Abstract
A series of pyridazino[4,5-b]indole derivatives containing alkyl-, benzyl- and phenacyl-substituted 1,2,3-triazolylmethyl units was synthesized using click chemistry approach. All 30 compounds of the series were screened in vitro against four cancer cell lines, viz. breast cancer cells MDA-MB-231 and MCF 7, human primary glioblastoma U-87 and human neuroblastoma IMR-32 cell lines. Most of the compounds exhibited potent cancer cell growth inhibition activity at very low micromolar concentrations. The IC50 value of compounds 7v and 7x against human neuroblastoma IMR-32 cell line is 0.07 and 0.04 µM, respectively. Among the tested compounds, ten compounds showed IC50 value less than 1 µM against MDA-MB-231 cells, whereas against IMR-32 cells, nine compounds and, against U-87 cells, six compounds showed similar inhibition activity. Further, these molecules exhibited prominent binding affinity and docking scores in the molecular simulation study with the target enzyme PI3 kinase.
Graphical Abstract
This paper illustrates the synthesis of new fused indole–pyridazinone derivatives containing substituted 1,2,3-triazoles via click chemistry approach. Most of the compounds exhibited potent cancer cell growth inhibition activity at very low micromolar concentrations.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
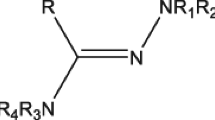
The pyridazino[4,5-b]indole structural unit (Fig. 1) has extensive applications in medicinal chemistry because of its bio-isosterism with β-carboline and γ-carboline. The pharmacological significance of pyridazino[4,5-b]indole systems has been well explored in terms of antiarrhythmic (Lerch and Kaiser, 1982), antihypertensive (Monge et al., 1991b), blood platelet aggregation and inotropics (Monge et al., 1991a), thromboxane A2 synthetase inhibitor (Monge et al., 1987), serotonin antagonistic (Nantka-Namirski and Ozdowska, 1972), MAO inhibitor (Monge et al., 1980), anxiolytic (Evanno et al., 1999), antihistaminic and HIV-1 reverse transcriptase inhibitor (Font et al., 1995) activities. The naturally occurring β-carboline alkaloid, harmine (Fig. 1), inhibits the dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) activity, which is associated with the neurodegenerative diseases such as Down syndrome and the Alzheimer disease, by interacting with residues Leu241 (hinge region) and Lys188 in the ATP-binding pocket through two hydrogen bonds. The methoxy group and the nitrogen atom of the pyridine ring are involved in the formation of these two hydrogen bonds (Adayev et al., 2011; Ogawa et al., 2010). It is proposed that fused indole–pyridazinone system could be a proper molecular structure to provide an effective interaction with the hinge fraction of the target proteins. This is further supported by the fact that 5-benzylated 4-oxo-3,4-dihydro-5H-pyridazino[4,5-b]indoles are highly selective toward phosphatidylinositol-3-kinases (PI3Ks) (Bruel et al., 2012). On the other hand, 1,2,3-triazole and its derivatives find great importance in medicinal chemistry research due to their important biological actions in addition to their synthetic applications (Agalave et al., 2011; Thirumurugan et al., 2013). Triazole is a core structural moiety found in some important drugs like tazobactam, cefatrizine, carboxyamidotriazole and rufinamide. The most significant and current studies have discovered that 1,2,3-triazole derivatives exhibit a broad spectrum of pharmacological activity such as antimicrobial (Kushwaha et al., 2014), anti-inflammatory (Shafi et al., 2012), anticonvulsant (Ulloora et al., 2013), antituberculosis (Addla et al., 2014; Patpi et al., 2012; Yempala et al., 2014) and, in particular, anticancer (Akselsen et al., 2012; Howell et al., 2009; Pagliai et al., 2006) activity. Owing to the drug-like properties of 1,2,3-triazole derivatives and also based on the preliminary structure–activity information (Güven and Jones, 1993), we have planned to incorporate 1,2,3-triazole unit into the pyridazino[4,5-b]indole structure with an expectation that the amalgamation of these two active pharmacophores in a single molecular framework would enhance the efficacy of the hybrid molecule. With this background, we have synthesized a library of 30 indole-fused pyridazinone derivatives containing substituted 1,2,3-triazoles at the indole nitrogen via click chemistry approach and explored in vitro anticancer activity of these hybrid molecules.
Results and discussion
Chemistry
The basic indole moiety was built by precise and proficient three-step synthesis following Fischer indole synthetic protocol (Humphrey and Kuethe, 2006) as given in Scheme 1. The indole-2-carboxylic ester intermediate (3) was alkylated using propargyl bromide to get compound 4. The intermediate 4 was then acylated at position 3 by following Friedel–Craft acylation protocol using AlCl3 as the Lewis acid catalyst to get acyl derivative (5). The indole-fused pyridazinone ring system (6) was then constructed by refluxing intermediate 5 with hydrazine hydrate in ethanol. In all the steps, the products were isolated with excellent yield (>80 %). Finally, 1,2,3-triazole system was introduced by following the conventional click chemistry protocol with the propargylated intermediate (6) using various alkyl, benzyl and phenacyl azides (Scheme 2). We primarily focused on scrutinizing the variation in the activity of molecules by the incorporation of alkyl, benzyl and phenacyl substitutions with various electron-withdrawing and electron-donating groups on the triazole fragment of the hybrid structure. Hence, a library of 30 molecules was synthesized. The structural specifics of the synthesized compounds are depicted in Table 1. All the intermediate and target compounds were characterized using spectral techniques as well as by elemental analysis. The 1H NMR spectrum of 4a displayed singlet peaks at δ 5.79 and 2.64 ppm due to methylene (CH2) and alkyne (≡CH) protons, respectively, of the propargyl group which confirms the successful execution of the N-propargylation reaction. The signals at δ 4.42 and 1.44 ppm in the spectrum correspond to methylene and methyl protons, respectively, of the ester group. Further, the spectrum showed multiplet signals in the region δ 7.01–7.71 ppm corresponding to aromatic protons. The acetylation of 4a to give 5a was evident by the 1H NMR spectrum of 5a which displayed a singlet peak at δ 2.61 ppm due to methyl protons of the acetyl group (–COCH3) along with other characteristic signals. In the 13C NMR spectrum, the carbonyl and methyl carbons of the acetyl group appeared at δ 171.2 and 30.2 ppm, respectively. The conversion of 5a to 6a with the formation of indole-fused pyridazinone ring is confirmed by the appearance of a singlet, corresponding to ring –CONH proton, at δ 12.72 ppm in the 1H NMR spectrum of 6a. Also the signals due to the ester group disappeared in the spectrum which further confirms the formation of the pyridazinone ring. In the 1H NMR spectra of the final compounds, the presence of a singlet peak at δ ~6.0 ppm due to protons of methylene (which links the indole nitrogen and C-4 of the triazole ring) group and disappearance of signals due to the propargyl group confirm the successful completion of the click reaction and the formation of 1,2,3-triazole ring system in these molecules. Further, 13C NMR and mass spectral analysis data are in agreement with the chemical structure of these target molecules. The 13C spectrum of 7a shows peaks at δ 54.1, 40.5 and 11.0 corresponding to the benzylic carbon, methylene carbon linked to indole nitrogen and the methyl group present in the pyridazinone ring, respectively, which confirms the structure of the product. The characterization data are given in the experimental part, and some representative spectra are given in the supporting information.
In vitro anticancer activity
All thirty compounds were subjected to in vitro cell proliferation assay study against a panel of four cancer cell lines, viz. breast cancer cells MDA-MB-231 and MCF7, human primary glioblastoma U-87 and human neuroblastoma IMR-32 cell lines, in order to investigate the cytotoxic nature of the molecules. The MTT assay, with doxorubicin as the standard drug, was used for the screening of compounds. The growth corresponding to the control group (with cells and medium alone) was considered as 100 % (or 0 % inhibition). The percentage inhibition with respect to the compound-treated cells was calculated relative to the control group. It was observed that most of the synthesized molecules are active against IMR-32 and U-87 cells at a test concentration of 10 µM (Fig. 2). A few molecules are potent enough to inhibit the growth close to 90 % against the tested cancer cell lines, in particular against U-87 cells. Compounds 7h, 7k, 7u, 7v, 7w and 7aa exhibited prominent antiproliferative activity (>80 % growth inhibition), and most of the other compounds exhibited moderate activity (>50 % growth inhibition) against U-87 cells. U-87, derived from human malignant gliomas, is the most commonly investigated glioblastoma cell line. Glioblastoma is the most common malignant primary brain tumor in adults and is one of the most fatal of all cancers. Compounds 7n, 7o, 7q, 7z, 7aa and 7ac showed a significant growth inhibition activity (>80 %) against IMR-32 cell line. Interestingly, all other compounds of the series demonstrated moderate activity (nearly 60 % growth inhibition) against the same cell line. IMR-32 is associated with a childhood tumor of the nervous system that expresses remarkable clinical heterogeneity. Some of the test compounds, viz. 7j, 7n, 7u, 7v and 7y, possess promising antiproliferative activity (nearly 70 % growth inhibition) against breast cancer cells MCF-7, whereas compounds 7f, 7m, 7n, 7o, 7s, 7t, 7u, 7v, 7x, 7y, 7z, 7aa, 7ab, 7ac and 7ad showed admirable activity against MDA-MB-231 cells. Further, most of the compounds of this series are non-toxic to non-cancerous HEK 293 cells (Fig. 3) which signifies the specificity of the compounds toward cancerous cells. Only four compounds, viz. 7aa, 7ab, 7ac and 7ad, show toxic nature against the non-cancerous cell line in which 7ac and 7ad effect significantly the growth of normal cell (nearly 50 % growth inhibition). Both compounds contain an isopropyl group on the 1,2,3-triazole ring, and the presence of this group may be responsible for the observed toxicity.
The IC50 value was determined for compounds which have shown a significant inhibitory activity against all the tested cell lines, and the values are given in Table 2. From the data, it is evident that all compounds show prominent inhibition response against IMR-32 cells with very low IC50 values. Compound 5-[(1-cyclohexyl-1H-1,2,3-triazol-4-yl)methyl]-8-fluoro-1-methyl-3,5-dihydro-4H-pyridazino[4,5-b]indol-4-one (7x) is the most potent among the tested molecules (IC50 = 0.04 µM). It was assumed that inhibition of the PI3K pathway may improve the ability of TRAIL (TNF-related apoptosis-inducing ligand) to induce apoptosis in neuroblastoma cells (Efron et al., 2003). Compounds 7n, 7t, 7u, 7v, 7x and 7aa containing 4-fluorophenacyl, 4-nitrobenzyl, 4-fluorobenzyl, 2-fluorobenzyl, cyclohexyl and 4-nitrophenacyl, respectively, exhibit good activity against U-87 cells with IC50 values less than 0.3 µM. The PI3K/Akt oncogenic pathway is also vital in glioblastomas. The malfunctioning of tumor suppressor gene phosphatase and tensin homolog (PTEN), a regulator of the PI3K pathway or activated PI3K/Akt pathway that impel increased proliferation, survival, neovascularization, glycolysis and invasion is reported in 70–80 % of malignant gliomas. Thus, PI3K inhibitors could be an attractive therapeutic target for treating malignant glioma. Compounds 7f, 7n, 7o, 7v, 7x and 7ac demonstrated good activity against MDA-MB-231 and 7aa against MCF-7 with IC50 value less than 0.5 µM. Hence, these molecules with excellent inhibition activity against the cancerous cells at very low micromolar concentrations are promising lead molecules for further biological investigation and to develop efficient antitumor drugs.
The phosphatidylinositol-3-kinases play an important role in controlling various characteristics of the malignant phenotypes, including proliferation, survival and apoptosis, adhesion and mobility, angiogenesis and cell size. The PI3K pathway is activated by the breakdown of PTEN gene and also by extension and mutation of the PIK3CA protein, which encodes the p110α PI3K isoform. The PI3K pathway is reported to be the most frequently activated pathway in periodic human tumors. It has been anticipated that mutation in one or more PI3K pathway components is responsible for up to 30 % of all human cancers. In order to study probable binding mode for all the compounds (7a–7ad) inside the ATP-binding site of PI3K, molecular modeling studies using the accessible crystal structure of p110 gamma isoform in complex with a known inhibitor 1E7V (PDB code, 1E7V) (Fig. 4) with a resolution of 2.4 Å (Walker et al., 2000) were carried out. The docking score, number of hydrogen bonds and details of the interacting amino acid for the target molecules are given in Table 3. The docking pose of the lead molecules and 1E7V with the enzyme is illustrated in Fig. 5.
Table 3 and Fig. 5 illustrate that almost all compounds show interaction with Val882, implying that the presence of pyridazine ring adjacent to Val882 contributes a favorable role in forming extra hydrogen bonds as compared to 1E7V crystal structure. In addition, the presence of fused indole and pyridazine ring system makes the head part of the compounds more rigid that makes the structures more favorable to fit into the pocket aligning adjacent to the val882. The triazole extension attached to the indole ring acts as a tail freely orienting into the pocket. The pocket of this protein to which the ligand binds is quite deep enough and the amino acid interacting with the ligands is at the rim of the pocket (Val882, Ser806), which may not allow the ligands to escape, further making the ligands to fix firmly into the pocket.
Conclusions
We illustrate a simple, mild and efficient synthesis of pyridazino[4,5-b]indole ring systems containing alkyl-, benzyl- and phenacyl-substituted 1,2,3-triazolylmethyl units using Fisher indole, Friedel–Craft acylation and click chemistry protocols. The preliminary antiproliferative examination of the molecules was carried out against four cancer cell lines, viz. breast cancer cells MDA-MB-231 and MCF-7, human primary glioblastoma U-87 cell line and human neuroblastoma IMR-32 cell line. Most of the compounds showed a significant activity against all the tested cell lines with IC50 values ranging in lower micromolar concentrations. Compound 7x containing cyclohexyl substitution at the 1,2,3-triazole unit exhibited outstanding activity against three among four tested cancer cell lines with very low IC50 values, particularly against IMR-32 cell with an IC50 value of 0.04 µM. Compound 7v containing 2-fluorobenzyl-substituted 1,2,3-triazole unit happens to be the next prominent active molecule which exhibited an IC50 value of 0.07 µM against IMR-32 cell line. However, it was observed that replacement of a 2-fluorobenzyl group with a 4-fluorobenzyl group reduces the activity comparatively. Among 4-cyanobenzyl, 4-nitrobenzyl and 4-fluorobenzyl substitutions on the triazole segment, 4-fluoro substitution was found to be important in enhancing the activity. It was also noted that molecules with a fluorine atom on the indole nucleus show improved activity when compared to their non-fluorine analogues. The molecular docking study was carried out for all target compounds inside the ATP-binding site of PI3K with the crystal structure of p 110 isoform with a known inhibitor 1E7V. The docking scores and the binding modes of the molecules with the receptor suggest that these compounds could be potential lead pharmacophores to inhibit the overexpression of PI3 kinases, as PI3K pathway is known to be the most frequently activated pathway in human tumors. Hence, these molecules, with a significant in vitro activity and docking score, are promising lead molecules to be considered for further in vivo biological evaluations.
Experimental
Materials and instruments
The chemicals and solvents used for the synthesis were procured from Sigma-Aldrich (Germany), Merck (India) and Spectrochem Chemicals Pvt. Ltd. All the solvents were distilled and dried before usage. The progress of the reactions was monitored by TLC using pre-coated aluminum sheets with 60 F254 silica gel (Merck KGaA). Silica gel with a mesh size of 230–400 was used as the stationary phase in column chromatography purification. The melting point of the synthesized compounds was recorded by a Stuart SMP3 melting point apparatus. The 1H NMR spectra of the intermediates and final compounds were recorded using a Bruker 300 MHz/400 MHz NMR spectrometer using TMS as internal standard. The 13C NMR spectra of the compounds were recorded with a 100-MHz spectrometer. The elemental analysis was done using a Thermo Electron Corporation EA-112 series C, H, N, S analyzer. The mass spectra were recorded using a Waters micromass Q-Tof microspectrometer. The LC–MS analysis was performed using an Agilent 1200 series mass spectrometer.
General procedure for the synthesis of ethyl 1-(prop-2-ynyl)-1H-indole-2-carboxylates (4a and 4b)
To a solution of ethyl-1H-indole-2-carboxylate intermediate (3a/3b) in DMF, K2CO3 was added and the reaction mass was kept for stirring at RT for 15 min. Propargyl bromide (80 % solution in toluene) was then charged into the reaction mass followed by catalytic amount of n-tetrabutyl ammonium bromide, and the reaction mass was stirred at room temperature (RT) for 6 h. The reaction was monitored using TLC, and after the completion of the reaction, the reaction mass was added to cold water with stirring. The product was extracted with ethyl acetate, and the organic layer was given thorough water wash followed by brine wash. The organic layer was then dried using Na2SO4 and concentrated to get the crude product. The crude product was then purified using column chromatography using ethylacetate/pet ether (1:1) as the eluent to get the pure product (4a/4b).
Ethyl 1-(prop-2-ynyl)-1H-indole-2-carboxylate (4a)
The above-mentioned procedure was followed for compound 3a (7 g, 37.04 mmol) in DMF (35 mL) using K2CO3 (12.8 g, 92.6 mmol) and propargyl bromide (44.44 mmol) to get the intermediate (4a) as red viscous liquid. Yield: 7.4 g (88 %); 1 H NMR (300 MHz, CDCl3): δ 7.01–7.71 (5H, m, Ar–H), 5.79 (2H, s, N–CH2), 4.42 (2H, q, J = 7.4 Hz, CO2CH2), 2.64 (1H, s, –C≡CH), 1.44 (3H, t, J = 7.4 Hz, CO2CH2 CH 3 ); 13 C NMR (100 MHz, CDCl3): δ 164.2 (C, C=O), 149.5 (C, C-8), 145.3 (C, C-9), 138.4 (C, C-2), 125.6 (CH, C-5), 123.1 (CH, C-6), 120.5 (CH, C-4), 120.2 (CH, C-7), 110.9 (CH, C-3), 75.1 (C, C≡CH), 69.3 (CH, C≡CH), 61.2 (CH2, OCH2), 38.7 (CH2, NCH2), 10.9 (CH3). Anal. calculated for C14H13NO2; C, 73.99; H, 5.77; N, 6.16. Found: C, 74.28; H, 5.00; N, 6.22.
Ethyl 5-fluoro-1-(prop-2-ynyl)-1H-indole-2-carboxylate (4b)
The above-mentioned procedure was followed for compound 3b (7g, 33.82 mmol) in DMF (35 mL), using K2CO3 (11.67 g, 84.54 mmol) and propargyl bromide (40.58 mmol) to afford the intermediate (4b) as reddish brown viscous mass. Yield: 7 g (84 %); 1 H NMR (300 MHz, CDCl3): δ 6.97–7.59 (4H, m, Ar–H), 5.80 (2H, s, N–CH2), 4.41 (2H, q, J = 7.4 Hz, CO2CH2), 2.66 (1H, s, –C≡CH), 1.45 (3H, t, J = 7.4 Hz, CO2CH2 CH 3 ); 13 C NMR (100 MHz, CDCl3): δ 163.6 (C, C=O), 159.0 (C, C-5), 147.6 (C, C-8), 143.4 (C, C-9), 139.6 (C, C-2), 124.8 (CH, C-6), 122.1 (CH, C-4), 118.9 (CH, C-7), 111.6 (CH, C-3), 74.8 (C, C≡CH), 65.3 (CH, C≡CH), 59.6 (CH2, OCH2), 38.5 (CH2, NCH2), 11.2 (CH3). Anal. calculated for C14H12FNO2; C, 68.56; H, 4.93; N, 5.71. Found: C, 68.29; H, 4.98; N, 5.62.
General procedure for the synthesis of ethyl 3-acetyl-1-(prop-2-ynyl)-1H-indole-2-carboxylates (5a and 5b)
Anhydrous AlCl3 was taken in dry CH2Cl2 in a RB flask. The content of the flask was cooled to 0 °C, and calculated amount of acetyl chloride was added dropwise into the reaction mass. After the addition, the reaction mass was kept for stirring at 0 °C for about 15 min. A solution of intermediate (4a/4b) in CH2Cl2 was then charged dropwise into the reaction mass at the same temperature. After the addition, the reaction mass was refluxed at 40 °C for 2 h. The reaction was monitored using TLC. After the completion of the reaction, the reaction mass was poured into a mixture of ice-cold water and dichloromethane. The resulted mass was filtered through Celite, and the organic layer was separated. The organic layer was washed with water then with brine solution, dried using Na2SO4 and concentrated. The crude product was then purified using column chromatography using ethyl acetate/pet ether (1:1) as the eluent.
Ethyl-3-acetyl-1-(prop-2-ynyl)-1H-indole-2-carboxylate (5a)
Intermediate 5a was synthesized by carrying out the above-mentioned procedure for 4a (6.5 g, 28.63 mmol) with AlCl3 (4.2 g, 31.49 mmol), acetyl chloride (2.5 g, 31.49 mmol) to get the pure product as greenish yellow oily compound. Yield: 6.3 g (82 %); 1 H NMR (300 MHz, CDCl3): δ 7.26–7.56 (4H, m, Ar–H), 5.76 (2H, s, N–CH2), 4.46 (2H, q, J = 7.8 Hz, CO2CH2), 2.68 (1H, s, –C≡CH), 2.61 (3H, s, COCH3), 1.44 (3H, t J = 7.8 Hz, CO2CH2 CH 3 ); 13 C NMR (100 MHz, CDCl3): δ 171.2 (C, COMe), 162.3 (C, CO2Et), 157.5 (C, C-3), 148.7 (C, C-8), 142.0 (C, C-9), 137.8 (C, C-2), 126.0 (CH, C-5), 123.6 (CH, C-6), 119.1 (CH, C-4), 109.6 (CH, C-7), 75.2 (C, C≡CH), 68.0 (CH, C≡CH), 60.6 (CH2, OCH2), 38.3 (CH2, NCH2), 30.2 (CH3, COCH3), 12.0 (CH3); Anal. calculated for C16H15NO3; C, 71.36; H, 5.61; N, 5.20. Found: C, 71.57; H, 5.69; N, 5.12.
Ethyl-3-acetyl-5-fluoro-1-(prop-2-ynyl)-1H-indole-2-carboxylate (5b)
The acylated intermediate 5b was synthesized by following the above procedure for intermediate 4b (6 g, 24.46 mmol) with AlCl3 (3.6 g, 26.91 mmol), acetyl chloride (2.1 g, 26.91 mmol) to obtain the pure product as yellow oily compound. Yield: 5.9 g (84 %); 1 H NMR (300 MHz, CDCl3): δ 7.14–7.56 (3H, m, Ar–H), 5.78 (2H, s, N–CH2), 4.41 (2H, q, J = 7.8 Hz, CO2CH2), 2.64 (1H, s, –C≡CH), 2.60 (3H, s, COCH3), 1.42 (3H, t, J = 7.8 Hz, CO2CH2 CH 3 ); 13 C NMR (100 MHz, CDCl3): δ 172.3 (C, COMe), 163.8 (C, CO2Et), 159.6 (C, C-5), 157.1 (C, C-3), 146.9 (C, C-8), 142.1 (C, C-9), 138.1 (C, C-2), 125.1 (CH, C-6), 120.9 (CH, C-4), 110.1 (CH, C-7), 75.9 (C, C≡CH), 68.2 (CH, C≡CH), 61.6 (CH2, OCH2), 39.9 (CH2, NCH2), 30.0 (CH3, COCH3), 12.0 (CH3); Anal. calculated for C16H14FNO3; C, 66.89; H, 4.91; N, 4.88. Found: C, 66.75; H, 5.01; N, 4.92.
General procedure for the synthesis of 1-methyl-5-(prop-2-ynyl)-3H-pyridazino[4,5-b]indol-4(5H)-one (6a and 6b)
To a solution of intermediate 5a/5b in ethanol, hydrazine hydrate was added and the reaction mass was heated to 80 °C for 6 h and kept for stirring at RT for 16 h. The reaction was monitored using TLC, and after the completion of the reaction, ethanol was evaporated under reduced pressure and the residue was poured into cold dilute HCl with stirring. The precipitated product was filtered, was given thorough water wash followed by diethyl ether wash and then dried in an vacuum oven to get the pure product.
1-Methyl-5-(prop-2-ynyl)-3H-pyridazino[4,5-b]indol-4(5H)-one (6a)
The indole–pyridazinone-fused intermediate 6a was synthesized by following the above-described procedure for 5a (5 g, 18.57 mmol) using ethanol (25 mL) and hydrazine hydrate (37.13 mmol) to yield the product as white solid. Yield: 3.2 g (73 %); mp > 250 °C; 1 H NMR (300 MHz, [D6]DMSO): δ 12.72 (1H, s, NH), 7.44–8.17 (4H, m, Ar–H), 5.79 (2H, s, NCH2), 2.72 (3H, s, Ar–CH3), 2.66 (1H, s, C≡CH); 13 C NMR (100 MHz, CDCl3): δ 158.1 (C, C=O), 147.8 (C, C-1), 145.9 (C, C-12), 131.2 (C, C-13), 130.1 (C, C-11), 125.6 (C, C-10), 121.5 (CH, C-8), 120.1 (CH, C-7), 119.4 (CH, C-9), 110.8 (CH, C-6), 76.2 (C, C≡CH), 69.8 (CH, C≡CH), 39.0 (CH2, NCH2), 22.1 (CH3); Anal. calculated for C14H11N3O; C, 70.87; H, 4.67; N, 17.71. Found: C, 71.12 H, 4.62; N, 17.79.
8-Fluoro-1-methyl-5-(prop-2-ynyl)-3H-pyridazino[4,5-b]indol-4(5H)-one (6b)
Intermediate 6b was synthesized by carrying out the above-illustrated procedure for 5b (5 g, 17.40 mmol) using ethanol (25 mL) and hydrazine hydrate (34.81 mmol) to yield the product as white solid. Yield: 3.2 g (72 %); mp > 250 °C; 1 H NMR (300 MHz, [D6]DMSO): δ 12.74 (1H, s, NH), 7.39–8.11 (3H, m, Ar–H), 5.76 (2H, s, NCH2), 2.70 (3H, s, Ar–CH3), 2.68 (1H, s, C≡CH); 13 C NMR (100 MHz, CDCl3): δ 157.6 (C, C=O), 156.8 (C, C-8), 149.8 (C, C-1), 145.1 (C, C-12), 132.7 (C, C-13), 131.9 (C, C-11), 124.2 (C, C-10), 121.0 (CH, C-7), 120.1 (CH, C-9), 111.2 (CH, C-6), 77.4 (C, C≡CH), 70.2 (CH, C≡CH), 38.5 (CH2, NCH2), 22.9 (CH3); Anal. calculated for C14H10FN3O; C, 65.88; H, 3.95; N, 16.46. Found: C, 65.99; H, 3.87; N, 16.51.
General procedure for the synthesis of final compounds (7a–z, 7aa–ad)
A calculated amount of alkyl/benzyl/phenacyl bromide was taken in tert-butanol/H2O (1:1) solvent system, to which calculated amount (1.1 equivalents) of NaN3 was added and the reaction mass was stirred at 80 °C for about 1 h. After the formation of alkyl/benzyl/phenacyl azide, the temperature was brought back to RT. The propargylated pre-final intermediate, 6a/6b, (1 equivalent), was then introduced to the reaction mass followed by the addition of CuI (catalytic amount) and a pinch of alumina (as support to the copper catalyst). The reaction mass was stirred at room temperature overnight. After the completion of the reaction, the solvent was evaporated under reduced pressure and the product was extracted with ethyl acetate. The copper residue present in the organic layer was removed by washing it with ammonia. The organic layer was then dried using Na2SO4 and concentrated under reduced pressure. The crude product was then purified by column chromatographic technique using petroleum ether/ethyl acetate (1:1) as the mobile phase.
The structural characterization data of the final compounds are listed below.
5-[(1-Benzyl-1H-1,2,3-triazol-4-yl)methyl]-1-methyl-3H-pyridazino[4,5-b]indol-4(5H)-one (7a)
1 H NMR (400 MHz, CDCl3) δ 7.16–7.84 (10H, m, Ar–H), 6.00 (2H, s, NCH2-Ar), 5.41 (2H, s, NCH2-Ph), 2.63 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 162.9 (C, C=O), 159.9 (C, C-5 of triazole ring), 144.7, 139.1, 134.5, 129.0, 128.9, 128.0, 127.9, 127.7, 126.9, 125.1, 122.9, 122.0, 121.8, 121.2, 111.5, 107.1 (Ar–C), 54.1 (CH2, CH2Ph), 40.5 (CH2, CH2N<), 11.0 (CH3); Anal. calculated for C21H18N6O; C, 68.09; H, 4.90; N, 22.69. Found: C, 68.21; H, 4.83; N, 22.60.
5-{[1-(4-Methoxybenzyl)-1H-1,2,3-triazol-4-yl]methyl}-1-methyl-3H-pyridazino[4,5-b]indol-4(5H)-one (7b)
1 H NMR (400 MHz, CDCl3) δ 6.81–7.68 (9H, m, Ar–H), 6.14 (2H, s, NCH2-Ar), 5.42 (2H, s, NCH2-Ph), 3.77 (3H, s, OCH3), 2.63 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 163.0 (C, C=O), 160.9 (C, PhOMe), 158.7, 138.6, 136.2, 129.9, 127.8, 126.3, 126.0, 124.5, 123.1, 121.9, 120.1, 119.8, 119.0, 113.6, 110.1, 107.4 (Ar–C), 60.3 (CH3, OMe), 55.9 (CH2, CH2Ph), 41.2 (CH2, CH2N<), 11.0 (CH3); Anal. calculated for C22H20N6O2; C, 65.99; H, 5.03; N, 20.99. Found: C, 65.90; H, 5.08; N, 21.14.
5-{{1-[4-(Trifluoromethyl)benzyl]-1H-1,2,3-triazol-4-yl}methyl}-1-methyl-3H-pyridazino [4,5-b]indol-4(5H)-one (7c)
1 H NMR (400 MHz, CDCl3) δ 7.17–7.83 (9H, m, Ar–H), 6.0 (2H, s, NCH2-Ar), 5.47 (2H, s, NCH2-Ph), 2.63 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 163.0 (C, C=O), 159.9, 145.1, 139.1, 138.5, 131.4, 130.7, 128.1, 126.9, 126.1 (Ar–C), 125.1 (C, CF3), 123.1, 122.4, 122.0, 121.8, 121.2, 111.4, 109.2, 107.1 (Ar–C), 54.2 (CH2, CH2Ph), 41.0 (CH2, CH2N<), 10.7 (CH3); Anal. calculated for C22H17F3N6O; C, 60.27; H, 3.91; N, 19.17. Found: C, 60.36; H, 3.88; N, 19.14.
5-{{1-[4-(Trifluoromethoxy)benzyl]-1H-1,2,3-triazol-4-yl}methyl}-1-methyl-3H-pyridazino [4,5-b]indol-4(5H)-one (7d)
1 H NMR (300 MHz, CDCl3) δ 7.16–7.88 (9H, m, Ar–H), 6.02 (2H, s, NCH2-Ar), 5.43 (2H, s, NCH2-Ph), 2.66 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 161.8 (C, C=O), 160.4 (C, PhOCF3), 158.6, 145.2, 141.6, 136.1, 134.2, 133.6, 131.2, 131.4, 130.1, 129.2, 125.1, 124.4, 123.4, 122.1, 122.0 (Ar–C), 121.1 (C, OCF3), 119.6 (Ar–C), 54.8 (CH2, CH2Ph), 40.6 (CH2, CH2N<), 10.9 (CH3); MS (m/z) 455.1 (M + H)+; Anal. calculated for C22H17F3N6O2; C, 58.15; H, 3.77; N, 18.49. Found: C, 58.05; H, 3.80; N, 18.54.
4-{{4-[(1-Methyl-4-oxo-3,4-dihydropyridazino[4,5-b]indol-5-yl)methyl]-1H-1,2,3-triazol-1-yl}methyl}benzonitrile (7e)
1 H NMR (300 MHz, CDCl3) δ 7.17–7.99 (9H, m, Ar–H), 6.29 (2H, s, NCH2-Ar), 5.32 (2H, s, NCH2-Ph), 2.66 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 161.6 (C, C=O), 160.1, 146.7, 145.1, 139.2, 136.2, 132.1, 132.0, 130.1, 130.0, 127.6, 125.5, 124.9, 122.9, 121.8, 119.6 (Ar–C), 117.0 (C, CN), 112.0, 106.8 (Ar–C), 54.7 (CH2, CH2Ph), 40.2 (CH2, CH2N<), 10.8 (CH3); Anal. calculated for C22H17N7O; C, 66.82; H, 4.33; N, 24.80. Found: C, 66.96; H, 4.28; N, 24.83.
5-{[1-(4-Nitrobenzyl)-1H-1,2,3-triazol-4-yl]methyl}-1-methyl-3H-pyridazino[4,5-b]indol-4(5H)-one (7f)
1 H NMR (300 MHz, CDCl3) δ 7.20–8.21 (9H, m, Ar–H), 6.03 (2H, s, NCH2-Ar), 5.55 (2H, s, NCH2-Ph), 2.66 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 162.7 (C, C=O), 160.9, 158.7, 146.1, 142.7, 138.1, 133.1, 132.0, 131.0, 129.2, 128.5, 126.1, 124.6, 122.2, 122.0, 121.1, 111.6, 107.0 (Ar–C), 54.6 (CH2, CH2Ph), 40.2 (CH2, CH2N<), 10.6 (CH3); Anal. calculated for C21H17N7O3; C, 60.72; H, 4.12; N, 23.60. Found: C, 60.96; H, 4.24; N, 23.75.
5-{[1-(4-Fluorobenzyl)-1H-1,2,3-triazol-4-yl]methyl}-1-methyl-3H-pyridazino[4,5-b]indol-4(5H)-one (7g)
1 H NMR (400 MHz, CDCl3) δ 6.97–7.84 (9H, m, Ar–H), 5.99 (2H, s, NCH2-Ar), 5.36 (2H, s, NCH2-Ph), 2.64 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 164.0 (C, C-F), 162.9 (C, C=O), 161.5, 159.9, 144.8, 139.1, 130.4, 129.9, 129.8, 126.9, 125.1, 122.8, 122.0, 121.2, 116.1, 115.9, 111.5, 107.0 (Ar–C), 53.3 (CH2, CH2Ph), 40.5 (CH2, CH2N<), 11.0 (CH3); Anal. calculated for C21H17FN6O; C, 64.94; H, 4.41; N, 21.64. Found: C, 64.80; H, 4.48; N, 21.71.
5-{[1-(2-Fluorobenzyl)-1H-1,2,3-triazol-4-yl]methyl}-1-methyl-3H-pyridazino[4,5-b]indol-4(5H)-one (7h)
1 H NMR (400 MHz, CDCl3) δ 7.03–7.84 (9H, m, Ar–H), 6.01 (2H, s, NCH2-Ar), 5.47 (2H, s, NCH2-Ph), 2.64 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 162.9 (C, C-F), 161.6 (C, C=O), 159.9, 159.1, 139.1, 130.8, 130.3, 126.9, 125.0, 124.8, 123.1, 122.1, 121.8, 121.2, 115.8, 115.6, 111.5, 107.1 (Ar–C), 47.6 (CH2, CH2Ph), 40.5 (CH2, CH2N<), 11.0 (CH3); Anal. calculated for C21H17FN6O; C, 64.94; H, 4.41; N, 21.64. Found: C, 65.13; H, 4.48; N, 21.70.
5-[(1-Cyclopentyl-1H-1,2,3-triazol-4-yl)methyl]-1-methyl-3H-pyridazino[4,5-b]indol-4(5H)-one (7i)
1 H NMR (300 MHz, CDCl3) δ 7.11–7.88 (5H, m, Ar–H), 5.99 (2H, s, NCH2-Ar), 4.79–4.84 (1H, m, NCH<), 2.68 (3H, s, Ar–CH3), 1.67–2.67 (8H, m, CH2 of cyclopentyl); 13 C NMR (100 MHz, CDCl3) δ 163.0 (C, C=O), 160.0, 144.0, 139.1, 126.9, 125.0, 122.1, 121.7, 121.4, 121.1, 111.6, 107.0 (Ar–C), 61.8 (CH, C-1 of cyclopentyl ring), 40.6 (CH2, CH2N<), 33.3 (CH2, C-2/C-5 of cyclopentyl ring), 24.0 (CH2, C-3/C-4 of cyclopentyl ring), 11.0 (CH3); Anal. calculated for C19H20N6O; C, 65.50; H, 5.79; N, 24.12. Found: C, 65.71; H, 5.71; N, 24.14.
5-[(1-Cyclohexyl-1H-1,2,3-triazol-4-yl)methyl]-1-methyl-3H-pyridazino[4,5-b]indol-4(5H)-one (7j)
1 H NMR (300 MHz, CDCl3) δ 7.17–7.89 (6H, m, Ar–H), 6.03 (2H, s, NCH2-Ar), 4.76–4.85 (1H, m, NCH<), 2.68 (3H, s, Ar–CH3), 1.86–2.23 (10H, m, CH2 of cyclohexyl); 13 C NMR (100 MHz, CDCl3) δ 162.9 (C, C=O), 160.0, 144.0, 139.1, 126.9, 125.0, 122.1, 121.7, 121.4, 121.1, 111.6, 107.0 (Ar–C), 61.8 (CH, C-1 of cyclohexyl ring), 40.6 (CH2, CH2N<), 33.3 (CH2, C-2/C-6 of cyclohexyl ring), 28.5 (CH2, C-4 of cyclohexyl ring), 24.0 (CH2, C-3/C-5 of cyclohexyl ring), 11.0 (CH3); Anal. calculated for C20H22N6O; C, 66.28; H, 6.12; N, 23.19. Found: C, 66.40; H, 6.18; N, 23.08.
1-Methyl-5-{{1-[2-oxo-2-(p-tolyl)ethyl]triazol-4-yl}methyl}-3H-pyridazino[4,5-b]indol-4-one (7k)
1 H NMR (400 MHz, CDCl3) δ 7.14–8.01 (9H, m, Ar–H), 6.11 (2H, s, NCH2-Ar), 5.75 (2H, s, NCH2COPh), 2.66 (3H, s, Ar–CH3), 2.45 (3H, s, CH3Ph); 13 C NMR (100 MHz, CDCl3) δ 187.2 (C, COPh), 165.6 (C, C=O), 161.9, 160.8, 159.8, 141.9, 135.7, 133.3, 130.2, 129.6, 124.4, 124.2, 123.0, 122.0, 121.2, 116.8, 116.1, 112.6, 109.4 (Ar–C) 52.8 (CH2, CH2COPh), 43.9 (CH2, CH2N<), 21.1 (CH3, PhCH3), 11.2 (CH3); Anal. calculated for C23H20N6O2; C, 66.98; H, 4.89; N, 20.38. Found: C, 66.87; H, 4.93; N, 20.44.
5-{{1-[2-(4-Methoxyphenyl)-2-oxo-ethyl]triazol-4-yl}methyl}-1-methyl-3H-pyridazino[4,5-b]indol-4-one (7l)
1 H NMR (400 MHz, CDCl3) δ 7.24–7.99 (10H, m, Ar–H), 6.18 (2H, s, NCH2-Ar), 5.78 (2H, s, NCH2COPh), 3.79 (3H, s, OCH3), 2.65 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 185.2 (C, COPh), 163.1 (C, C=O), 160.82, 159.9, 159.0, 159.0, 140.9, 133.7, 132.1, 129.2, 124.9, 124.2, 123.0, 122.0, 121.2, 116.8, 116.1, 112.6, 109.4 (Ar–C), 58.2 (CH3, OCH3), 50.3 (CH2, CH2COPh), 40.5 (CH2, CH2N<), 10.9 (CH3); MS (m/z) 429.1 (M + H)+; Anal. calculated for C23H20N6O2; C, 66.98; H, 4.89; N, 20.38. Found: C, 67.13; H, 4.81; N, 20.49.
5-{{1-[2-(4-Nitrophenyl)-2-oxo-ethyl]triazol-4-yl}methyl}-1-methyl-3H-pyridazino[4,5-b]indol-4-one (7m)
1 H NMR (400 MHz, CDCl3) δ 7.26–8.21 (10H, m, Ar–H), 6.19 (2H, s, NCH2-Ar), 5.74 (2H, s, NCH2COPh), 2.64 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 185.3 (C, COPh), 162.6 (C, C=O), 161.1, 160.7, 158.6, 144.3, 138.7, 133.1, 130.6, 125.7, 125.0, 125.0, 124.5, 123.0, 122.2, 118.1, 117.5, 114.1, 111.2 (Ar–C), 51.7 (CH2, CH2COPh), 41.7 (CH2, CH2N<), 10.8 (CH3); MS (m/z) 444.1 (M + H)+; Anal. calculated for C22H17N7O4; C, 59.59; H, 3.86; N, 22.11. Found: C, 59.67; H, 3.80; N, 22.17.
5-{{1-[2-(4-Fluorophenyl)-2-oxo-ethyl]triazol-4-yl}methyl}-1-methyl-3H-pyridazino[4,5-b]indol-4-one (7n)
1 H NMR (400 MHz, CDCl3) δ 7.19–7.87 (10H, m, Ar–H), 6.25 (2H, s, NCH2-Ar), 5.79 (2H, s, NCH2COPh), 2.63 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 184.9 (C, COPh), 161.0 (C, C=O), 160.5, 159.7, 158.6, 148.7, 141.5, 139.3, 135.6, 128.5, 124.1, 123.0, 122.7, 122.0, 121.1, 120.3, 119.3, 116.7, 114.3 (Ar–C), 50.6 (CH2, CH2COPh), 40.9 (CH2, CH2N<), 10.4 (CH3); Anal. calculated for C22H17FN6O2; C, 63.46; H, 4.11; N, 20.18. Found: C, 63.65; H, 4.02; N, 20.24.
5-[(1-Benzyl-1H-1,2,3-triazol-4-yl)methyl]-8-fluoro-1-methyl-3H-pyridazino[4,5-b]indol-4(5H)-one (7o)
1 H NMR (400 MHz, CDCl3) δ 7.11–7.82 (10H, m, Ar–H), 6.11 (2H, s, NCH2-Ar), 5.44 (2H, s, NCH2-Ph), 2.64 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 161.0 (C, C=O), 160.8, 157.3, 145.9, 140.4, 136.1, 130.8, 129.2, 128.9, 127.5, 127.0, 126.6, 124.9, 122.8, 122.3, 121.6, 110.9, 108.3 (Ar–C), 54.8 (CH2, CH2Ph), 40.7 (CH2, CH2N<), 10.0 (CH3); Anal. calculated for C21H17FN6O; C, 64.94; H, 4.41; N, 21.64. Found: C, 65.10; H, 4.32; N, 21.60.
5-{[1-(4-Methoxybenzyl)-1H-1,2,3-triazol-4-yl]methyl}-8-fluoro-1-methyl-3H-pyridazino [4,5-b]indol-4(5H)-one (7p)
1 H NMR (400 MHz, CDCl3) δ 7.08–7.92 (9H, m, Ar–H), 5.95 (2H, s, NCH2-Ar), 5.34 (2H, s, NCH2-Ph), 3.77 (3H, s, OCH3), 2.64 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 163.0 (C, C=O), 159.8, 159.6, 157.4, 144.3, 135.7, 129.5, 127.1, 127.0, 126.5, 123.4, 122.6, 114.4, 114.0, 113.7, 112.7, 106.6, 106.2 (Ar–C), 58.3 (CH3, OMe), 53.7 (CH2, CH2Ph), 40.6 (CH2, CH2N<), 11.0 (CH3). Anal. calculated for C22H19FN6O2; C, 63.15; H, 4.58; N, 20.08. Found: C, 63.29; H, 4.49; N, 20.11.
5-{{1-[4-(Trifluoromethyl)benzyl]-1H-1,2,3-triazol-4-yl}methyl}-8-fluoro-1-methyl-3H-pyridazino[4,5-b]indol-4(5H)-one (7q)
1 H NMR (300 MHz, CDCl3) δ 7.16–7.87 (8H, m, Ar–H), 5.99 (2H, s, NCH2-Ar), 5.51 (2H, s, NCH2-Ph), 2.65 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 163.1 (C, C=O), 159.8, 159.6, 157.4, 144.8, 138.4, 135.7, 128.1, 127.1, 127.0, 126.0 (Ar–C), 124.4 (C, CF3), 123.3, 123.2, 114.1, 113.9, 112.6, 106.7, 106.0 (Ar–C), 53.4 (CH2, CH2Ph), 40.6 (CH2, CH2N<), 11.0 (CH3); MS (m/z) 457.1 (M + H)+; Anal. calculated for C22H16F4N6O; C, 57.90; H, 3.53; N, 18.41. Found: C, 57.79; H, 3.58; N, 18.47.
5-{{1-[4-(Trifluoromethoxy)benzyl]-1H-1,2,3-triazol-4-yl}methyl}-8-fluoro-1-methyl-3H-pyridazino[4,5-b]indol-4(5H)-one (7r)
1 H NMR (400 MHz, CDCl3) δ 7.11–7.84 (8H, m, Ar–H), 5.97 (2H, s, NCH2-Ar), 5.42 (2H, s, NCH2-Ph), 2.64 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 163.1 (C, C=O), 159.8, 157.4, 149.4, 144.6, 135.7, 134.3, 133.2, 131.3, 130.9, 129.8, 128.8, 123.3, 125.6, 123.1, 121.6, 121.5 (Ar–C), 121.2 (C, OCF3), 119.1, 53.2 (CH2, CH2Ph), 40.6 (CH2, CH2N<), 10.9 (CH3); Anal. calculated for C22H16F4N6O2; C, 55.94; H, 3.41; N, 17.79. Found: C, 55.82; H, 3.49; N, 17.83.
4-{{4-[(8-Fluoro-1-methyl-4-oxo-3,4-dihydropyridazino[4,5-b]indol-5-yl)methyl]-1H-1,2,3-triazol-1-yl}methyl}benzonitrile (7s)
1 H NMR (300 MHz, CDCl3) δ 7.13–7.87 (8H, m, Ar–H), 5.99 (2H, s, NCH2-Ar), 5.51 (2H, s, NCH2-Ph), 2.66 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 163.2 (C, C=O), 159.8, 157.4, 144.9, 139.6, 135.7, 132.8, 128.4, 127.1, 127.0, 123.4, 123.3 (Ar–C), 118.1 (C, CN), 114.2, 113.9, 112.7, 112.5, 106.7, 106.1 (Ar–C), 53.4 (CH2, CH2Ph), 40.5 (CH2, CH2N<), 11.0 (CH3); MS (m/z) 414.1 (M + H)+; Anal. calculated for C22H16FN7O; C, 63.92; H, 3.90; N, 23.72. Found: C, 64.04; H, 3.94; N, 23.63.
5-{[1-(4-Nitrobenzyl)-1H-1,2,3-triazol-4-yl]methyl}-8-fluoro-1-methyl-3H-pyridazino[4,5-b] indol-4(5H)-one (7t)
1 H NMR (300 MHz, CDCl3) δ 7.13–8.21 (9H, m, Ar–H), 5.99 (2H, s, NCH2-Ar), 5.55 (2H, s, NCH2-Ph), 2.66 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 162.0 (C, C=O), 160.7, 159.6, 157.1, 145.1, 141.9, 138.1, 132.2, 131.6, 131.1, 130.9, 130.1, 127.6, 127.1, 125.6, 123.6, 120.6, 119.6 (Ar–C), 54.5 (CH2, CH2Ph), 40.1 (CH2, CH2N<), 10.6 (CH3); Anal. calculated for C21H16FN7O3; C, 58.20; H, 3.72; N, 22.62. Found: C, 58.28; H, 3.69; N, 22.58.
5-{[1-(4-Fluorobenzyl)-1H-1,2,3-triazol-4-yl]methyl}-8-fluoro-1-methyl-3H-pyridazino[4,5-b] indol-4(5H)-one (7u)
1 H NMR (400 MHz, CDCl3) δ 7.11–7.94 (9H, m, Ar–H), 6.01 (2H, s, NCH2-Ar), 5.50 (2H, s, NCH2-Ph), 2.63 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 165.6 (C, C-F), 163.0 (C, C=O), 162.8, 160.6, 159.1, 146.6, 142.3, 132.8, 130.9, 130.1, 128.5, 126.6, 124.1, 123.2, 122.4, 122.0, 116.0, 112.6 (Ar–C), 52.5 (CH2, CH2Ph), 40.9 (CH2, CH2N<), 10.3 (CH3); Anal. calculated for C21H16F2N6O; C, 62.06; H, 3.97; N, 20.68. Found: C, 62.19; H, 3.90; N, 20.62.
5-{[1-(2-Fluorobenzyl)-1H-1,2,3-triazol-4-yl]methyl}-8-fluoro-1-methyl-3H-pyridazino[4,5-b]indol-4(5H)-one (7v)
1 H NMR (400 MHz, CDCl3) δ 7.11–7.94 (9H, m, Ar–H), 6.08 (2H, s, NCH2-Ar), 5.56 (2H, s, NCH2-Ph), 2.61 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 165.0 (C, C-F), 163.6 (C, C=O), 161.6, 161.1, 159.0, 140.2, 133.1, 132.8, 129.3, 128.4, 124.8, 124.0, 123.9, 122.7, 117.1, 116.3, 113.2, 109.8 (Ar–C), 47.7 (CH2, CH2Ph), 40.5 (CH2, CH2N<), 10.8 (CH3); Anal. calculated for C21H16F2N6O; C, 62.06; H, 3.97; N, 20.68. Found: C, 62.18; H, 4.00; N, 20.60.
5-[(1-Cyclopentyl-1H-1,2,3-triazol-4-yl)methyl]-8-fluoro-1-methyl-3H-pyridazino[4,5-b] indol-4(5H)-one (7w)
1 H NMR (300 MHz, CDCl3) δ 7.08–7.79 (4H, m, Ar–H), 6.05 (2H, s, NCH2-Ar), 4.81–4.86 (1H, m, NCH<), 2.66 (3H, s, Ar–CH3), 1.65–2.66 (8H, m, CH2 of cyclopentyl); 13 C NMR (100 MHz, CDCl3) δ 164.7 (C, C=O), 162.9, 161.2, 146.7, 144.1, 129.8, 124.9, 123.9, 122.1, 121.1, 113.8, 109.1 (Ar–C), 61.8 (CH, C-1 of cyclopentyl ring), 40.6 (CH2, CH2N<), 34.3 (CH2, C-2/C-5 of cyclopentyl ring), 25.1 (CH2, C-3/C-4 of cyclopentyl ring), 10.9 (CH3); MS (m/z) 367.1 (M + H)+; Anal. calculated for C19H19FN6O; C, 62.28; H, 5.23; N, 22.94. Found: C, 62.37; H, 5.18; N, 22.89.
5-[(1-Cyclohexyl-1H-1,2,3-triazol-4-yl)methyl]-8-fluoro-1-methyl-3H-pyridazino[4,5-b] indol-4(5H)-one (7x)
1 H NMR (400 MHz, CDCl3) δ 7.09–7.81 (5H, m, Ar–H), 6.04 (2H, s, NCH2-Ar), 4.79–4.85 (1H, m, NCH<), 2.66 (3H, s, Ar–CH3), 1.86–2.23 (10H, m, CH2 of cyclohexyl); 13 C NMR (100 MHz, CDCl3) δ 164.6 (C, C=O), 162.8, 161.3, 145.9, 141.5, 128.3, 127.3, 123.6, 122.0, 122.0, 113.3, 109.7 (Ar–C), 60.8 (CH, C-1 of cyclohexyl ring), 40.8 (CH2, CH2N<), 33.4 (CH2, C-2/C-6 of cyclohexyl ring), 28.6 (CH2, C-4 of cyclohexyl ring), 24.9 (CH2, C-3/C-5 of cyclohexyl ring), 10.7 (CH3); Anal. calculated for C20H21FN6O; C, 63.14; H, 5.56; N, 22.09. Found: C, 63.24; H, 5.52; N, 21.99.
8-Fluoro-1-methyl-5-{{1-[2-oxo-2-(p-tolyl)ethyl]triazol-4-yl}methyl}-3H-pyridazino[4,5-b] indol-4-one (7y)
1 H NMR (300 MHz, CDCl3) δ 7.12–8.01 (8H, m, Ar–H), 6.06 (2H, s, NCH2-Ar), 5.74 (2H, s, NCH2COPh), 2.64 (3H, s, Ar–CH3), 2.45 (3H, s, CH3Ph); 13 C NMR (100 MHz, CDCl3) δ 184.1 (C, COPh), 163.0 (C, C=O), 161.1, 160.9, 155.6, 146.8, 142.2, 137.2, 135.7, 131.4, 127.5, 126.9, 124.5, 123.0, 121.0, 119.6, 118.3, 114.8, 112.8 (Ar–C), 52.7 (CH2, CH2COPh), 40.2 (CH2, CH2N<), 19.1 (CH3, PhCH3), 10.9 (CH3); Anal. calculated for C23H19FN6O2; C, 64.18; H, 4.45; N, 19.52. Found: C, 64.29; H, 4.39; N, 19.47.
8-Fluoro-5-{{1-[2-(4-methoxyphenyl)-2-oxo-ethyl]triazol-4-yl}methyl}-1-methyl-3H-pyridazino[4,5-b]indol-4-one (7z)
1 H NMR (300 MHz, CDCl3) δ 7.12–7.94 (9H, m, Ar–H), 6.09 (2H, s, NCH2-Ar), 5.79 (2H, s, NCH2COPh), 3.77 (3H, s, OCH3), 2.64 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 184.1 (C, COPh), 164.6 (C, C=O), 162.8, 161.6, 160.7, 155.5, 142.4, 134.7, 133.9, 131.0, 126.7, 126.0, 125.9, 123.8, 121.1, 119.7, 118.3, 115.1, 112.3 (Ar–C), 57.7 (CH3, OMe), 51.3 (CH2, CH2COPh), 40.9 (CH2, CH2N<), 10.9 (CH3); Anal. calculated for C23H19FN6O3; C, 61.88; H, 4.29; N, 18.82. Found: C, 61.80; H, 4.34; N, 18.86.
8-Fluoro-5-{{1-[2-(4-nitrophenyl)-2-oxo-ethyl]triazol-4-yl}methyl}-1-methyl-3H-pyridazino[4,5-b]indol-4-one (7aa)
1 H NMR (400 MHz, CDCl3) δ 7.17–7.97 (9H, m, Ar–H), 6.21 (2H, s, NCH2-Ar), 5.76 (2H, s, NCH2COPh), 2.66 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 185.2 (C, COPh), 163.3 (C, C=O), 162.9, 162.0, 160.6, 155.1 146.2, 140.6, 135.1, 133.1, 130.4, 126.6, 125.2, 124.0, 123.3, 120.1, 119.6, 117.0, 113.5 (Ar–C), 52.6 (CH2, CH2COPh), 40.1 (CH2, CH2N<), 11.0 (CH3); Anal. calculated for C22H16FN7O4; C, 57.27; H, 3.50; N, 21.25. Found: C, 57.20; H, 3.57; N, 21.29.
8-Fluoro-5-{{1-[2-(4-fluorophenyl)-2-oxo-ethyl]triazol-4-yl}methyl}-1-methyl-3H-pyridazino[4,5-b]indol-4-one (7ab)
1 H NMR (400 MHz, CDCl3) δ 7.15–7.91 (9H, m, Ar–H), 6.22 (2H, s, NCH2-Ar), 5.77 (2H, s, NCH2COPh), 2.61 (3H, s, Ar–CH3); 13 C NMR (100 MHz, CDCl3) δ 186.0 (C, COPh), 162.5 (C, C=O), 162.1, 160.9, 159.1, 158.6, 150.5, 145.5, 142.2, 140.2, 130.1, 128.9, 126.9, 125.5, 122.3, 121.7, 120.9, 120.7, 118.3 (Ar–C), 51.9 (CH2, CH2COPh), 41.6 (CH2, CH2N<), 10.7 (CH3); Anal. calculated for C22H16F2N6O2; C, 60.83; H, 3.71; N, 19.35. Found: C, 60.94; H, 3.67; N, 19.29.
5-[(1-Isopropyl-1H-1,2,3-triazol-4-yl)methyl]-1-methyl-3H-pyridazino[4,5-b]indol-4(5H)-one (7ac)
1 H NMR (300 MHz, CDCl3) δ 7.11–7.87 (5H, m, Ar–H), 5.80 (2H, s, NCH2-Ar), 4.01–4.15 (1H, m, NCH(Me)2), 2.63 (3H, s, Ar–CH3), 1.58 (6H, d, CH3); 13 C NMR (100 MHz, CDCl3) δ 163.6 (C, C=O), 146.8, 145.3, 143.3, 133.9, 124.1, 123.1, 122.1, 121.1, 118.5, 118.1, 115.4 (Ar–C), 55.1 (CH, CH(CH3)2), 41.7 (CH2, CH2N<), 23.9 (2 CH3, CH(CH3)2), 10.8 (CH3); Anal. calculated for C17H18N6O; C, 63.34; H, 5.63; N, 26.07. Found: C, 63.43; H, 5.67; N, 25.98.
8-Fluoro-5-[(1-isopropyl-1H-1,2,3-triazol-4-yl)methyl]-1-methyl-3H-pyridazino[4,5-b]indol-4(5H)-one (7ad)
1 H NMR (300 MHz, CDCl3) δ 7.14–7.82 (4H, m, Ar–H), 5.77 (2H, s, NCH2-Ar), 4.11–4.18 (1H, m, NCH(Me)2), 2.66 (3H, s, Ar–CH3), 1.61 (6H, d, CH3); 13 C NMR (100 MHz, CDCl3) δ 165.0 (C, C=O), 157.7, 148.1, 147.2, 145.8, 139.5, 128.6, 125.1, 122.0, 120.6, 117.34, 114.8 (Ar–C), 53.8 (CH, CH(CH3)2), 41.8 (CH2, CH2N<), 22.9 (2 CH3, CH(CH3)2), 10.7 (CH3); Anal. calculated for C17H17FN6O; C, 59.99; H, 5.03; N, 24.69. Found: C, 60.11; H, 5.08; N, 24.60.
Cell proliferation assay study
Cell lines and culture conditions
Human breast cancer cells (MDA-MB 231), MCF-7, human primary glioblastoma cell line U-87 and human neuroblastoma IMR-32 cell line were procured from National Center for Cell Sciences, Pune, India. All cells were grown in RPMI-1640 supplemented with 10 % heat-inactivated fetal bovine serum (FBS), 100 IU/mL penicillin, 100 mg/mL streptomycin and 2 mM glutamine. Cultures were maintained in a humidified atmosphere with 5 % CO2 at 37 °C. The cells were subcultured twice each week, seeding at a density of about 2 × 103 cells/mL.
MTT assay
The cell viability was determined by (4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells (5 × 103 cells/well) were seeded to 96-well culture plate and cultured with or without compounds at 10 µM concentration for 72 h in a final volume of 200 µL. After treatment, the medium was removed and 20 µl of MTT (5 mg/ml in PBS) was added to the fresh medium. After 2-h incubation at 37 °C, 100 µl of DMSO was added to each well and plates were agitated for 1 min. Absorbance was read at 570 nm on a multi-well plate reader (VICTOR3, PerkinEmler). Percent inhibition of proliferation was calculated as a fraction of control (without compound). The experiment was performed in triplicate.
Docking studies
The crystal structure of PI3 kinase alpha with inhibitor (pdb id: 1E7V) having resolution of 2.4 Å was retrieved from the Protein Data Bank, and it was prepared using protein preparation wizard (Maestro, v9.2, Schrodinger, LLC, New York). Initially formal charges and bond orders were added for heterogroups, and later hydrogens were added to the system. After removing water molecules in all structures, the structure was energy-minimized. The compounds that were synthesized were drawn and prepared using Ligprep (LigPrep v2.2, Schrodinger LLC, New York) with Epik (Epik v1.6, Schrodinger, LLC, New York) to expand protonation and tautomeric states at 7.0 ± 2.0 pH units. The conformational sampling was performed on all database molecules using the ConfGen search algorithm. The ConfGen with force field (OPLS_2005) was employed, and duplicate poses within 1.0 Å rmsd were eliminated to remove repeated conformers. The glide energy grid was generated, and it was defined by a rectangular box surrounding the crystal ligand in X-ray structure. The prepared ligands were then docked using Glide 5.0 module (Glide v5.0 Schrodinger, LLC, New York). The binding site created earlier was chosen, and XP (extra precision) docking was performed, which gives precise protein–ligand binding affinities.
References
Adayev T, Wegiel J, Hwang YW (2011) Harmine is an ATP-competitive inhibitor for dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A). Arch Biochem Biophys 507:212–218
Addla D, Jallapally A, Gurram D, Yogeeswari P, Sriram D, Kantevari S (2014) Rational design, synthesis and antitubercular evaluation of novel 2-(trifluoromethyl) phenothiazine-[1,2,3]triazole hybrids. Bioorg Med Chem Lett 24:233–236
Agalave SG, Maujan SR, Pore VS (2011) Click chemistry: 1,2,3-triazoles as pharmacophores. Chem Asia J 6:2696–2718
Akselsen QW, Odlo K, Cheng J-J, Maccari G, Botta M, Hansen TV (2012) Synthesis, biological evaluation and molecular modeling of 1,2,3-triazole analogs of combretastatin A-1. Bioorg Med Chem 20:234–242
Bruel A, Logé C, De-Tauzia ML, Ravache M, Guevel RL, Guillouzo C, Lohier JF, Santos JSO, Lozach O, Meijer L, Ruchaud S, Bénédetti H, Robert JM (2012) Synthesis and biological evaluation of new 5-benzylated 4-oxo-3,4-dihydro-5H-pyridazino[4,5-b]indoles as PI3Kα inhibitors. Eur J Med Chem 57:225–233
Efron PA, Chen MK, Dai W, Langham MR Jr, Beierle EA (2003) Inhibition of the PI3K/AKT pathway enhances trail induced apoptosis in neuroblastoma cells. J Surg Res 114:308
Evanno Y, Dubois L, Sevrin M, Marguet F, Froissant J, Bartsch R, Gille C (1999) 4-oxo-3,5-dihydro-4H-pyridazino[4,5-b]indole-1-acetamide derivatives (CA 2298522 A1). Chem Abstr 130:168385
Font M, Monge A, Cuartero A, Elorriaga A, Martínez-Irujo JJ, Alberdi E, Santiago E, Prieto I, Lasarte JJ, Sarobe P, Borrás F (1995) Indoles and pyridazino[4,5-b]indoles as nonnucleoside analog inhibitors of HIV-1 reverse transcriptase. Eur J Med Chem 30:963–971
Güven A, Jones RA (1993) Potentially tautomeric 1,2,3,4-tetrahydro-1,4-dioxo-5H-pyridazino[4,5-b]indole. Tetrahedron 49:11145–11154
Howell LA, Howman A, O’Connell MA, Mueller A, Searcey M (2009) Synthesis and evaluation of 9-aminoacridines derived from benzyne click chemistry. Bioorg Med Chem Lett 19:5880–5883
Humphrey GR, Kuethe JT (2006) Practical methodologies for the synthesis of indoles. Chem Rev 106:2875–2911
Kushwaha K, Kaushik N, Lata JainSC (2014) Design and synthesis of novel 2H-chromen-2-one derivatives bearing 1,2,3-triazole moiety as lead antimicrobials. Bioorg Med Chem Lett 24:1795–1801
Lerch U, Kaiser J (1982) DE 3121137; (1983) Chem Abstr 98:126140
Monge A, Palop JA, Martinez MT, Fernandez-Alvarez E (1979) An Quim 77:889; (1980) Chem Abstr 93:2873
Monge A, Aldana I, Erro A, Parrado P, Font M, Alvarez T, Rocha E, Fernandez-Alvarez E (1985) An R Acad Farm 51:485; (1987) Chem Abstr 107:254
Monge A, Aldana I, Alvarez T, Font M, Santiago E, Latre JA, Bermejillo MJ, Lopez-Unzu MJ, Fernandez-Alvarez E (1991a) New 5H-pyridazino[4,5-b]indole derivatives. Synthesis and studies as inhibitors of blood platelet aggregation and inotropics. J Med Chem 34:3023
Monge A, Aldana I, Alvarez T, Losa MJ, Font M, Cenarruzabeitia E, Lasheras B, Frechilla D, Castiella E, Fernandez-Alvarez E (1991b) 1-Hydrazino-4-(3,5-dimethyl-1-pyrazolyl)-5H-pyridazino[4,5-b]indole. A new antihypertensive agent. Eur J Med Chem 26:655–658
Nantka-Namirski P, Ozdowska Z (1972) 2-Carbethoxyindole derivatives. I. Synthesis of 8-alkoxy- and 8,9-benzo-3H-pyridazino(4,5-b) indol-4-ones. Acta Pol Pharm 29:9–15
Ogawa Y, Nonaka Y, Goto T, Ohnishi E, Hiramatsu T, Kii I, Yoshida M, Ikura T, Onogi H, Shibuya H, Hosoya T, Ito N, Hagiwara M (2010) Development of a novel selective inhibitor of the down syndrome-related kinase Dyrk1A. Nat Commun 1:86
Pagliai F, Pirali T, Grosso ED, Brisco RD, Tron GC, Sorba G, Genazzani AA (2006) Rapid synthesis of triazole-modified resveratrol analogues via click chemistry. J Med Chem 49:467–470
Patpi SR, Pulipati L, Yogeeswari P, Sriram D, Jain N, Sridhar B, Murthy R, Kalivendi SV, Kantevari S (2012) Design, synthesis, and structure–activity correlations of novel dibenzo[b, d]furan, dibenzo[b, d]thiophene, and N-methylcarbazole clubbed 1,2,3-triazoles as potent inhibitors of mycobacterium tuberculosis. J Med Chem 55:3911–3922
Shafi S, Alam MM, Mulakayala N, Mulakayala C, Vanaja G, Kalle AM, Pallu R, Alam MS (2012) Synthesis of novel 2-mercapto benzothiazole and 1,2,3-triazole based bis-heterocycles: Their anti-inflammatory and anti-nociceptive activities. Eur J Med Chem 49:324–333
Thirumurugan P, Matosiuk D, Jozwiak K (2013) Click chemistry for drug development and diverse chemical–biology applications. Chem Rev 113:4905–4979
Ulloora S, Shabaraya R, Adhikari AV (2013) Facile synthesis of new imidazo[1,2-a]pyridines carrying 1,2,3-triazoles via click chemistry and their antiepileptic studies. Bioorg Med Chem Lett 23:3368–3372
Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL (2000) Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol Cell 6:909–919
Yempala T, Sridevi JP, Yogeeswari P, Sriram D, Kantevari S (2014) Rational design and synthesis of novel dibenzo[b, d]furan-1,2,3-triazole conjugates as potent inhibitors of Mycobacterium tuberculosis. Eur J Med Chem 71:160–167
Acknowledgments
N.P. thanks NITK, Surathkal, for the financial support. The authors thank Dr. Reddy’s Institute of Life Sciences, Hyderabad, for NMR facility and Anthem Biosciences, Bangalore, for mass spectral facility.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Panathur, N., Gokhale, N., Dalimba, U. et al. Synthesis of novel 5-[(1,2,3-triazol-4-yl)methyl]-1-methyl-3H-pyridazino[4,5-b]indol-4-one derivatives by click reaction and exploration of their anticancer activity. Med Chem Res 25, 135–148 (2016). https://doi.org/10.1007/s00044-015-1473-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-015-1473-y