.44 (s, 3H, CH3), 4.73 (s, 2H, NH2), 4.91 (s, 2H, OCH2), 6.71–7.73 (m, 12H, Ar-H), 9.27 (s, 1H, NH). LC-MS m/z 361 (M + 1). Anal. Cal. for C22H20N2O3 (360): C, 73.32; H, 5.59; N, 7.77. Found: C, 73.20; H, 5.55; N, 7.69%.
N-(2-Amino-phenyl)-2-(4-benzoyl-2,5-dimethyl-phenoxy)-acetamide 8b
Yield: 80%. M.P.: 147–150 °C. IR (KBr) νmax (cm−1): 1685 (C=O), 1755 (amide, C=O), 3140–3260 (NH), 3320–3420 (NH2). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.35 (s, 6H, 2CH3), 4.67 (s, 2H, NH2), 4.88 (s, 2H, OCH2), 6.84–7.76 (m, 11H, Ar–H), 9.32 (s, 1H, NH). LC-MS m/z 375 (M + 1). Anal. Cal. for C23H22N2O3 (374): C, 73.78; H, 5.92; N, 7.48. Found: C, 73.67; H, 5.80; N, 7.45%.
N-(2-Amino-phenyl)-2-[4-(4-bromo-benzoyl)-2-methyl-phenoxy]-acetamide 8c
Yield: 94%. M.P.: 186–188 °C. IR (KBr) νmax (cm−1): 1680 (C=O), 1750 (amide, C=O), 3150–3265 (NH), 3305–3405 (NH2). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.32 (s, 3H, CH3), 4.75 (s, 2H, NH2), 4.93 (s, 2H, OCH2), 6.65–7.87 (m, 11H, Ar-H), 9.21 (s, 1H, NH). LC-MS m/z 438 (M+), 440 (M + 2). Anal. Cal. for C22H19BrN2O3 (438): C, 60.15; H, 4.36; N, 6.38. Found: C, 60.08; H, 4.30; N, 6.27%.
N-(2-Amino-phenyl)-2-[2-chloro-4-(4-chloro-benzoyl)-6-fluoro-phenoxy]-acetamide 8d
Yield: 84%. M.P.: 158–160 °C. IR (KBr) νmax (cm−1): 1655 (C=O), 1705 (amide, C=O), 3140–3240 (NH), 3310–3410 (NH2). 1H NMR (400 MHz) (CDCl3) δ (ppm): 4.55 (s, 2H, NH2), 4.84 (s, 2H, OCH2), 6.82–7.95 (m, 10H, Ar–H), 9.72 (s, 1H, NH). LC-MS m/z 432 (M+), 434 (M + 2), 436 (M + 4). Anal. Cal. for C21H15Cl2FN2O3 (432): C, 58.22; H, 3.49; N, 6.47. Found: C, 58.10; H, 3.45; N, 6.36%.
N-(2-Amino-phenyl)-2-[2-methyl-4-(4-methyl-benzoyl)-phenoxy]-acetamide 8e
Yield: 91%. M.P.: 180–182 °C. IR (KBr) νmax (cm−1): 1685 (C=O), 1750 (amide, C=O), 3150–3250 (NH), 3315–3415 (NH2). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.66 (s, 6H, 2CH3), 4.32 (s, 2H, NH2), 4.71 (s, 2H, OCH2), 6.83–7.86 (m, 11H, Ar-H), 9.62 (s, 1H, NH). LC-MS m/z 375 (M + 1) Anal. Cal. for C23H22N2O3 (374): C, 73.78; H, 5.92; N, 7.48. Found: C, 73.80; H, 5.95; N, 7.43%.
N-(2-Amino-phenyl)-2-[2-methyl-4-(2-methyl-benzoyl)-phenoxy]-acetamide 8f
Yield: 92%. M.P.: 190–192 °C. IR (KBr) νmax (cm−1): 1660 (C=O), 1725 (amide, C=O), 3115–3225 (NH), 3310–3410 (NH2). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.42 (s, 6H, 2CH3), 4.64 (s, 2H, NH2), 4.95 (s, 2H, OCH2), 6.67–7.52 (m, 11H, Ar-H), 9.33 (s, 1H, NH). LC-MS m/z 375 (M + 1). Anal. Cal. for C23H22N2O3 (374): C, 73.78; H, 5.92; N, 7.48. Found: C, 73.76; H, 5.85; N, 7.35%.
General procedure for the synthesis of 2-(4-benzoylphenoxy)-N-{2-[2-(4-benzoyl-phenoxy)-acetylamino]-phenyl}-acetamide 9a–l
To a solution of compounds 8a–f (0.003 mol) in dry DCM (20 mL), substituted (4-benzoyl-phenoxy)-acetic acids 6a–f were added at 25–30 °C, followed by the addition of lutidine (0.003 mol). The reaction mixture was stirred at room temperature for 30–40 min. The reaction mixture was cooled to 0–5 °C and TBTU (0.001 mol) was added over a period of 30 min while maintaining the temperature below 5 °C. The reaction allowed to stir overnight and monitored by TLC using hexane and ethyl acetate (4:1). After completion of reaction, the reaction mixture was diluted with 30 mL of DCM and treated with 10% sodium bicarbonate solution (3 × 30 mL). The organic layer was washed with water (3 × 20 mL), dried over anhydrous sodium sulfate and concentrated to afford compounds 9a–l.
2-(4-Benzoyl-2-methyl-phenoxy)-N-{2-[2-(4-benzoyl-2-methyl-phenoxy)-acetylamino]-phenyl}-acetamide 9a
Yield: 80%. M.P.: 88–90 °C; IR (KBr) νmax (cm−1): 1665 (C=O), 1740 (amide, C=O), 3100–3210 (NH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.29 (s, 6H, 2CH3), 4.76 (s, 4H, 2OCH2), 6.95–7.65 (m, 20H, Ar-H), 9.67 (s, 2H, 2NH). 13C NMR (DMSO-d6) δ: 194.8 (2C=O), 167.0 (2CONH), 159.7 (2C, C1, C19), 138.2 (2C, C7, C25), 132.5 (2C, C13, C14), 132.4 (2CH, C10, C28), 130.7 (2CH, C3, C21), 130.4 (2CH, C8, C26), 130.1 (2CH, C12, C30), 129.6 (2C, C4, C22), 129.6 (2CH, C9, C25), 128.8 (2CH, C11, C29), 128.7 (2CH, C5, C23), 127.0 (2CH, C16, C17), 126.3 (2C, C2, C20), 126.0 (2CH, C15, C18), 111.2 (2CH, C6, C24), 67.5 (2OCH2), 16.5 (2CH3). LC-MS m/z 613 (M + 1). Anal. Cal. for C38H32N2O6 (612): C, 74.49; H, 5.26; N, 4.57. Found: C, 74.37; H, 5.19; N, 4.40%.
2-(4-Benzoyl-2,5-dimethyl-phenoxy)-N-{2-[2-(4-benzoyl-2,5-dimethyl-phenoxy)-acetyl amino]-phenyl}-acetamide 9b
Yield: 75%. M.P.: 92–94 °C. IR (KBr) νmax (cm−1): 1685 (C=O), 1740 (amide, C=O), 3110–3230 (NH).1H NMR (400 MHz) (CDCl3) δ (ppm): 2.45 (s, 12H, 4CH3), 4.78 (s, 4H, 2OCH2), 6.58–7.69 (m, 18H, Ar–H), 9.38 (s, 2H, 2NH). 13C NMR (DMSO-d6) δ: 194.0 (2C=O), 165.5 (2CONH), 162.8 (2C, C1, C19), 139.5 (2C, C7, C25), 138.5 (2C, C5, C23), 135.2 (2C, C13, C14), 132.4 (2CH, C10, C28), 131.7 (2CH, C3, C21), 129.2 (2C, C4, C22), 128.3 (2CH, C8, C26), 127.5 (2CH, C12, C30), 126.0 (4CH, C9, C11, C27, C29), 124.7 (2CH, C16, C17), 123.5 (2CH, C15, C18), 122.1 (2C, C2, C20), 117.6 (2CH, C6, C24), 68.7 (2OCH2), 19.0 (2CH3), 16.0 (2CH3). LC-MS m/z 641 (M + 1). Anal. Cal. for C40H36N2O6 (640): C, 74.49; H, 5.26; N, 4.57. Found: C, 74.40; H, 5.21; N, 4.49%.
N-{2-[2-(4-Benzoyl-2-methylphenoxy)-acetylamino]-phenyl}-2-[2-methyl-4-(4-methyl-benzoyl)-phenoxy]-acetamide 9c
Yield: 71%. M.P.: 90–92 °C. IR (KBr) νmax (cm−1): 1675 (C=O), 1750 (amide, C=O), 3140–3260 (NH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.33 (s, 9H, 3CH3), 4.84 (s, 4H, 2OCH2), 6.73–7.87 (m, 19H, Ar–H), 9.77 (s, 2H, 2NH). 13C NMR (DMSO-d6) δ: 193.4 (2C=O), 166.3 (2CONH), 161.4 (2C, C1, C19), 140.4 (2C, C7, C25), 136.5 (2C, C13, C14), 133.8 (2CH, C8, C26), 133.3 (2CH, C12, C30), 132.5 (2CH, C3, C21), 131.4 (2CH, C9, C27), 130.8 (2CH, C11, C29), 129.5 (2C, C4, C22), 128.6 (2C, C10, C28), 127.4 (2CH, C5, C23), 126.4 (2CH, C16, C17), 123.6 (2C, C2, C20), 122.7 (2CH, C15, C18), 119.8 (2CH, C6, C24), 69.4 (2OCH2), 16.5 (2CH3). LC-MS m/z 627 (M + 1). Anal. Cal. for C39H34N2O6 (626): C, 74.74; H, 5.47; N, 4.47. Found: C, 74.71; H, 5.35; N, 4.37%.
N-{2-[2-(4-Benzoyl-2-methylphenoxy)-acetylamino]-phenyl}-2-[2-methyl-4-(2-methyl-benzoyl)-phenoxy]-acetamide 9d
Yield: 90%. M.P.: 94–96 °C. IR (KBr) νmax (cm−1): 1665 (C=O), 1730 (amide, C=O), 3110–3230 (NH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.35 (s, 9H, 3CH3), 4.67 (s, 4H, 2OCH2), 6.57–7.76 (m, 19H, Ar–H), 9.76 (s, 2H, 2NH). 13C NMR (DMSO-d6) δ: 193.7 (2C=O), 168.5 (2CONH), 164.3 (C, C1), 163.1 (C, C19), 140.5 (2C, C7, C25), 138.5 (C, C23), 137.4 (2C, C13, C14), 135.6 (2CH, C10, C28), 133.4 (CH, C3), 132.4 (CH, C21), 131.7 (C, C22), 130.3 (4CH, C8, C12, C26, C30), 129.6 (C, C4), 128.3 (4CH, C9, C11, C27, C29), 127.4 (CH, C5), 127.6 (2CH, C6, C17), 127.6 (C, C2), 124.3 (2CH, C15, C18), 122.8 (C, C20), 121.6 (CH, C24), 119.3 (CH, C6), 70.6 (2OCH2), 19.7 (CH3), 16.1 (2CH3). LC-MS m/z 627 (M + 1). Anal. Cal. for C39H34N2O6 (626): C, 74.74; H, 5.47; N, 4.47. Found: C, 74.68; H, 5.41; N, 4.39%.
N-{2-[2-(4-Benzoyl-2,5-dimethylphenoxy)-acetylamino]-phenyl}-2-[4-(4-bromo-benzoyl)-2-methyl-phenoxy]-acetamide 9e
Yield: 83%. M.P.: 91–93 °C. IR (KBr) νmax (cm−1): 1675 (C=O), 1730 (amide, C=O), 3120–3220 (NH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.75 (s, 9H, 3CH3), 4.72 (s, 4H, 2OCH2), 6.43–7.52 (m, 18H, Ar–H), 9.72 (s, 2H, 2NH). 13C NMR (DMSO-d6) δ: 194.3 (2C=O), 166.2 (CONH), 165.3 (CONH), 164.4 (C, C19), 142.5 (C, C1), 140.6 (C, C6), 138.7 (C, C25), 137.3 (C, C10), 136.2 (C, C7), 133.5 (C, C4), 132.6 (2C, C13, C14), 131.7 (CH, C21), 130.8 (2CH, C8, C12), 128.9 (2CH, C26, C30), 127.4 (C, C22), 126.7 (2CH, C9, C11), 123.5 (2CH, C27, C29), 122.2 (CH, C23), 121.2 (CH, C3), 120.7 (2CH, C16, C17), 120.6 (C, C20), 119.3 (C, C2), 118.5 (2CH, C15, C18), 116.8 (CH, C5), 115.3 (CH, C24), 69.7 (2OCH2), 18.8 (CH3), 15.6 (2CH3). LC-MS m/z 706 (M+), 708 (M + 2). Anal. Cal. for C39H33BrN2O6 (706): C, 66.39; H, 4.71; N, 3.97. Found: C, 66.12; H, 4.68; N, 3.82%.
2-[4-(4-Bromo-benzoyl)-2-methylphenoxy]-N-(2-{2-[4-(4-bromo-benzoyl)-2-methyl-phenoxy]-acetylamino}-phenyl)-acetamide 9f
Yield: 79%. M.P.: 87–89 °C. IR (KBr) νmax (cm−1): 1685 (C=O), 1770 (amide, C=O), 3160–3240 (NH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.54 (s, 6H, 2CH3), 4.76 (s, 4H, 2OCH2), 6.74–7.85 (m, 18H, Ar–H), 9.58 (s, 2H, 2NH). 13C NMR (DMSO-d6) δ: 194.7 (2C=O), 165.5 (2CONH), 162.4 (2C, C1, C19), 139.7 (2C, C7, C25), 134.9 (4CH, C8, C12, C26, C30), 132.8 (2C, C4, C22), 130.0 (2CH, C3, C21), 129.3 (4CH, C9, C11, C27, C29), 128.5 (2C, C13, C14), 128.6 (2CH, C5, C23), 126.8 (2C, C10, C28), 124.5 (2CH, C15, C18), 123.0 (2CH, C16, C17), 121.8 (2C, C2, C20), 116.8 (2CH, C6, C24), 65.8 (2OCH2), 15.1 (2CH3). LC-MS m/z 770 (M+), 772 (M + 2), 774 (M + 4). Anal. Cal. for C38H30Br2N2O6 (770): C, 59.24; H, 3.92; N, 3.64. Found: C, 59.20; H, 3.81; N, 3.53%.
N-{2-[2-(4-Benzoyl-3-methylphenoxy)-acetylamino]-phenyl}-2-[4-(4-bromo-benzoyl)-2-methyl-phenoxy]-acetamide 9g
Yield: 73%. M.P.: 84–86 °C; IR (KBr) νmax (cm−1): 1685 (C=O), 1740 (amide, C=O), 3130–3240 (NH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.13 (s, 6H, 2CH3), 4.64 (s, 2H, 2OCH2), 6.53–7.62 (m, 19H, Ar–H), 9.59 (s, 2H, 2NH). 13C NMR (DMSO-d6) δ: 193.3 (2C=O), 167.3 (2CONH), 166.3 (2C, C1, C19), 139.5 (C, C10), 138.5 (C, C25), 136.6 (C, C7), 135.1 (2C, C13, C14), 134.4 (CH, C28), 133.3 (2CH, C3, C21), 132.3 (2CH, C26, C30), 131.2 (C, CH C4, C12), 130.4 (C, CH, C8, C22), 129.2 (2CH, C9, C11), 125.3 (2CH, C27, C29), 124.3 (2CH, C5, C23), 123.6 (2CH, C16, C17), 122.1 (2C, C2, C20), 120.6 (2CH, C15, C18), 119.3 (2CH, C6, C24), 65.2 (2OCH2), 20.5 (CH3), 16.7 (2CH3). LC-MS m/z 692 (M+), 694 (M + 2). Anal. Cal. for C38H31N2 BrO6 (692): C, 66.00; H, 4.52; N, 4.05. Found: C, 65.91; H, 4.19; N, 4.02%.
N-{2-[2-(4-Benzoyl-2,5-dimethylphenoxy)-acetylamino]-phenyl}-2-(4-benzoyl-2-methyl-phenoxy)-acetamide 9h
Yield: 80%. M.P.: 95–97 °C. IR (KBr) νmax cm−11665 (C=O), 1720 (amide, C=O), 3120–3230 (NH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.44 (s, 9H, 3CH3), 4.76 (s, 4H, 2OCH2), 6.89–7.66 (m, 19H, Ar–H), 9.67 (s, 2H, 2NH). 13C NMR (DMSO-d6) δ: 194.5 (2C=O), 165.4 (CONH), 162.6 (CONH), 161.7 (2C, C1, C19), 139.8 (C, C7), 138.9 (C, C25), 136.4 (2C, C13, C14), 135.9 (2CH, C26, C30), 134.3 (CH, C10), 132.2 (2CH, C3, C21), 131.7 (2CH, C27, C29), 131.2 (2CH, C8, C12), 128.5 (2C, C4, C22), 126.3 (2CH, C9, C11), 124.3 (2CH, C5, C23), 122.8 (C, C28), 121.9 (2CH, C16, C17), 120.0 (2C, C2, C20), 118.3 (2CH, C15, C18), 114.2 (2CH, C6, C24), 73.2 (2OCH2), 20.6 (CH3), 16.6 (2CH3). LC-MS m/z 627 (M + 1). Anal. Cal. for C39H34N2O6 (626): C, 74.74; H, 5.47; N, 4.47; O. Found: C, 74.40; H, 5.21; N, 4.49%.
N-{2-[2-(4-Benzoyl-2-methylphenoxy)-acetylamino]-phenyl}-2-[2-chloro-4-(4-chloro-benzoyl)-6-fluoro-phenoxy]-acetamide 9i
Yield: 88%. M.P.: 81–83 °C. IR (KBr) νmax (cm−1): 1685 (C=O), 1750 (amide, C=O), 3130–3230 (NH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.43 (s, 3H, CH3), 4.76 (s, 4H, 2OCH2), 6.78–7.76 (m, 18H, Ar–H), 9.86 (s, 2H, 2NH). 13C NMR (DMSO-d6) δ: 194.7 (2C=O), 164.5 (2CONH), 163.3 (2C, C1, C19), 152.3 (C, C19), 145.8 (C, C1), 137.5 (C, C7), 136.2 (C, C5), 135.4 (C, C25), 133.9 (2C, C13, C14), 133.5 (2CH, C26, C30), 132.4 (CH, C10), 131.7 (CH, C21), 130.4 (CH, C3), 129.8 (2CH, C27, C29), 128.7 (C, C4), 127.9 (2CH, C8, C12), 126.2 (C, C22), 124.2 (2CH, C9, C11), 123.5 (CH, C23), 122.4 (CH, C23),122.0 (2CH, C16, C17), 121.2 (C, C20), 120.8 (2CH, C15, C18), 119.4 (C, C2), 117.6 (CH, C6), 116.4 (CH, C24), 65.2 (2OCH2), 20.4 (CH3), 15.3 (2CH3). LC-MS m/z 686 (M+), 688 (M + 2), 690 (M + 4). Anal. Cal. for C37H27Cl2FN2O6 (686): C, 68.83; H, 3.97; N, 4.09. Found: C, 68.75; H, 3.87; N, 4.02%.
N-{2-[2-(4-Benzoyl-2,5-dimethylphenoxy)-acetylamino]-phenyl}-2-[4-(2-methyl-benzoyl)-2-methyl-phenoxy]-acetamide 9j
Yield: 76%. M.P.: 86–88 °C. IR (KBr) νmax (cm−1): 1665 (C=O), 1750 (amide, C=O), 3160–3260 (NH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.35 (s, 12H, 4CH3), 4.81 (s, 4H, 2OCH2), 6.83–7.75 (m, 18H, Ar–H), 9.86 (s, 2H, 2NH). 13C NMR (DMSO-d6) δ: 195.2 (2C=O), 167.7 (2CONH), 161.6 (C, C19), 154.9 (C, C1), 147.4 (C, C6), 136.9 (C, C10), 134.0 (C, C25), 133.5 (C, C7), 133.1 (C, C4), 132.5 (2C, C13, C14), 132.7 (2CH, C26, C30), 131.8 (CH, C21), 130.5 (2CH, C8, C27), 130.7 (2CH, C12, C29), 130.9 (C, C22), 129.0 (2CH, C9, C11), 128.5 (CH, C23), 127.4 (CH, C3), 127.2 (2CH, C16, C17), 126.3 (C, C28), 124.2 (C, C20), 118.7 (C, C2), 115.2 (2CH, C15, C18), 111.6 (CH, C5), 110.5 (CH, C24), 68.3 (OCH2), 67.5 (OCH2), 16.7 (3CH3). LC-MS m/z 641 (M + 1). Anal. Cal. for C40H36N2O6 (640): C, 74.98; H, 5.66; N, 4.37. Found: C, 74.88; H, 5.64; N, 4.32%.
N-(2-{2-[4-(4-Bromo-benzoyl)-2-methylphenoxy]-acetylamino}-phenyl)-2-[2-methyl-4-(2-methyl-benzoyl)-phenoxy]-acetamide 9k
Yield: 87%. M.P.: 94–96 °C. IR (KBr) νmax (cm−1): 1675 (C=O), 1710 (amide, C=O), 3130–3250 (NH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.44 (s, 9H, 3CH3), 4.69 (s, 4H, 2OCH2), 6.82–7.71 (m, 18H, Ar–H), 9.71 (s, 2H, 2NH). 13C NMR (DMSO-d6) δ: 194.4 (2C=O), 166.3 (2CONH), 164.6 (2C, C1, C19), 155.7 (C, C8), 150.4 (C, C7), 143.5 (C, C25), 138.7 (2C, C13, C14), 136.8 (2CH, C26, C30), 134.9 (CH, C10), 133.3 (2CH, C3, C21), 132.1 (2CH, C27, C29), 131.3 (2C, C4, C22), 130.7 (CH, C12), 129.9 (C, C4), 128.2 (CH, C9), 124.2 (2CH, C5, C23), 122.7 (C, C28), 121.6 (CH, C11), 120.5 (2CH, C16, C17), 119.8 (2C, C2, C20), 117.5 (2CH, C15, C18), 116.3 (2CH, C6, C24), 67.9 (2OCH2), 20.3 (CH3), 16.6 (2CH3). LC-MS m/z 706 (M+), 708 (M + 2). Anal. Cal. for C39H33BrN2O6 (706): C, 66.39; H, 4.71; N, 3.97. Found: C, 66.32; H, 4.63; N, 3.86%.
N-(2-{2-[4-(4-Bromo-benzoyl)-2-methylphenoxy]-acetylamino}-phenyl)-2-[2-methyl-4-(4-methyl-benzoyl)-phenoxy]-acetamide 9l
Yield: 72%. M.P.:102–104 °C. IR (KBr) νmax (cm−1): 1635 (C=O), 1750 (amide, C=O), 3160–3250 (NH). 1H NMR (400 MHz) (CDCl3) δ (ppm): 2.43 (s, 9H, 3CH3), 4.81 (s, 4H, 2OCH2), 6.74–7.62 (m, 18H, Ar–H), 9.72 (s, 2H, 2NH). 13C NMR (DMSO-d6) δ: 194.2 (2C=O), 166.9 (2CONH), 165.2 (2C, C1, C19), 138.4 (C, C10), 136.4 (C, C25), 134.3 (C, C7), 133.6 (2C, C13, C14), 132.4 (2CH, C26, C30), 131.8 (2CH, C3, C27), 130.0 (2CH, C8, C12), 129.8 (2C, C4, C22), 128.6 (2CH, C9, C11), 127.1 (2CH, C5, C23), 126.1 (C, C28), 124.1 (2CH, C16, C17), 122.1 (2C, C2, C20), 121.6 (2CH, C15, C18), 116.4 (2CH, C6, C24), 65.2 (2OCH2), 19.2 (CH3), 15.3 (2CH3). LC-MS m/z 706 (M+), 708 (M + 2). Anal. Cal. for C39H33BrN2O6 (706): C, 66.39; H, 4.71; N, 3.97. Found: C, 66.35; H, 4.68; N, 3.93%.
General procedure for the synthesis of 2-(4-Benzoyl-phenoxy)-N-{2-[[2-(4-benzoyl-phenoxy)-acetyl]-(2-piperidin-1-yl-ethyl)-amino]-phenyl}-acetamide 11a–l
A mixture of compounds 9a–l (0.001 mol) and 1-(2-chloroethyl)piperidine hydrochloride 10 (0.001 mol) in the presence of potassium tertiary butoxide (0.005 mol) and DMSO (10 mL) was refluxed for 8–10 h then cooled. The reaction was monitored on TLC, after the completion of the reaction the residual mass was triturated with ice cold water to remove potassium tertiary butoxide and DMSO then, it was extracted with DCM (30 mL). The organic layer was washed with a saturated sodium chloride solution (3 × 20 mL), 10% sodium hydroxide solution (3 × 20 mL) followed by distilled water (3 × 30 mL) and then dried over anhydrous sodium sulfate. Finally the solvent was evaporated to dryness and recrystallized with ethanol to afford the title compounds 11a–l.
2-(4-Benzoyl-2-methylphenoxy)-N-{2-[2-(4-benzoyl-2-methylphenoxy)-acetylamino]-phenyl}-N-(2-piperidin-1-yl-ethyl)-acetamide 11a
Yield: 89%. M.P.: 102–104 °C; IR (KBr) νmax (cm−1): 1685 (C=O), 1760 (amide, C=O), 3160–3270 (NH). 1H NMR (400 MHz, DMSO-d6) δ (ppm): 2.21–2.31 (m, 6H, ring-3CH2), 2.37 (t, 4H, ring-2NCH2), 2.72 (s, 3H, CH3), 3.29 (s, 3H, CH3), 3.58 (t, 2H, NCH2), 4.03 (t, 2H, CONCH2), 4.53 (s, 2H, OCH2), 4.72 (s, 2H, OCH2), 6.71–7.72 (m, 20H, Ar–H), 8.75 (s, 1H, NH). 13C NMR (DMSO-d6) δ: 184.8 (2C=O), 167.0 (CONH), 164.9 (CO–N), 162.5 (2C, C1, C19), 133.0 (2C, C7, C25), 132.4 (2C, C13, C14), 130.5 (2CH, C10, C28), 130.3 (2CH C3, C21), 129.8 (4CH, C8, C12, C26, C30), 129.6 (2C, C4, C22), 128.7 (4CH, C9, C11, C27, C29), 128.4 (2CH, C5, C23), 128.2 (2CH, C16, C17), 127.0 (2C, C2, C20), 118.2 (2CH, C15, C18), 111.3 (2CH, C6, C24), 81.4 (CH2, OCH2), 78.2 (CH2, OCH2), 56.2 (CH2, NCH2), 51.7 (CH2, NCH2 of morphiline ring), 46.0 (CH2, CO-NCH2), 33.1 (2CH2, CCH2), 31.3 (CH2, CCH2), 15.5 (2CH3). LC-MS m/z 724.12 (M + 1). Anal. Cal. for C45H45N3O6 (723.33): C, 74.67; H, 6.27; N, 5.81. Found: C, 74.68; H, 6.26; N, 5.78%.
2-(4-Benzoyl-2,5-dimethylphenoxy)-N-{2-[2-(4-benzoyl-2,5-dimethylphenoxy)-acetylamino]-phenyl}-N-(2-piperidin-1-yl-ethyl)-acetamide 11b
Yield: 90%. M.P.: 97–99 °C. IR (KBr) νmax (cm−1): 1655 (C=O), 1740 (amide, C=O), 3190–3270 (NH).1H NMR (400 MHz, DMSO-d6) δ (ppm): 1.51–1.83 (m, 6H, ring-3CH2), 2.13 (t, 4H, ring-2NCH2), 2.31 (s, 6H, 2CH3), 2.35 (s, 3H, CH3), 2.49 (s, 3H, CH3), 2.73 (t, 2H, NCH2), 3.54 (t, 2H, CONCH2), 4.75 (s, 2H, OCH2), 4.89 (s, 2H, OCH2), 6.56–7.62 (m, 18H, Ar-H), 9.37 (s, 1H, NH). 13C NMR (DMSO-d6) δ: 186.8 (2C=O), 168.6 (CONH), 166.6 (CO-N), 164.8 (2C, C1, C19), 139.7 (2C, C7, C25), 138.5 (2C, C5, C23), 136.6 (2C, C13, C14), 135.5 (2CH, C10, C28), 133.5 (2CH, C3, C21), 132.4 (2C, C4, C22), 131.3 (4CH, C8, C12, C26, C30), 129.1 (4CH, C9, C11, C27, C29), 126.6 (2CH, C15, C18), 124.5 (2C, C2, C20), 122.2 (2CH, C16, C17), 117.8 (2CH, C6, C24), 76.5 (CH2, OCH2), 74.6 (CH2, OCH2), 55.4 (CH2, NCH2), 52.6 (CH2, ring-NCH2), 48.6 (CH2, CO–NCH2), 26.4 (2CH2, CCH2), 23.2 (CH2, CCH2), 16.2 (2CH3), 12.4 (2CH3). LC-MS m/z 752.36 (M + 1). Anal. Cal. for C47H49N3O6 (751.36): C, 75.08; H, 6.57; N, 5.59. Found: C, 74.98; H, 6.47; N, 5.48%.
N-{2-[2-(4-Benzoyl-2-methylphenoxy)-acetylamino]-phenyl}-2-[2-methyl-4-(4-methyl-benzoyl)-phenoxy]-N-(2-piperidin-1-yl-ethyl)-acetamide 11c
Yield: 78%. M.P.: 92–94 °C. IR (KBr) νmax (cm−1): 1685 (C=O), 1710 (amide, C=O), 3140–3250 (NH). 1H NMR (400 MHz, DMSO-d6) δ (ppm): 1.60–1.88 (m, 6H, ring-3CH2), 2.16 (t, 4H, ring-2NCH2), 2.39 (s, 3H, 3CH3), 2.44 (s, 3H, CH3), 2.62 (s, 3H, CH3),2.75 (t, 2H, NCH2), 3.57 (t, 2H, CONCH2), 4.81 (s, 2H, OCH2), 4.87 (s, 2H, OCH2), 6.72–7.82 (m, 18H, Ar-H), 9.54 (s, 1H, NH). 13C NMR (DMSO-d6) δ: 187.3 (2C=O), 168.5 (CONH), 166.2 (CO-N), 142.4 (C, C10), 139.8 (C, C25), 137.5 (C, C7), 135.3 (2C, C13, C14), 134.3 (C, C28), 133.2 (2CH, C3, C21), 132.4 (2CH, C26, C30), 130.6 (4CH, C4, C8, C12, C22), 129.5 (2CH, C9, C11), 128.3 (2CH, C27, C29), 126.1 (2CH, C5, C23), 125.8 (2CH, C16, C17), 124.5 (2CH, C2, C20), 121.1 (2CH, C15, C18), 116.8 (2CH, C6, C24), 76.9 (CH2, OCH2), 74.8 (CH2, OCH2), 56.4 (CH2, NCH2), 54.0 (CH2, ring-NCH2), 49.5 (CH2, CO–NCH2), 26.4 (2CH2, CCH2), 25.3 (CH2, CCH2), 21.2 (CH3), 12.4 (2CH3). LC-MS m/z 738.35 (M + 1). Anal. Cal. for C46H47N3O6 (737.35): C, 74.88; H, 6.42; N, 5.69. Found: C, 74.79; H, 6.46; N, 5.67%.
N-{2-[2-(4-Benzoyl-2-methylphenoxy)-acetylamino]-phenyl}-2-[2-methyl-4-(2-methyl-benzoyl)-phenoxy]-N-(2-piperidin-1-yl-ethyl)-acetamide 11d
Yield: 89%. M.P.: 98–100 °C. IR (KBr) νmax cm−11695 (C=O), 1780 (amide, C=O), 3150–3260 (NH). 1H NMR (400 MHz, DMSO-d6) δ (ppm): 1.56–1.81 (m, 6H, ring-3CH2), 2.18 (t, 4H, ring-2NCH2), 2.35 (s, 3H, 3CH3), 2.42 (s, 3H, CH3), 2.55 (s, 3H, CH3), 3.21 (t, 2H, NCH2), 3.64 (t, 2H, CONCH2), 4.79 (s, 2H, OCH2), 4.90 (s, 2H, OCH2), 6.8–7.6 (m, 19H, Ar-H), 9.6 (s, 1H, NH). 13C NMR (DMSO-d6) δ: 187.2 (2C=O), 167.8 (CONH), 165.6 (CO–N), 139.9 (C, C8), 138.4 (C, C7), 136.3 (C, C25), 135.7 (2C, C13, C14), 133.8 (C, C28), 132.4 (CH, C10), 131.7 (2CH, C3, C21), 131.3 (2CH, C26, C30), 130.6 (C, 2CH, C4, C12, C22), 128.8 (CH, C9, 126.7 (2CH, C27, C29), 124.3 (2CH, C5, C23), 123.9 (CH, C11), 122.3 (2CH, C16, C17), 121.6 (2C, C2, C20), 120.7 (2CH, C15, C18), 114.2 (2CH, C6, C24), 76.9 (CH2, OCH2), 73.2 (CH2, OCH2), 55.7 (CH2, NCH2), 53.3 (CH2, ring-NCH2), 48.9 (CH2, CO-NCH2), 27.6 (2CH2, CCH2), 24.3 (CH2, CCH2), 16.3 (CH3), 11.0 (2CH3). LC-MS m/z 738.35 (M + 1). Anal. Cal. for C46H47N3O6 (737.35): C, 74.88; H, 6.42; N, 5.69. Found C, 74.79; H, 6.34; N, 5.58%.
2-(4-Benzoyl-2,5-dimethylphenoxy)-N-(2-{2-[4-(4-bromo-benzoyl)-2-methylphenoxy]-acetylamino}-phenyl)-N-(2-piperidin-1-yl-ethyl)-acetamide 11e
Yield: 82%. M.P.: 90–92 °C. IR (KBr) νmax (cm−1): 1665 (C=O), 1725 (amide, C=O), 3150–3250 (NH). 1H NMR (400 MHz, DMSO-d6) δ (ppm): 1.61–1.88 (m, 6H, ring-3CH2), 2.15 (t, 4H, ring-2NCH2), 2.31 (s, 3H, CH3), 2.49 (s, 3H, CH3), 2.64 (s, 3H, CH3), 3.13 (t, 2H, NCH2), 3.57 (t, 2H, CONCH2), 4.80 (s, 2H, OCH2), 4.97 (s, 2H, OCH2), 6.67–7.74 (m, 18H, Ar-H), 9.56 (s, 1H, NH). 13C NMR (DMSO-d6) δ: 189.2 (2C=O), 166.4 (CONH), 164.3 (CO–N), 162.8 (C, C19), 145.6 (C, C1), 139.5 (C, C7), 138.3 (C, C5), 137.6 (C, C25), 136.3 (2C, C13, C14), 134.8 (2CH, C26, C30), 134.4 (CH, C10), 133.4 (CH, C21), 133.6 (CH, C3), 132.7 (2CH, C27, C29), 131.4 (C, C4), 130.4 (2CH, C8, C12), 130.1 (C, C22), 128.5 (2CH, C9, C11), 127.7 (CH, C23), 126.2 (C, C28), 124.6 (2CH, C16, C17), 123.5 (C, C20), 121.3 (2CH, C18, C15), 120.2 (C, C2), 116.3 (CH, C6), 113.4 (CH, C24), 76.2 (CH2, OCH2), 74.3 (CH2, OCH2), 56.5 (CH2, NCH2), 53.8 (CH2, ring-NCH2), 48.1 (CH2, CO–NCH2), 27.7 (2CH2, CCH2), 25.8 (CH2, CCH2), 15.7 (CH3), 11.4 (2CH3). LC-MS m/z 815.23 (M+), 817.29 (M + 2). Anal. Cal. for C46H46BrN3O6 (815.26): C, 67.64; H, 5.68; N, 5.14. Found): C, 67.69; H, 5.61; N, 5.18%.
2-[4-(4-Bromo-benzoyl)-2-methylphenoxy]-N-(2-{2-[4-(4-bromo-benzoyl)-2-methyl phenoxy]-acetylamino}-phenyl)-N-(2-piperidin-1-yl-ethyl)-acetamide 11f
Yield: 88%. M.P.: 95–97 °C. IR (KBr) νmax (cm−1): 1695 (2C=O), 1750 (amide, C=O), 3140–3250 (NH). 1H NMR (400 MHz, DMSO-d6) δ (ppm): 1.53–1.86 (m, 6H, ring-3CH2), 2.16 (t, 4H, ring-2NCH2), 2.31 (s, 3H, CH3), 2.42 (s, 3H, CH3), 2.73 (t, 2H, NCH2), 3.54 (t, 2H, CONCH2), 4.75 (s, 2H, OCH2), 4.89 (s, 2H, OCH2), 6.62–7.83 (m, 18H, Ar–H), 9.42 (s, 1H, NH). 13C NMR (DMSO-d6) δ: 187.8 (2C=O), 168.5 (CONH), 165.7 (CO-N), 162.3 (2C, C1, C19), 138.6 (2C, C7, C25), 137.4 (2C, C13, C14), 135.8 (4CH, C8, C12, C26, C30), 132.2 (2CH, C3, C21), 131.6 (4CH, C9, C11, C27, C29), 130.3 (2C, C4, C22), 129.6 (2CH, C5, C23), 128.3 (2C, C10, C28), 127.6 (2CH, C16, C17), 124.3 (2C, C2, C20), 121.5 (2CH, C15, C18), 119.5 (2CH, C16, C24), 77.5 (CH2, OCH2), 74.8 (CH2, OCH2), 54.9 (CH2, NCH2), 52.5 (CH2, ring-NCH2), 49.5 (CH2, CO–NCH2), 26.7 (2CH2, CCH2), 24.2 (CH2, CCH2), 16.6 (2CH3). LC-MS m/z 879.15 (M + 1), 881.19 (M + 2), 883.21 (M + 4). Anal. Cal. for C45H43Br2N3O6 (879.15): C, 61.30; H, 4.92; N, 4.77. Found C, 61.29; H, 4.86; N, 4.74%.
2-(4-Benzoyl-2-methylphenoxy)-N-(2-{2-[4-(4-bromo-benzoyl)-2-methylphenoxy]-acetyl amino}-phenyl)-N-(2-piperidin-1-yl-ethyl)-acetamide 11g
Yield: 79%. M.P.: 102–104 °C. IR (KBr) νmax (cm−1): 1665 (C=O), 1735 (amide, C=O), 3140–3240 (NH). 1H NMR (400 MHz, DMSO-d6) δ (ppm): 1.43–1.79 (m, 6H, ring-3CH2), 2.26 (t, 4H, ring-2NCH2), 2.38 (s, 3H, CH3), 2.47 (s, 3H, CH3), 2.85 (t, 2H, NCH2), 3.65 (t, 2H, CONCH2), 4.62 (s, 2H, OCH2), 4.83 (s, 2H, OCH2), 6.73–7.79 (m, 19H, Ar–H), 9.83 (s, 1H, NH). 13C NMR (DMSO-d6) δ: 193.4 (2C=O), 168.8 (CONH), 166.4 (CO–N), 164.5 (2C, C1, C19), 139.5 (C, C7), 137.4 (C, C25), 136.4 (2C, C13, C14), 134.8 (2CH, C26, C30), 133.2 (CH, C10), 132.1 (2CH, C3, C2), 131.5 (2CH, C27, C29), 130.8 (2CH, C8, C12), 130.9 (2C, C4, C22), 129.6 (2CH, C11, C9), 128.6 (2CH, C5, C23), 127.5 (C, C28), 123.0 (2CH, C10, C15), 122.7 (2C, C2, C20), 119.6 (2CH, C15, C18), 113.3 (2CH, C6, C24), 77.5 (CH2, OCH2), 75.5 (CH2, OCH2), 53.4 (CH2, NCH2), 52.8 (CH2, ring-NCH2), 49.4 (CH2, ring-NCH2), 23.3 (2CH2, CCH2), 21.2 (CH2, CCH2), 11.4 (2CH3). LC-MS m/z 801.24 (M+), 803.21 (M + 2). Anal. Cal. for C45H44BrN3O6 (801.24): C, 67.33; H, 5.52; N, 5.23. Found: C, 67.30; H, 5.44; N, 5.28%.
N-{2-[2-(4-Benzoyl-2,5-dimethylphenoxy)-acetylamino]-phenyl}-2-(4-benzoyl-2-methyl phenoxy)-N-(2-piperidin-1-yl-ethyl)-acetamide 11h
Yield: 82%. M.P.: 107–109 °C; IR (KBr) νmax (cm−1): 1670 (C=O), 1720 (amide, C=O), 3225–3325 (NH). 1H NMR (400 MHz, DMSO-d6) δ (ppm): 1.72–1.83 (m, 6H, ring-3CH2), 2.22 (t, 4H, ring-2NCH2), 2.45 (s, 3H, CH3), 2.62 (s, 6H, 2CH3), 2.82 (t, 2H, NCH2), 3.61 (t, 2H, CONCH2), 4.75 (s, 2H, OCH2), 4.90 (s, 2H, OCH2), 6.62–7.73 (m, 19H, Ar–H), 9.84 (s, 1H, NH). 13C NMR (DMSO-d6) δ: 190.4 (2C=O), 171.6 (CONH), 169.3 (CO–N), 168.4 (C, C1), 143.6 (C, C19), 139.5 (2C, C7, C25), 138.7 (C, C23), 136.7 (2C, C13, C14), 134.9 (2CH, C10, C28), 133.8 (CH, C3), 132.5 (CH, C21), 130.4 (C, C22), 128.8 (4CH, C8, C12, C26, C30), 127.4 (C, C4), 125.6 (4CH, C9, C11, C27, C29), 124.2 (CH, C5), 121.3 (2CH, C16, C17), 120.5 (C, C2), 120.4 (2CH, C15, C18), 119.7 (C, C20), 118.0 (CH, C24), 113.8 (CH, C6), 82.1 (CH2, OCH2), 75.5 (CH2, OCH2), 58.9 (CH2, NCH2), 55.4 (CH2, ring-NCH2), 52.3 (CH2, CO-NCH2), 28.6 (2CH2, CCH2), 26.2 (CH2, CCH2), 14.7 (CH3), 12.0 (2CH3). LC-MS m/z 738.35 (M + 1). Anal. Cal. for C46H47N3O6 (737.35) C, 74.88; H, 6.42; N, 5.69. Found: C, 74.75; H, 6.32; N, 5.51%.
N-{2-[2-(4-Benzoyl-2-methylphenoxy)-acetylamino]-phenyl}-2-[2-chloro-4-(4-chloro-benzoyl)-6-fluorophenoxy]-N-(2-piperidin-1-yl-ethyl)-acetamide 11i
Yield: 76%. M.P.: 123–125 °C. IR (KBr) νmax (cm−1): 1670 (C=O), 1720 (amide, C=O), 3110–3210 (NH). 1H NMR (400 MHz, DMSO-d6) δ (ppm): 1.39–1.73 (m, 6H, ring-3CH2), 2.18 (t, 4H, ring-2NCH2), 2.36 (s, 3H, CH3), 2.73 (t, 2H, NCH2), 3.46 (t, 2H, CONCH2), 4.69 (s, 2H, OCH2), 4.73 (s, 2H, OCH2), 6.42–7.69 (m, 18H, Ar–H), 9.77 (s, 1H, NH). 13C NMR (DMSO-d6) δ: 193.7 (2C=O), 170.2 (CONH), 168.5 (CO–N), 157.2 (C, C19), 150.5 (C, C1), 142.7 (C, C6), 140.8 (C, C25), 138.7 (C, C10), 137.7 (C, C7), 136.6 (C, C4), 133.9 (2C, C13, C14), 132.6 (CH, C28), 130.5 (CH, C21), 129.6 (2CH, C8, C12), 128.2 (2CH, C26, C30), 127.2 (C, C22), 126.7 (2CH, C11, C9), 125.3 (2CH, C27, C29), 124.2 (CH, C23), 123.2 (CH, C3), 122.1 (2CH, C16, C17), 121.5 (C, C20), 120.7 (C, C2), 118.5 (2CH, C15, C18), 115.6 (CH, C5), 113.9 (CH, C24), 83.5 (CH2, OCH2), 81.1 (CH2, OCH2), 69.7 (CH2, NCH2), 68.4 (CH2, ring-NCH2), 53.3 (CH2, CO-NCH2), 27.6 (2CH2, CCH2), 25.3 (CH2, CCH2), 14.6 (2CH3). LC-MS m/z 795.23 (M+), 797.22 (M + 2), 799.28 (M + 4). Anal. Cal. for C44H40Cl2FN3O6 (795.23): C, 66.33; H, 5.06; N, 5.27. Found: C, 66.22; H, 4.96; N, 5.13%.
N-{2-[2-(4-Benzoyl-2,5-dimethylphenoxy)-acetylamino]-phenyl}-2-[2-methyl-4-(2-methyl-benzoyl)phenoxy]-N-(2-piperidin-1-yl-ethyl)-acetamide 11j
Yield: 89%. M.P.: 112–114 °C. IR (KBr) νmax (cm−1): 1675 (C=O), 1710 (amide, C=O), 3220–3320 (NH). 1H NMR (400 MHz, DMSO-d6) δ (ppm): 1.49–1.78 (m, 6H, ring-3CH2), 2.23 (t, 4H, ring-2NCH2), 2.39 (s, 3H, CH3), 2.45 (s, 6H, 2CH3), 2.54 (s, 3H, CH3), 3.24 (t, 2H, NCH2), 3.51 (t, 2H, CONCH2), 4.71 (s, 2H, OCH2), 4.86 (s, 2H, OCH2), 6.81–7.72 (m, 18H, Ar-H), 9.69 (s, 1H, NH). 13C NMR (DMSO-d6) δ: 196.8 (2C=O), 173.1 (CONH), 169.2 (CO-N), 168.5 (C, C1), 147.5 (C, C19), 144.7 (C, C8), 141.5 (C, C7), 140.9 (C, C28), 135.4 (C, C23), 133.6 (2C, C14), 132.7 (CH, C10), 131.3 (CH, C3), 130.2 (CH, C21), 130.2 (2CH, C30), 130.1 (C, CH, C4, C12), 129.2 (CH, C9), 129.5 (2CH, C27, C29), 128.4 (CH, C5), 127.5 (CH, C11), 126.3 (2CH, C16, C17), 124.6 (C, C2), 123.8 (2CH, C15, C18), 122.7 (1C, C20), 120.8 (CH, C24), 119.7 (CH, C6), 78.73 (CH2, OCH2), 76.8 (CH2, OCH2), 68.9 (CH2, NCH2), 67.5 (CH2, ring-NCH2), 55.1 (CH2, CO-NCH2), 30.5 (2CH2, CCH2), 28.9 (CH2, CCH2), 18.4 (CH3), 17.53 (CH3), 13.22 (2CH3). LC-MS m/z 752.35 (M + 1). Anal. Cal. for C47H49N3O6 (751.36): C, 75.08; H, 6.57; N, 5.59. Found: C, 74.98; H, 6.47; N, 5.44%.
N-(2-{2-[4-(4-Bromo-benzoyl)-2-methylphenoxy]-acetylamino}-phenyl)-2-[2-methyl-4-(2-methyl-benzoyl)-phenoxy]-N-(2-piperidin-1-yl-ethyl)-acetamide 11k
Yield: 85%. M.P.: 109–111 °C. IR (KBr) νmax (cm−1): 1670 (C=O), 1735 (amide, C=O), 3125–3225 (NH). 1H NMR (400 MHz, DMSO-d6) δ (ppm): 1.42–1.73 (m, 6H, ring-3CH2), 2.19 (t, 4H, ring-2NCH2), 2.30 (s, 3H, CH3), 2.35 (s, 3H, CH3), 2.51 (s, 3H, CH3), 2.80 (t, 2H, NCH2), 3.49 (t, 2H, CONCH2), 4.79 (s, 2H, OCH2), 4.85 (s, 2H, OCH2), 6.81–7.79 (m, 18H, Ar–H), 9.63 (s, 1H, NH). 13C NMR (DMSO-d6) δ: 195.1 (2C=O), 170.3 (CONH), 166.5 (CO–N), 164.5 (2C, C1, C19) 145.4 (2C, C1, C19), 141.3 (C, C8), 138.4 (C, C7), 135.6 (C, C25), 134.5 (2C, C13, C14), 131.7 (2CH, C26, C30), 132.3 (CH, C10), 131.2 (2CH, C3, C21), 130.6 (2CH, C27, C29), 129.5 (2C, CH, C4, C22, C12), 129.1 (CH, C9), 128.6 (2CH, C5, C23), 127.6 (C, C28), 125.5 (CH, C11), 124.3 (2CH, C16, C17), 122.8 (2C, C2, C20), 121.3 (2CH, C15, C18), 116.6 (2CH, C6, C24), 78.7 (CH2, OCH2), 76.5 (CH2, OCH2), 60.4 (CH2, NCH2), 59.9 (CH2, ring-NCH2), 52.2 (CH2, CO–NCH2), 31.7 (2CH2, CCH2), 29.1 (CH2, CCH2), 19.4 (CH3), 15.1 (2CH3). LC-MS m/z 815.25 (M+), 817.23 (M + 2). Anal. Cal. for C46H46BrN3O6 (815.26): C, 67.64; H, 5.68; N, 5.14. Found C, 67.55; H, 5.58; N, 5.03%.
N-(2-{2-[4-(4-Bromo-benzoyl)-2-methylphenoxy]-acetylamino}-phenyl)-2-[2-methyl-4-(4-methyl-benzoyl)-phenoxy]-N-(2-piperidin-1-yl-ethyl)-acetamide 11l
Yield: 75%. M.P.:102–104 °C. IR (KBr) νmax (cm−1): 1675 (C=O), 1730 (amide, C=O), 3120–3220 (NH). 1H NMR (400 MHz, DMSO-d6) δ (ppm): 1.51–1.83 (m, 6H, ring-3CH2), 2.13 (t, 4H, ring-2NCH2), 2.34 (s, 9H, CH3), 2.44 (s, 3H, CH3), 2.65 (s, 3H, CH3), 2.73 (t, 2H, NCH2), 3.54 (t, 2H, CONCH2), 4.75 (s, 2H, OCH2), 4.89 (s, 2H, OCH2), 6.71–7.62 (m, 18H, Ar–H), 9.73 (s, 1H, NH). 13C NMR (DMSO-d6) δ: 194.8 (2C=O), 166.6 (CONH), 165.3 (CO–N), 152.4 (2C, C1, C19), 138.5 (C, C10), 136.2 (C, C25), 134.4 (C, C7), 133.6 (2C, C13, C14), 132.4 (2CH, C26, C30), 131.4 (2CH, C3, C21), 130.7 (2CH, C27, C29), 129.8 (2C, C4, C22), 128.4 (2CH, C8, C12), 127.3 (2CH, C9, C11), 126.2 (2CH, C5, C23), 124.1 (C, C28), 123.1 (2CH, C16, C17), 122.6 (2C, C2, C22), 120.7 (2CH, C15, C18), 118.5 (2CH, C6, C24), 82.6 (CH2, OCH2), 78.5 (CH2, OCH2), 61.2 (CH2, NCH2), 59.3 (CH2, ring-NCH2), 49.5 (CH2, CO-NCH2), 27.2 (2CH2, CCH2), 24.3 (CH2, CCH2), 19.8 (CH3), 15.1 (2CH3). LC-MS m/z 815.21 (M+), 817.24 (M + 2). Anal. Cal. for C46H46BrN3O6 (815.26): C, 67.64; H, 5.68; N, 5.14. Found: C, 66.26; H, 4.58; N, 5.02%.
Biology
Animal Cell culture and growth inhibition studies
The A549, MCF-7, A375, HepG2, ACHN, B16F10 cells were cultured in DMEM, enriched with 10% FBS with penicillin and streptomycin and maintained in a 98% humidified atmosphere containing 5% CO2 at 37 °C. The ability of the series of compounds piperidine conjugated with benzophenone 11a–l to inhibit cancer cell proliferation was assessed by tryphan blue dye exclusion, MTT assay and lactate LDH release assay as reported previously (Thirusangu et al. 2017) and IC50 values were calculated. In brief, cells were seeded at 4 × 103 cells per well into 96-well plate, cultured overnight and treated with increasing concentration of compounds 11a–l (0, 2, 5, 10, 25, and 50 µM in DMSO) diluted in a medium containing 0.5% FBS for 48 h.
In vitro treatment of compound 11f
The B16F10 cells were cultured, serum starved for 2 h and treated with varied concentration of compound 11f (0, 4, and 8 μM) for 48 h along with appropriate vehicle control wherever necessary for further analysis.
Annexin V staining
The cultured B16F10 cells were treated with compound 11f (0, 4, and 8 µM) for 48 h and stained with Annexin V staining-FITC Fluorescence Microscopy Kit (Malojirao et al. 2018) as per manufacturer’s instruction and images were documented.
TUNEL assay
The B16F10 cells were cultured on poly-L-lysine pre coated cover slips and exposed with a lead compound 11f (0, 4, and 8 µM) for 48 h. Cells were fixed with 4% paraformaldehyde for 15 min and permeabilized by 0.25% triton X-100 for 20 min and TUNEL was performed as per manufacturer’s instruction (Malojirao et al. 2018).
Cell fractionation and preparation of cell lysates
The whole cell lysate was prepared as mentioned before (Mohammed et al. 2018). In brief, cell treated with or without compound 11f (0, 4, and 8 μM), were harvested and whole cell extract was prepared using Radio immune precipitation assay buffer (RIPA buffer) (100 mM tris pH-7.5, 1% triton X-100, 0.1% sodium dodecyl sulfate (SDS), 140 mM NaCl, 0.5% sodium deoxycholate, 5 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail. All cellular fractioned proteins were quantified in Nanodrop- biospectrophotometer (Eppendorf, Germany).
Immunoblot (IB) analysis
The cell lyastes were separated on a precast SDS polyacrylamide gels, transferred to polyvinylidene difluoride membrane, probed with appropriate antibodies and detected with the BCIP-NBT chromogen (Thirusangu et al. 2017). The bands were densitometrically analyzed and documented.
Animals and ethics
The BALB/c mice (27–30 g) were used throughout the study and maintained as per The Committee for the Purpose of control and supervision of experiments on animals guidelines with ethical clearance (NCP/IAEC/CL/101/05/2013-14).
Melanoma tumor model development and treatment
The murine melanoma B16F10 cell lines cultured as procedure depicted earlier (Overwijk and Restifo 2001). The cultured B16F10 cells were harvested by trypsinization using 0.5% trypsin. The cell were centrifuged at 3000 rpm for 5 min and washed with DMEM media thrice. A melanoma tumor model was developed by injecting B16F10 cells (1 × 106 cells) into the skin of male mice subcutaneously (s.c.). The tumor growth was analyzed by the black spot size on the skin of each mices (Overwijk and Restifo 2001). The animals were segregated into three groups with n = 6. One group served as control and the other two groups received compound 11f (10 and 20 mg/kg body weight) thrice a week into the peripheral sites of tumors for 3 weeks. At the termination of the experiments, all animals were sacrificed, tumors were photographed, excised and weighed, and the volumes [0.5 (length × breadth2)] were calculated and a part of tumor tissue was used for lysate preparation.
Molecular modeling studies
The molecular operating environment (MOE) 2015 was used for in silco studies which were performed on windows 2010 version. The protein coding gene (Bcl-2 and XIAP) which is an apoptosis regulator was imported from the protein data bank (PDB ID: 1gjh and 2opz) and the protein was visualized using sequence option and non interacting water molecules were removed. The partial charge of the protein was adjusted, using the force field method AMBER 99. Later, the protein was subjected to 3D protonation at cut off 12.0, and further hydrogen was added according to standard geometry and the enzyme was energy minimized using force field MMFF94x at 0.01 KJ per mole gradients. The ligand preparation was done by drawing the structure of ligand by using a marvin Sketch, and adjusting the partial charges using Hamilton MMFF94 force field method and subsequently 3D protonation and hydrogen addition was performed according to standard geometry. Ligands were energy minimized at cut off 12 using MMFF94x force field at 0.01 KJ per mole gradient. Docking was performed using the option simulation followed by dock on selected active site amino acids using sequence option, and further docked with setting options such as: receptor and solvent, selected residues, alpha triangle, affinity dG, force field refinement, and best 10 pose. After obtaining docking results, out of the 10 best posed resulted for each chemical structure, best pose was retained. The resultant best pose score values in the series were used for analysis of docking and interaction.
Statistical analysis
Values were represented as mean ± standard deviation (SD). Statistical significance was evaluated by one-way analysis of variance (ANOVA) followed by two-tailed 13 Student’s t-test. MS excel 8.1 version software was used for data analysis and statistical significant values were expressed as *p < 0.05 and **p < 0.01.
Results and discussion
Chemistry
The synthesis of the title compounds 11a–l was accomplished by a synthetic procedure as shown in Scheme 1. All the synthesized compounds were established by IR, 1H NMR, 13C NMR, and mass spectral data. First, the benzoylated products 3a–f were synthesized by the benzoylation of substituted phenols 1a–c with substituted benzoyl chlorides 2a–d under low temperature. Fries rearrangement of compounds 3a–f, using anhydrous aluminum chloride as a catalyst was then carried out under neat condition, to afford hydroxy benzophenones 4a–f. The compounds 4a–f on etherification with ethyl chloroacetate using dry acetone as a solvent gave substituted ethyl 2-(4-benzoylphenoxy)acetates 5a–f. Further, the compounds 5a–f on refluxing with aqueous sodium hydroxide in ethanol gave (4-benzoyl-phenoxy)-acetic acids 6a–f. The compounds 6a–f were coupled with o-phenylenediamine 7 in the presence of 2,6 lutidine and TBTU as a coupling agent and dichloromethane (DCM) as a solvent to yield substituted N-(2-amino-phenyl)-2-(4-benzoyl-phenoxy)-acetamides 8a–f. Then, all the substituted compounds 6a–f, on treatment with the compounds 8a–f using coupling agent TBTU and lutidine as a catalyst, afforded the expected products 9a–l in a good yield. Finally, the condensation of 9a–l with 1-(2-chloroethyl)piperidine hydrochloride 10 for 6 h in the presence of potassium tertiary butoxide and dimethyl sulphoxide (DMSO), furnished the title compounds 11a–l. Among 3a–f series the spectrum of compound 3a is taken as a representative example and the formation of this compound was confirmed by the appearance of the carbonyl stretching band for the ester group at 1715 cm−1 in the IR spectrum and the appearance of nine aromatic protons between 7.11 and 8.25 ppm in proton NMR spectrum. The mass spectrum of compound 3a gave significant stable M + 1 peak at m/z 213 which also evident for the formation of compound 3a. Further, the spectrum of compound 4a, was considered as a representative example of the series 4a–f. The IR spectrum of the compound 4a, was established by the disappearance of the carbonyl stretching band of the ester group of compound 3a and appearance of the OH stretching band at 3530–3630 cm−1. And also, the appearance of broad singlet for OH proton at δ 12.01 ppm and decrease in one aromatic proton between 6.73 and 7.68 ppm in proton NMR spectrum. The mass spectrum of compound 4a offered significant stable M + 1 peak at m/z 213 which is considered as additional evidence for the formation of this compound. Besides, the compound 5a was taken as a representative example for the 5a–f series, which was confirmed by the disappearance of the OH stretching of compound 4a and appearance of the carbonyl stretching band for the ester group at 1755 cm−1 in the IR absorption spectrum. The proton NMR observations of compound 5a revealed the disappearance of broad singlet for the OH proton of compound 4a and appearance of a triplet and quartet for CH3 and CH2 protons at δ 2.31 and 4.13 ppm, respectively. Also, mass spectrum gave significant stable M + 1 peak at m/z 299 which clearly affirmed the formation of compound 5a. Similarly the formation of compound 6a was confirmed with the appearance of carbonyl and OH groups stretching bands of carboxylic acid at 1720 and 3420–3520 cm−1 respectively, and disappearance of carbonyl stretching of ester group of compound 5a in the IR spectrum. In proton NMR, the appearance of singlet peak of COOH proton at δ 13.13 ppm and disappearance of triplet and quartet peaks for CH3 and CH2 protons, respectively, of compound 5a has confirmed the formation of the compound 6a. The mass spectrum of compound 6a gave significant stable M + 1 peak at m/z 271 which also affirmed the formation of the compound 6a. Likewise, the structure of compound 8a was confirmed by the disappearance of the carbonyl and OH stretching bands of carboxylic acid group of compound 6a and appearance of NH and NH2 stretching bands in the range 3110–3230 and 3315–3410 cm−1 respectively in the IR spectrum. Also, in proton NMR, the appearance of NH2 and NH protons at δ 4.73 and 9.27 ppm, respectively, and an increase in four aromatic protons confirmed the formation of the product 8a. The mass spectrum of compound 8a gave significant stable M + 1 peak at m/z 361 which also proves the formation of the compound 8a. Moreover, the spectrum of the compound 9a was considered as a representative example for the series 9a–l. This was supported by the disappearance of COOH and NH2 stretching of the compound 6a and 8a respectively, and the appearance of only NH stretching between 3100 and 3210 cm−1 in the IR spectrum. Interestingly, compound 9a is a symmetrical molecule, and it was also proved by the NMR spectrum by the disappearance of COOH proton of 6a and NH2 proton of 8a and appearance of singlet of NH proton at δ 9.67 ppm. Besides, there is an increase in aromatic protons in range δ 6.95–7.65 ppm and mass spectrum of compound 9a gave significant stable M + 1 peak at m/z 613 which clearly affirmed the formation of product 9a. Finally, in the title compounds series 11a–l, compound 11a is taken as a representative example to explain characterizations. The structure was confirmed by the appearance of 14 CH2 protons in upfield region and two types of methyl protons at δ 2.72 and 3.29 ppm and OCH2 protons at δ 4.53 and 4.72 ppm and the multiplet signal in the range δ 6.71–7.72 ppm for aromatic protons as well as a singlet at 8.75 for NH proton in proton NMR spectrum confirmed the formation of title product. The mass spectrum of compound 11a exhibited M + 1 peak at m/z 724 which also revealed the formation of the title compound 11a.
Synthesis of piperidine appended benzophenone analogs via amide linkage 2-(4-Benzoyl-phenoxy)-N-{2-[[2-(4-benzoyl-phenoxy)-acetyl]-(2-piperidin-1-yl-ethyl)-amino]-phenyl}-acetamide analogs (11a–l). Reaction conditions and yield: (i) Aq. NaOH, stirring 0–5 °C for 2–3 h, yield: 82–93%, (ii) Anhy. AlCl3, 150–170 °C for 2–3 h, yield: 72–85%, (iii) ClCH2COOC2H5/Dry Acetone, K2CO3, Reflux, 60 °C for 8–10 h, yield: 89–93%, (iv) Aq. NaOH/Ethanol, Reflux, for 5–6 h, yield: 84–94%, (v) TBTU/Lutidine, Dry DCM, Stirring 0–5 °C for 30 min then, overnight at RT, yield: 80–94%, (vi) TBTU/Lutidine, Dry DCM, Stirring 0–5 °C for 30 min then, overnight at RT, yield: 71–90%, (vii) Piperidine hydrochloride/DMSO, KOtBu, Reflux, 8–10 h, yield: 75–90%
Pharmacology
Compound 11f is a lead compound and its structure activity relationship (SAR)
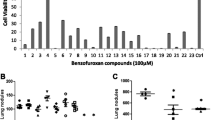
In the current decade, it’s been constantly observed that the increased occurrence of cancer worldwide and the immediate necessity of novel anti-neoplastic compounds target specific action. The piperidine derivatives are found to possess diverse pharmacological activities and form an essential part of the molecular structure of important drugs (Khanum et al. 2009; Hu et al. 2014; Wang et al. 2015; Xin-Hua et al. 2012). Similarly benzophenone analogs are also great importance fundamentally due to their significant biological activity both in vitro and in vivo (Al-Ghorbani et al. 2017; Khanum et al. 2005; Puttaswamy et al. 2018; Ranganatha et al. 2013; Revesz et al. 2004; Zabiulla et al. 2016). In the event of exploring the new set of anticancer molecules here we conjugated the two pharmacologically active backbones namely, benzophenone and piperdine, appended with amide bond linkage by substituting with various halogens and methyl groups at different position of the rings. The antiproliferative studies included the cancers of different origin viz, melanoma, lung adenocarcinoma, renal cell carcinoma, hepatocellular, and breast carcinoma with three independent assay types such as 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), lactate dehydrogenase (LDH) and trypan blue assay. The growth inhibition value inferred increasing cytotoxicity against B16F10 > A375 > A549 > ACHN > HepG2 > MCF7 cells (Table 1). The melanoma cell lines of both murine and human origin showed maximum sensitivity at 8 µM more or less in all three independent experiments.
Detailed SAR revealed that, the compound 11f with two bromo groups at the para position in rings A and E and two methyl groups at ortho position in rings B and D showed a maximum inhibitory effect of 8 and 10 µM on B16F10 and A375 cells respectively. The results were not significant against A549, MCF-7, ACHN, and HepG2 cell lines. The compounds 11e, 11g, 11k, and 11l with a single bromo group at the para position of ring E and more number of CH3 groups at different position did not show antiproliferative effect, but indicated the role of bromo and methyl groups in the biological activity, showing slight cytotoxicity (Table 1). Whereas the compound 11i with the other halogens like chloro, fluro and with methyl substituent has no activity. Further, compounds 11a, 11b, 11c, 11d, 11h, and 11j with only methyl substituent has also did not displayed activity. The overall inference is the compound 11f with two bromo and two methyl groups at different position in the rings is responsible for the biological activity and hence chosen as a lead compound for elucidation of molecular mechanisms in detail (Table 1).
Compound 11f triggers cellular apoptotic hallmarks
Understanding of cellular process that contribute to cancer, has increased substantially since Hanahan and Weinberg detailed the hallmark of cancer, including emerging ideas and failure to activate the same results in anticancer response (Hanahan and Weinberg 2011). Thus, one can foresee a future paradigm where activation of specific pathways of cell death is still intact and predict the response to therapies (Ricci and Zong 2006; Plati et al. 2011). Realizing this point, herewith we have provided an experimental evidence of novel pharmacophore compound 11f which can target melanoma through the activation of cell death pathways in a systemic investigation. The mechanism of cytotoxicity was explored for cellular and molecular changes in B16F10 cells treated with or without compound 11f (0, 4, and 8 µM) by annexin V staining and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay. Translocation of phosphatidyl serine from inner to outer leaflet of the cell membrane is typical hallmarks of apoptotic event which can be measured by Annexin V-FITC staining. Flip-flop of B16F10 cell membrane was evident in a concentration dependent manner which absolutely absent in untreated cells. The intensity of the fluorescence is a direct measure of the exposed annexin molecules in 11f treated cells, which was observed under fluorescence microscopy (Fig. 1a). Framing to more supportive evidence for apoptotic cell damage, DNA damage was accomplished by TUNEL assay, which detects specific DNA breaks by end labeling. Upon 11f (0, 4, and 8 µM) treatment to B16F10 cells, control cells did not show any significant TUNEL positive nuclei, whereas the percentage of the same was 31.3% and 46.2% respectively, at two different concentrations (Fig. 1b). The efficacy was compared with positive control DNAse treatment and counterstained with Hoechst stain for specific nuclear localization.
Compound 11f induces apoptotic hallmarks. The B16F10 cell lines treated with compound 11f at 0, 3 and 5 µM for 48 h and used for further experiments. a Annexin V immunostaining images of bright field and fluorescent images. b TUNEL positive cells of control, DNase treated (positive control) and compound 11f treated fluorescence. Hoechst counter stains and merged images
Compound 11f induces double inhibition of X-linked IAP (XIAP) and Bcl-2 and elevates pro-apoptotic gene expression
Apoptosis is mainly regulated in two predominant pathways; intrinsic and extrinsic apoptotic pathway. Which includes a series of biochemical changes such as disruption of the mitochondrial membrane and release of apoptogenic molecules to cytosol, activation of caspase cascade, induction of DNA fragmentation. The members of the Bcl-2 family mainly regulate the intrinsic apoptotic signaling pathway, which comprises of both pro-apoptotic (Bax and Bad) and anti-apoptotic (Bcl-2) proteins (Thomadaki and Scorilas 2006). Balance between these two groups decides the fate of the cells. On the induction of apoptotic stimuli, the balance shifts towards the pro-apoptoic proteins and facilitates the release of apoptogenic molecules. This results in proteolytic cleavage and maturation of caspases (Caspase-3) to execute cellular fatality. On the other hand, the majority of the tumors tend to express the higher basal level of active caspase-3 without apoptotic induction. The intrinsic drive to apoptosis is restrained by concomitant upregulation of IAP (XIAP) which counteracts the caspase activity in tumors (Yang and Cao 2003). Considering the facts, targeting anti-apoptotic (Bcl-2 and XIAP) in cancer cells might increase the therapeutic effect by shifting the balance in favor of pro-apoptotic molecules.
To confirm that the mitochondrial pathway is involved in compound 11f apoptosis, the protein expression of pro-apoptotic Bax and Bad and anti-apoptotic Bcl-2 protein was examined in in vitro system. As shown in Fig. 2, treatment of B16F10 cells with compound 11f for 48 h resulted in increased levels of Bax and Bad, counteracting Bcl-2 protein expression. In addition to measuring expression levels of members of the Bcl-2 family, the changes in expression of IAP family proteins were also investigated. The results show that the expression of two IAP family proteins, namely XIAP, were reduced following treatment with higher doses of compound 11f, as shown in Fig. 2. This eventually leads to the increased expression of caspase-3 in a concentration dependent manner. Based on the results, the conclusion drawn is, compound 11f induces dual inhibition of XIAP and Bcl-2 and eventually upregulates pro-apoptotic gene expression such as Bax, Bad, and Caspase-3.
Compound 11f modulating Bcl-2 gene related proteins in vitro: The B16F10 cell lines treated with compound 11f at 0, 3, and 5 µM for 48 h. Whole cell lysate fractions were used for IB. The differential expression of Bcl-2, XIAP, Bax, Bad, Caspase-3, and β actin. Note: Statistically significant values expressed as *p < 0.05 and **p < 0.01
Compound 11f regresses murine melanoma by dual inhibition of XIAP and Bcl-2 in vivo
Malignant melanoma is a highly aggressive cancer and most consistent model for metastasis studies (Miller and Mihm 2006; Madan et al. 2010). In the present investigation B16F10 melanoma cells were used to develop a melanoma tumor model in mice. After the onset of tumor development, six doses of compound 11f (0, 10, and 20 mg/kg.b.w) given on every alternate day, resulting in reduction in tumor growth in a dose dependant manner, along with a notable reduction in the physical morphology of tumor bearing animals. The dissected tumor size depicted the tumor regression with 2.2 to 4.6 folds (Fig. 3a). To explore the molecular mechanism of compound 11f in vivo, whole cell lysate of tumor tissue was subjected to immunoblot analysis. The results inferred that compound 11f modulates the Bcl-2 related proteins by specifically down regulating the Bcl-2 and XIAP protein. As a consequence proapototic protein such as Bax and Bad got upregulated leading to activation of apoptotic inducing protease casapse-3 which eventually damages the DNA as observed in the TUNEL assay in vitro (Fig. 1b). Taken together 11f is promising molecule against the skin cancer and which can be developed as an anticancer drug for skin cancer in near future.
Compound 11f regresses murine melanoma by activating modulating Bcl-2 gene related proteins in vivo: The B16F10 cells (1 × 106) were tail vein injected into the BALB/c mice. The mice received 0, 10, and 20 mg/kg/ b. wt compound 11f every-other day after 15 days of induction. a Anatomy of skin exhibiting tumor colonization and quantification of the tumor nodules in the skin. b IB studies signifying the altered translational expression of genes such as Bcl-2, XIAP, Bax, Bad, caspase-3, and β-actin
Compound 11f interacts strongly with Bcl-2 in docking studies
The molecular docking was performed as per the standard procedure (Mohammed et al. 2017; Mohammed and Khanum 2018) and the analysis revealed that, the compound 11f interacts strongly with Bcl-2 and XIAP proteins and 3D structure shows the interaction compound 11f in the active site C-terminal trans activation in Bcl-2 and in the active site of XIAP. The hydrogen-bond interaction view of the ligand molecule 11f with AlA43 and Glu42 in Bcl-2 protein with GLN399 in XIAP protein as shown in Fig. 4. Further, the interaction of compound 11f at the pocket site and residue amino acid of the complex Bcl2-11f and complex XIAP-11f is predicted (Fig. 5) with also the 3D interactions of 11f with hydrophobic amino acids, which represented by yellow and red color in Bcl-2 and XIAP protein respectively (Fig. 6). The sequences of complex Bcl-2-11f and XIAP-11f are arranged to their target sites is detected using MOE 2015 program (Fig. 7). The molecular mapping is the 3D plot of the electrostatic potentials relating to the electron density mapped onto the isoelectronic surface. It is helpful to understand the sites intended for the electrophillic and nucleophiliic reaction for the hydrogen bond. As well as, the size, shape and electrostatic potential value. The mapping, molecular electrostatic potentials using ribbon models, and conformer explorer of compound 11f with amino acid residue of Bcl-2 and XIAP proteins has predicted (Fig. 8). Similarly, we have performed, pose organizer viewer of the compound 11f with residue amino acid in Bcl-2 and XIAP and it was displays electrostatic interactions of Bcl-2 and XIAP proteins with the compound 11f with hydrogen view. The compound 11f displays hydrophobic and hydrophilic groups as green and orange respectively in both Bcl-2 and XIAP proteins (Figs 9 and 10). Ramachandran plot showing the phi/psi values of the modeled Bcl2 and XIAP proteins. Green color is the most favorable regions, red color region is allowed region, pale yellow is generously allowed region and white color is disallowed regions. The Graph presenting the rotamer energy plots, residue clashing and contact energy profile of amino acid residues the Bcl-2 and XIAP proteins as shown in Figs 11 and 12. Further, we have predicted the bond angle, bond length and dihedral angle of complex Bcl2-11f and XIAP-11f (Figs 13–15), the docking domains indicated a strong binding at the catalytic sites of proteins. The docking studies showed that the compound 11f can be taken for further study. Moreover, the possible binding conformation and orientations were analyzed by clustering methods, embedded in MOE 2015. From the Tables 2 and 3, the compound 11f showed good binding energies at −8.9876 to −8.3910 and −7.4863 to −6.8148 for Bcl-2 and XIAP proteins respectively.
Compound 11f interacts strongly with Bcl-2 and XIAP proteins. a 3D structure of the compound 11f in the active site C-terminal trans activation Bcl-2 and hydrogen-bond interaction view of the ligand molecule 11f with AlA43 an Glu42 in Bcl-2 protein. b 3D structure of the compound 11f in the active site of XIAP and hydrogen-bond interaction view of the ligand molecule 11f with GLN399 in XIAP protein
a The 3D interactions of the compound 11f with hydrophobic amino acids, which represented by yellow color in Bcl-2 protein. b The 3D interactions of the compound 11f with hydrophobic amino acids by red colour in XIAP protein. c The 2D interactions analysis of the compound 11f with Bcl-2 protein. d The 2D interactions analysis of the compound 11f with XIAP protein (color figure online)
Mapping molecular electrostatic potentials of compound 11f with Bcl-2 and XIAP: a Separated conformers and molecular surfaces using ribbon models and conformer explorer of the compound 11f with amino acid residue in Bcl-2. b Separated conformers and molecular surfaces using ribbon models and conformer explorer of the compound 11f with amino acid residue in XIAP
a Pose organizer viewer of the compound 11f with residue amino acid. b Displays electrostatic interactions of Bcl-2 proteins with the compound 11f. c Preparation view changed hydrogen view. d Displays hydrophobic groups: green is hydrophobic, orange is hydrophilic of Bcl-2 protein with the compound 11f (color figure online)
a Pose organizer viewer of the compound 11f with residue amino acid. b Displays electrostatic interactions of the XIAP protein with the compound 11f. c Preparation view changed hydrogen view. d Displays hydrophobic groups: green is hydrophobic, orange is hydrophilic of XIAP protein with the compound 11f (color figure online)
a Ramachandran plot showing the phi/psi values of the modeled Bcl2 protein. Green color- most favorable regions, red color region- allowed region, pale yellow- generously allowed region and white color- disallowed regions. b Graph presenting the rotamer energy plots of the Bcl-2 protein. c Residue clashing profile of amino acid residues between this system in production stage run. d The energy score (kcal mol−1) of the complex Bcl2-11f with lowest binding energy −8.9 kcal mol−1(color figure online)
a Ramachandran plot showing the phi/psi values of the modeled XIAP protein. Green color- most favorable regions, red color region- allowed region, pale yellow- generously allowed region and white color- disallowed regions. b Graph presenting the rotamer energy plots of the XIAP protein. c Residue clashing profile of amino acid residues between this system in production stage run. d The energy score (kcal mol−1) of the complex XIAP-11f with lowest binding energy −7.48 kcal mol−1 (color figure online)
Conclusion
In summary, a novel series of piperidine conjugated benzophenone analogs with amide linkage 11a–l were synthesized by incorporating, fluoro, chloro, bromo, and methyl groups at different position of the aromatic rings and evaluated against cancers of different origin through cell based assay system using B16F10, A375, A549, HepG2, ACHN, and MCF7 cells. From the current investigation, SAR of these compounds suggests that the position and the type of substituent on the aromatic ring in 11a–l are important for the activity. Experimental evidences postulated that compound 11f with two bromo groups at the para position in rings A and E and two methyl groups at ortho position in rings B and D shows target specific action against melanoma. In addition, compound 11f evokes the apoptotic cellular event leading to cell death. Also, compound 11f turned out to be a dual inhibitor of Bcl-2 and XIAP eventually causing the up regulation of Bax and Bad. Overall, the compound 11f is a promising anticancer molecule for the treatment of skin cancer with selective target against melanoma by inducing apoptogenic effect, which is also correlated with the molecular docking result.
Change history
22 July 2019
The original version of this article unfortunately contained an error in author group. The first author name was repeated twice.
References
Al-Ghorbani M, Thirusangu P, Gurupadaswamy HD, Vigneshwaran V, Mohammed YHE, Prabhakar BT, Khanum SA (2017) Synthesis of novel morpholine conjugated benzophenone analogues and evaluation of antagonistic role against neoplastic development. Bioorg Chem 71:55–66
Al-Ghorbani M, Thirusangu P, Gurupadaswamy HD, Girish V, Neralagundi HGN, Prabhakar BT, Khanum SA (2016) Synthesis and antiproliferative activity of benzophenone tagged pyridine analogues towards activation of caspase activated DNase mediated nuclear fragmentation in Dalton’s lymphoma. Bioorg Chem 65:73–81
Balasubramanyam K, Altaf M, Radhika AV, Swaminathan V, Aarthi R, Parag P, Sadhal Kundu TP (2004) Polyisoprenylated benzophenone, garcinol, a natural HAT inhibitor represses chromatin transcription and alters global gene expression. J Biol Chem 279:33716–33726
Eberle J, Hossini AM (2008) Expression and function of Bcl-2 proteins in melanoma. Curr Genomics 9:409–419
Elmore S (2007) Apoptosis: a review of programmed cell death. Toxicol Pathol 35:495–516
Frenzel A, Grespi F, Chmelewskij W, Villunger A (2009) Bcl-2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis, an international journal on programmed cell death 14:584–596
Gupta AK, Bharadwaj M, Mehrotra R (2016) Skin cancer concerns in people of color: risk factors and prevention. Asian Pac J Cancer Prev 17:5257–5264
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Henry GE, Jacobs H, Carrington CMS, Mclean S, Reynolds WF (1999) Prenylated benzophenone derivatives from Caribbean Clusia species (Guttiferae). Plukenetiones B-G and xerophenone A. Tetrahedron 55:1581–1596
Hu S, Gu Q, Wang Z, Weng Z, Cai Y, Dong X, Hu Y, Liu T, Xie X (2014) Design, synthesis, and biological evaluation of novel piperidine-4-carboxamide derivatives as potent CCR5 inhibitors. Eur J Med Chem 71:259–266
Iyer S, Chaplin DJ, Rosenthal DS, Boulares AH, Li LY, Smulson ME (1998) Induction of apoptosis in proliferating human endothelial cells by the tumour-specific antiangiogenesis agent combretastatin A-4. Cancer Res 58:4510–4514
Khanum SA, Shashikanth S, Umesha S, Kavitha R (2005) Synthesis and antimicrobial study of novel heterocyclic compounds from hydroxyl benzophenones. Eur J Med Chem 40:1156–1162
Khanum SA, Girish V, Suparshwa SS, Khanum NF (2009) Benzophenone-N-ethyl piperidine ether analogues-synthesis and efficacy as anti-inflammatory agent. Bioorg Med Chem Lett 19:1887–1891
Madan V, Lear JT, Szeimies RM (2010) Non-melanoma skin cancer. The Lancet 375:673–85
Malojirao VH, Vigneshwaran V, Thirusangu P, Mahmood R, Prabhakar BT (2018) The tumor antagonistic steroidal alkaloid Solanidine prompts the intrinsic suicidal signal mediated DFF-40 nuclear import and nucleosomal disruption. Life Sci 199:139–150
Miller AJ, Mihm Jr MC (2006) Melanoma. N Engl J Med 6:51–65
Mohammed YHE, Malojirao VH, Thirusangu P, Al-Ghorbani M, Prabhakar BT, Khanum SA (2018) The novel 4-phenyl-2-phenoxyacetamide thiazoles modulates the tumor hypoxia leading to the crackdown of neoangiogenesis and evoking the cell death. Eur J Med Chem 1:1826–1839
Mohammed YHE, Thirusangu P, Zabiulla, Vigneshwaran V, Prabhakar BT, Khanum SA (2017) The anti-invasive role of novel synthesized pyridazine hydrazide appended phenoxy acetic acid against neoplastic development targeting matrix metallo proteases. Biomed Pharmacother 95:375–386
Mohammed YHE, Khanum SA (2018) The critical role of novel benzophenone analogs on tumor growth inhibition targeting angiogenesis and apoptosis. Med Chem Comm 9:639–656
Overwijk WW, Restifo NP (2001) B16 as a mouse model for human melanoma. Curr Protoc Immunol 20:1–29
Ozkay Y, Isikdag I, Incesu Z, Akalin G (2010) Synthesis of 2-substituted-N-[4-(1- methyl-4,5-diphenyl-1H-imidazole-2-yl)phenyl]acetamide derivatives and evaluation of their anticancer activity. Eur J Med Chem 45:3320–3328
Parrish AB, Freel CD, Kornbluth S (2013) Cellular mechanisms controlling caspase activation and function. Cold Spring Harb Perspect Biol 5:1–24
Plati J, Bucur O, Khosravi-Far R (2011) Apoptotic cell signaling in cancer progression and therapy. Integr Biol 3:279–296
Puttaswamy N, Malojiao VH, Mohammed YHE, Sherapura A, Prabhakar BT, Khanum SA (2018) Synthesis and amelioration of inflammatory paw edema by novel benzophenone appended oxadiazole derivatives by exhibiting cyclooxygenase-2 antagonist activity. Biomed Pharmacother 103:1446–1455
Ranganatha VL, Begum BA, Prashanth T, Gurupadaswamy HD, Madhu SK, Shivakumar S, Khanum SA (2013) Synthesis and larvicidal properties of benzophenone comprise indole analogues against Culex quinquefasciatus. Drug invent. Today 5:275–280
Ranganatha VL, Zameer F, Meghashri S, Rekha ND, Girish V, Gurupadaswamy HD, Khanum SA (2013) Design, synthesis, and anticancer properties of novel benzophenone-conjugated coumarin analogs. Arch Pharm 346:1–11
Revesz L, Blum E, Di Padova FE, Buhl T, Feifel R, Gram H, Hiestand P, Manning U, Rucklin G (2004) SAR of benzoylpyridines and benzophenones as p38alpha MAP kinase inhibitors with oral activity. Bioorg Med Chem Lett 14:3601–3605
Ricci MS, Zong W (2006) Chemotherapeutic approaches for targeting cell death pathways. Oncologist 11:342–357
Schlitzer M, Bohm M, Sattler I (2002) Non-thiol farnesyltransferase inhibitors: structure–activity relationships of benzophenone-based bisubstrate analogue farnesyltransferase inhibitors. Bioorg Med Chem 10:615–620
Thirusangu P, Vigneshwaran V, Avin VBR, Rakesh H, Vikas HM, Prabhakar BT (2017) Scutellarein antagonizes the tumorigenesis by modulating cytokine VEGF mediated neoangiogenesis and DFF-40 actuated nucleosomal degradation. Biochem Biophys Res Commun 484:85–92
Thirusangu P, Vigneshwaran V, Prashanth T, Avin VBR, Malojirao VH, Rakesh H, Khanum SA, Mahmood R, Prabhakar BT (2017) BP-1T an antiangiogenic benzophenone-thiazole pharmacophore, counteracts HIF-1 signalling through p53/MDM2-mediated HIF-1α proteasomal degradation. Angiogenesis 20:55–71
Thirusangu P, Vigneshwaran V, Ranganatha VL, Avin VBR, Khanum SA, Riaz Mahmood R, Jayashree K, Prabhakar BT (2017) A tumoural angiogenic gateway blocker, Benzophenone-1B represses the HIF-1α nuclear translocation and its target gene activation against neoplastic progression. Biochem Pharmacol 125:26–40
Thomadaki H, Scorilas A (2006) Bcl2 family of apoptosis-related genes: functions and clinical implications in cancer. Crit Rev Clin Lab Sci 43:1–67
Wang P, Cai J, Chen J, Ji M (2015) Synthesis and anticancer activities of ceritinib analogs modified in the terminal piperidine ring. Eur J Med Chem 93:1–8
Xin-Hua L, Li J, Shi JB, Bao-An S, Xing-Bao Q (2012) Design and synthesis of novel 5-phenyl-N-piperidine ethanone containing 4,5-dihydropyrazole derivatives as potential antitumor agents. Eur J Med Chem 51:294–299
Yang D, An B, Wei W, Tian L, Huang B, Wang H (2015) Copper-catalyzed domino synthesis of nitrogen heterocycle-fused benzoimidazole and 1,2,4- benzothiadiazine 1,1-dioxide derivatives. A C S Comb Sci 17:113–119
Yang L, Cao ZH (2003) Coexistence of high levels of apoptotic signaling and inhibitor of apoptosis proteins in human tumor cells: implication for cancer specific therapy. Cancer Res 63:6815–6824
Zabiulla, Neralagundi HGS, Begum BA, Prabhakar BT, Khanum SA (2016) Design and synthesis of diamide-coupled benzophenones as potential anticancer agents. Eur J Med Chem 115:342–351
Acknowledgements
Zabiulla gratefully acknowledges the financial support provided by the Department of Science and Technology, New Delhi, Under INSPIRE-Fellowship scheme [IF140407]. YHEM is thankful to the University of Hajja, Yemen, for providing financial support. SAK thankfully acknowledges the financial support provided by VGST, Bangalore, under CISEE Programme [Project sanction order: No. VGST/ CISEE /282]. BTP gratefully acknowledges the grant extended by DBT (6242-P37/RGCB/PMD/DBT/PBKR/2015) and VGST (VGST/GRD231/CISEE/2013-14).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zabiulla, Z., Malojirao, V.H., Mohammed, Y.H.E. et al. Synthesis, molecular docking, and apoptogenic efficacy of novel N-heterocycle analogs to target B-cell lymphoma 2/X-linked inhibitors of apoptosis proteins to regress melanoma. Med Chem Res 28, 1132–1160 (2019). https://doi.org/10.1007/s00044-019-02357-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-019-02357-x