Abstract
Coastal wetlands are valuable aquatic ecosystems with high biological productivity, which provide services such as a reduction in nitrogen loading into coastal waters and storage of organic carbon acting as carbon dioxide sinks. The predicted rise of sea level or freshwater extractions, particularly in the arid Mediterranean biome, will salinize many coastal wetlands. However, there is considerable uncertainty about how salinization will affect microbial communities and biogeochemical processes. We determined the abundance of total prokaryotes, cyanobacteria, and viruses and quantified the heterotrophic production of prokaryotes sensitive- (predominantly Bacteria) and resistant- (predominantly Archaea) to erythromycin in 112 ponds from nine coastal wetlands. We explored the main drivers of prokaryotic abundance and heterotrophic production using generalized linear models (GLMs). The best GLM, including all the wetlands, indicated that the concentration of total dissolved nitrogen (TDN) positively affected the total abundance of prokaryotes and the heterotrophic erythromycin-resistant (ery-R) production. In contrast, heterotrophic erythromycin-sensitive (ery-S) production was negatively related to TDN. This negative relationship appeared to be mediated by salinity and virus abundance. Heterotrophic ery-S production declined as salinity and virus abundance increased. Consequently, we observed a switch from heterotrophic ery-S production towards ery-R production as salinity and virus abundance increased. Our results imply that microbial activity will change from heterotrophic bacterial-dominated processes to archaeal-dominated processes with anthropogenic nitrogen and salinization increases. However, more studies are required to link the mineralization rates of dissolved nitrogen and organic carbon with specific archaeal taxa to enable more accurate predictions on future scenarios in coastal wetlands.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Wetlands have high biological productivity and functional diversity (Mitsch and Gosselink 2000; Mitsch et al. 2015). Among the ecosystem services they provide, wetlands reduce nutrient loading from freshwater inflow (Verhoeven et al. 2006) and have been described as “the kidneys of the landscape” (Mitsch et al. 2015). Coastal wetlands, in particular, are considered global reservoirs of organic carbon because of their elevated primary productivity and high sedimentation rates that, in the long-term, minimize carbon dioxide emissions to the atmosphere, acting as carbon sinks (Roehm 2005; Chmura et al. 2003; Geertz-Hansen et al. 2011). Therefore, microbial activities such as denitrification and organic carbon production and mineralization mediate these wetland functions and services. These aquatic ecosystems are currently facing two main threats: eutrophication and salinization.
The production of nitrogen fertilizer has enabled increased food production but has drastically disrupted the nitrogen cycle by doubled its inputs to the Earth´s surface leading to eutrophication of many aquatic ecosystems (Gruber and Galloway 2008; Canfield et al. 2010). This change likely exceeds all other human effects on nutrient cycles (Gruber and Galloway 2008; Schlesinger 2009) but has received relatively less attention than the carbon cycle (Battye et al. 2017). A substantial fraction of this nitrogen, mainly derived from agricultural lands, is transported to rivers and groundwater through runoff and is lost through emissions of ammonia and other nitrogen compounds by denitrification (Schlesinger 2009; Battye et al. 2017). In many estuaries, natural and constructed wetlands modify the nitrogen loading into coastal zones, alleviating eutrophication (Walton et al. 2015). These transitional waters include saline wetlands such as natural marshes and multi-pond solar salterns (Razinkovas-Baziukas and Povilanskas 2012).
Multi-pond solar salterns are present along the coasts of the Mediterranean and Portugal since the Phoenicians, who used evaporation procedures dating from the Emperor Huang era about 2500 years B.C. to obtain salt (Baas-Becking 1931; García-Vargas and Martínez-Maganto 2006). They are excellent model systems to study microbial ecology along salinity gradients from that of seawater to salt saturation (Ventosa et al. 2014). Coastal saline wetlands also have great importance for waterbird conservation and sustainable aquaculture (Britton and Johnson 1987; Sánchez et al. 2006; Athearn et al. 2009; Walton et al. 2015). Most studies of microbial ecology in coastal wetlands have focused on extremophile microorganisms and their bioenergetic and diversity constraints (Antón et al. 1999; Pedrós-Alió et al. 2000; Oren 2001; Casamayor et al. 2002; Gasol et al. 2004; Ghai et al. 2011; Ventosa et al. 2014); but much less is known about moderately halophilic bacteria and archaea in oligo- to eu-saline waters (Hahn, 2006; Herbert et al. 2015).
The discovery that archaea are not exclusively extremophiles (DeLong 1992; Fuhrman et al. 1992; Karner et al. 2001) led to studies showing that archaea are abundant, ubiquitous, and diverse, with essential roles in both carbon and nitrogen cycling (Herndl et al. 2005; Justice et al. 2012; Offre et al. 2013), including mineralizing organic matter under oxic conditions in the ocean (Herndl et al. 2005; Ingalls et al. 2006). The organo-heterotrophic nature of some archaeal groups has been shown for specific carbohydrates (Lazar et al. 2016) and dissolved proteins (Orsi et al. 2016). Archaea also oxidize ammonia, and perform denitrification under anoxic or suboxic conditions (Francis et al. 2007; Offre et al. 2013) with potential implications for nitrogen removal in wetlands (You et al. 2009). However, the prevalence of heterotrophic metabolism in archaea and its biogeochemical relevance are poorly studied. Archaeal heterotrophic production patterns along gradients of dissolved organic carbon and nitrogen, and salinity, remain completely unexplored in the literature.
Currently, most wetlands are salinizing due to climatic warming or freshwater extraction (Herbert et al. 2015; Morrissey et al. 2014; Jeppesen et al. 2015). Coastal wetlands are particularly affected since the sea-level rise introduces seawater high up into estuaries (Herbert et al. 2015; Schuerch et al. 2018). Salinization alters water and sediment chemistry and changes biogeochemical reactions (Herbert et al. 2015). Although there is still considerable uncertainty about how salinization affects wetland biogeochemistry, it promotes the release of inorganic nitrogen (NH4+) and phosphorus (PO43−) with implications for internal wetland eutrophication (Ardón et al. 2013; Weston et al. 2006). Besides, salinization increases the generation of toxic sulfides (Lamers et al. 1998), which play an essential role in wetland N cycling, inhibiting the final steps of denitrification (Brunet and García-Gil 1996; Laverman et al. 2007). Overall, salinization disrupts many ecosystem services provided by wetlands, such as their capacity to store organic carbon (Weston et al. 2011; Luo et al. 2019) and remove nitrogen from water (Herbert et al. 2015; Franklin et al. 2017).
The projected scenario of global change for wetlands includes wetland salinization and more nitrogen inputs. These environmental risks can interact. Nitrogen inputs can boost organic carbon mineralization and reduce carbon storage in coastal marshes (Deegan et al. 2012), while salinization can affect microbial nitrogen removal (Franklin et al. 2017). Our work describes patterns in microbial abundances (total prokaryotes, cyanobacteria, and viruses) and heterotrophic production of prokaryotes sensitive (predominantly Bacteria domain) and resistant (predominantly Archaea domain) to erythromycin in a broad set of semi-natural and constructed wetlands covering gradients of salinity, dissolved organic carbon, and nitrogen along the Western Mediterranean. Furthermore, we identified the main drivers of these patterns and discussed potential changes in prokaryotic heterotrophic production under future scenarios associated with global change.
Materials and methods
Study sites
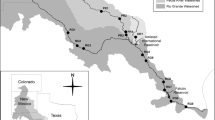
We sampled 112 saline ponds from nine coastal wetlands during the late spring-midsummer of 2011, 2012, and 2013 (Supplementary Table S1). The coastal wetlands from west to east were: (1) the Odiel marshes (OdielM) at the mouth of the Odiel river that included natural marshes and salterns, (2) the Veta la Palma (VPalma) fishponds at the mouth of the Guadalquivir river, (3) the Cabo de Gata (CGata) salterns, (4) El Hondo (Hondo) wetlands and (5) Santa Pola (SPola) salterns at the mouth of the Vinalopó river, and (6) the Ebro Delta (EbroD) marshes and salterns at the mouth of the Ebro river in Spain, (7) the Salin-de-Giraud and Saintes-Maries-de-la-Mer marshes and salterns at the mouth of the Rhône river (Camargue) in France, (8) the Molentargius, Santa Guilla, and Santa Caterina marshes and salterns (Sardinia) in Italy, and (9) Thyna solar salterns (Sfax) in Tunisia (Fig. 1a). All the study ponds are located in semiarid or arid areas, under typical Mediterranean climatic conditions, covering large gradients of salinity, from hypersaline (crystallizers in solar saltpans) to oligo-mesohaline (brackish and evaporation ponds) waters (Fig. 1b), and microbial heterogeneity (Fig. 1c). Multi-pond solar salterns are excellent model systems to study microbial ecology along salinity gradients from that of seawater to salt saturation (Ventosa et al. 2014). They are natural laboratories across which microbial communities can be compared. The brackish wetlands that we studied are also crucial for sustainable aquaculture and waterfowl conservation (Britton and Johnson 1987; Walton et al. 2015). Coordinate location, sampling dates, and physical–chemical and microbiological data for each pond are in Supplementary Table S1.
Map of the coastal wetlands included in the study obtained with ArcGIS a from West to East: Odiel marshes (OdielM), Veta la Palma, (VPalma), Cabo de Gata (CGata), El Hondo (Hondo), Santa Pola (SPola), Ebro Delta (EbroD), Saintes-Maries-de-la-Mer and Salin-de-Giraud (Camargue), Molentargius, Santa Guilla and Santa Caterine (Sardinia), and Sfax (Sfax). Imagery map credit: Esri, DigitalGlobe, Earthstar Geographics, CNES/Airbus DS, GeoEye, USDA FSA, USGS, Aerogrid, IGN, IGP, and the GIS User Community. b Orthophoto of the Odiel marshes including evaporation and crystallization ponds; Orthophoto credit: Junta de Andalucía. Consejería de Agricultura, Ganadería, Pesca y Desarrollo Sostenible (ámbito de Desarrollo Sostenible). http://www.juntadeandalucia.es/medioambiente/site/rediam/. c Picture of GF/F glass-fiber filters to illustrate the pigment diversity. Two top filters correspond to crystallizers and the bottom filter corresponds to evaporation ponds from water samples collected at the Odiel marshes
Chemical analyses
We recorded salinity with a multi-parameter probe (HANNA HI 9828). Water samples with salinity higher than 70 ppt were diluted with Milli-Q water until they were in the operating range of the probe. We measured total nutrient concentrations in unfiltered samples, while we filtered the samples through pre-combusted 0.7-μm pore-size Whatman GF/F glass-fiber filters for dissolved nutrient analysis. We measured total phosphorus (TP) and total dissolved phosphorus (TDP) using the molybdenum blue method (Murphy and Riley, 1962) after persulfate digestion (30 min, 120 °C). We analyzed total nitrogen (TN) and total dissolved nitrogen (TDN) by high–temperature catalytic oxidation (Álvarez-Salgado and Miller 1998) using a total nitrogen analyzer (Shimadzu TNM-1) and potassium-nitrate standards. We analyzed dissolved organic carbon (DOC) concentration as non-purgeable organic carbon by high–temperature catalytic oxidation using a total organic carbon analyzer (Shimadzu TOC-V CSH). DOC samples were pre-filtered through pre-combusted Whatman GF/F filters (2 h at 500 °C) and acidified with H3PO4 (final pH < 2). We calibrated the instrument using a four-point standard curve of potassium hydrogen phthalate. We purged the samples with phosphoric acid for 20 min, and we set up three to five injections for each sample.
Biological analyses
We determined chlorophyll-a (Chl-a) concentration by filtering 50–2000 ml of water through Whatman GF/F filters and extracting the pigments using 95% methanol in the dark for 24 h at 4 °C (APHA 1992). We measured the absorbance at 665 nm and 750 nm using a Perkin Elmer Lambda 40 spectrophotometer.
Samples for determining the abundances of total prokaryotes (PA) and cyanobacteria (CyA) by flow cytometry (Gasol and del Giorgio, 2000) were collected in cryovials, fixed with 1% paraformaldehyde and 0.05% glutaraldehyde in the dark (30 min at 4 °C), and frozen in liquid nitrogen and stored at − 80 °C until analysis. Once defrosted, the samples were diluted (≥ tenfold) with Milli-Q water to avoid the electronic coincidence of the prokaryotic cells. We counted the PA and CyA in a FACScalibur flow cytometer with a laser emitting at 448 nm and a suspension of yellow-green 1-μm latex beads (Polysciences) per sample as an internal standard. We stained the PA samples with a 10 μM DMSO solution of Sybr Green I stain (Molecular Probes) in the dark (10 min), run at low speed for 2 min, and detected in bivariate plots of side scatter (SSC) vs. FL1 (Green fluorescence). The CyA samples were run at high speed for 4 min and detected in bivariate plots of side scatter (SSC) vs. FL3 (Red fluorescence).
Samples for virus abundance (VA) were collected in cryovials, fixed with glutaraldehyde at a final concentration of 0.5% (15 min at 4 °C) in the dark, frozen in liquid nitrogen, and stored at − 80 °C until analysis by flow cytometry (Brussard et al., 2010). Before analysis, we diluted the samples ≥ 100-fold with TE-buffer pH 8.0 (10 mM Trishydroxymethyl-aminomethane, 1 mM ethylenediaminetetraacetic acid) to avoid the electronic coincidence in virus particle counts. We stained the VA samples with a working solution (1:200) of SYBR Green I (10,000 × concentrate in DMSO, Molecular Probes) for 10 min in the dark and then kept them at − 80° until counting. We added fluorescent microspheres (FluoSpheres carboxylate modified yellow-green fluorescent microspheres; 1.0 μm diameter) as an internal standard. We acquired the data in log mode and detected their signature in bivariate plots of side scatter (SSC) vs. FL1 (Green fluorescence). We processed the flow cytometry data using BD CellQuest Pro software.
We measured the heterotrophic prokaryotic production using the rate of 3H-Leucine incorporation into proteins (Smith and Azam, 1992). [3H]-Leucine incorporation by microbial eukaryotes appears irrelevant in short-term incubations even with high leucine concentrations (Buesing and Gessner 2003; Miranda et al. 2007). We assumed that, using erythromycin, we inhibit most bacterial production and determine the production mostly attributable to Archaea following the procedure proposed by Yokokawa et al. (2012). Although some authors have questioned the specificity of erythromycin to inhibit some bacteria phylum in the open ocean (Frank et al. 2016), this procedure appears to have high efficiencies (ca. 80%) in waters with salinities higher than the seawater (Oren et al. 1990; Pedrós-Alió et al. 2000) and for specific groups of bacteria (Du et al. 2016). For each pond, two sets of five samples (1.5 ml) (three replicate and two trichloroacetic-acid (TCA, 50%)-killed (final concentration10%) samples) were incubated at in situ temperature with 54.6 nM or 58.4 nM leucine (1:2 hot: cold v/v) for 2–5 h. In one of the sets of 5 samples, we also added 10 μg ml−1 of erythromycin (final concentration) to inhibit most of the bacterial production (hereafter, heterotrophic erythromycin-sensitive (ery-S) production). To obtain the production attributed mainly to archaeal and the few bacterial groups that were erythromycin-resistant (hereafter, heterotrophic erythromycin-resistant (ery-R) production), we subtracted from the total production (first set of samples) the ery-S production. We added TCA at a final concentration of 10% to end the incubations. In the laboratory, the samples were centrifuged (15,366g for 10 min), rinsed with TCA (5%), vortexed, and centrifuged again. Finally, we added 1.5 ml of liquid scintillation cocktail (Ecoscint A) to each sample and determined the incorporated radioactivity using an auto-calibrated scintillation counter (Beckman LS 6000 TA).
Statistical analyses
To determine the main drivers of the observed microbial patterns, we carried out four sets of generalized linear models (GLMs). In the first set of GLMs, the dependent microbial variables were prokaryotic abundance (PA), cyanobacterial abundance (CyA), heterotrophic erythromycin-sensitive (ery-S) production, and heterotrophic erythromycin-resistant (ery-R) production. The predictor variables selected were: site (i.e., each wetland complex as a categorical variable), and as continuous variables, salinity, total dissolved nitrogen (TDN), and total dissolved phosphorus (TDP). When necessary, log and square-root transformations were applied to the dependent and predictor variables to improve the fit to a normal distribution and avoid heteroscedasticity. In the second set of GLMs, we also included the concentration of DOC as a predictor variable but did not include the Camargue site since this variable was not measured there. In the third set of GLMs, we determined the best predictors of virus abundance (dependent variable) in the six sites (Odiel M, VPalma, CGata, SPola, Hondo, and EbroD) where this variable was determined. Nutrients were not included as predictors because viruses are mostly dependent on the host (bacterial or archaeal) density. We considered the study site a categorical variable and salinity, PA, and CyA as continuous variables. To determine the potential effect of viruses on the other microbial components, we performed the fourth set of GLMs in which VA is considered a predictor variable and ery-S production and ery-R production as dependent variables. We selected the model with the smallest value of the Akaike Information Criterion (AIC) as the best model for each dependent variable considered. More details of alternative models with ∆AIC < 2.0 are in the Supplementary Tables S2-S6. We performed all analyses using Statistica 7.0 software.
Results
The study ponds represent a wide range of environmental conditions. Salinity ranged from 0.22 to 343 ppt (Table 1), spanning from oligo- to hyper-saline conditions; the highest median value was in EbroD ponds, and the lowest median value in El Hondo ponds (Fig. 2a). DOC concentrations ranged from 0.24 to 5.76 mmol-C l−1 (Table1), with the highest median concentration in EbroD ponds (as for salinity) and the lowest in CGata ponds (Fig. 2c). TDN concentrations ranged from 0.02 to 0.62 mmol-N l−1 (Table 1), with EbroD ponds also showing the highest median TDN concentration and SPola saltpans the lowest values (Fig. 2b). TDP concentrations ranged from 0.50 to 40.09 μmol-P l−1 (Table 1), with the highest median value at OdielM wetlands and the lowest median at CGata saltpans (Fig. 2d).
Violin plots to visualize the data distribution of the salinity (a), total dissolved nitrogen (b), dissolved organic carbon (c), and total dissolved phosphorus (d) for each wetland site studied. White dots are the data, black lines are the median values, and the grey shadows are the data distributions for each wetland. Note panels (a) and (d) have log scales
Biological variables also ranged widely. Prokaryotic abundance (PA) ranged almost three orders of magnitude, from 0.35 × 106 to 252 × 106 cells ml−1 (Supplementary Table S1), with the highest median value in Tyna saltpans and the lowest in CGata saltpans (Fig. 3a). The variability in abundance of cyanobacteria was even higher, ranging from 0.01 to 38,105 (× 103) cells ml−1 (Supplementary Table S1; Fig. 3b). Virus abundance ranged from 0.02 to 3.07 (× 109) particles ml−1in the studied wetlands (Fig. 3c). Heterotrophic ery-S production ranged from 19 to 3007 pmol of leucine l−1 h−1 (Supplementary Table S1; Fig. 3d) with the highest median value at SPola saltpans and the lowest median at Thyna saltpans. Heterotrophic ery-R production ranged from 56 to 2290 pmol of leucine l−1 h−1, with the highest median value at EbroD ponds and the lowest at Camargue ponds (Supplementary Table S1; Fig. 3e). Chlorophyll-a concentrations ranged more than 10,000-fold from 0.04 to 617.41 μg l−1 (Table1), with the highest median value at Thyna saltpans and the lowest in the EbroD ponds (Fig. 3f).
Violin plots to visualize the data distribution of the abundance of prokaryotes (a), cyanobacteria (b), viruses (c), heterotrophic erythromycin-sensitive (predominantly bacteria) production (d), heterotrophic erythromycin-resistant (predominantly archaea) production (e) and chlorophyll concentration (f) for each wetland site studied. White dots are the data, black lines are the median values, and the grey shadows are the data distributions for each wetland. Note panels (a–c), and (f) have log scales
In the first set of GLMs, the best model for prokaryotic abundance included the TDN concentration and the wetland site (categorical) as predictors (Table 2). The residuals of prokaryotic abundance (once the site was controlled for) were significantly and positively related to TDN (Fig. 4a). Alternative (less explanatory) models also included salinity or TDP (Table supplementary S2). The best GLM for the cyanobacterial abundance included the site as a categorical variable and TDN and TDP as continuous variables, with a significant positive partial relationship with TDN and a negative partial relationship with TDP (Table 2). In the case of heterotrophic ery-S production, the best GLM included TDN and the wetland site as predictors (Table 2). The residuals of heterotrophic ery-S production (once the site was controlled for) were significantly and negatively related to TDN (Fig. 4b). Indeed, the higher the TDN concentration, the lower the heterotrophic ery-S production, irrespectively of the site. Alternative (less explanatory) models also included salinity or TDP (Supplementary Table S2). For heterotrophic ery-R production, the best GLM also included TDN and site as predictors (Table 2). In contrast to heterotrophic ery-S production, the residuals of heterotrophic ery-R production (once the site was controlled for) were significantly and positively related to TDN concentration (Fig. 4c). Indeed, the higher the TDN, the higher the production associated to archaea and bacterial groups resistant to erythromycin, irrespective of site. Alternative (less explanatory) models also included salinity or TDP (Supplementary Table S2).
Patterns of prokaryotic abundance and production and the concentration of total dissolved nitrogen. Partial effects for relationships between the residuals of prokaryotic abundance (a), the residuals of heterotrophic erythromycin-sensitive production (b) and the residuals of heterotrophic erythromycin-resistant production (c) and the concentration of total dissolved nitrogen, based on the best models shown in Table 2. In each case, the Y variable represents the residuals taken from the selected model after fitting the other predictor variables
In the second set of GLMs, we also included the concentration of DOC as a predictor variable, but excluded the Camargue site since these data were not available for that wetland. This predictor variable only affected the results previously exposed to the prokaryotic abundance and the heterotrophic ery-S production (Table supplementary S3). In this second analysis, prokaryotic abundance was affected negatively by salinity and positively by DOC concentration, while depending on the site; DOC concentration negatively affected heterotrophic ery-S production.
In the third set of GLMs, we determined the best GLM for the virus abundance, excluding nutrients as predictors (Table 3). This model included salinity as a continuous predictor and site as a categorical predictor. We observed a positive relationship between the residuals of virus abundance (once the site was controlled for) and salinity (Fig. 5). Alternative (less explanatory) models also included prokaryotic and cyanobacterial abundances (Table supplementary S4).
Partial effect for relationship between the residuals of virus abundance and salinity based on the best models shown in Table 3
Finally, in the fourth set of GLMs, we included viral abundance as a predictor of heterotrophic ery-S and ery-R production (Table supplementary S5), but only considered the sites where these data were available: Odiel M, VPalma, CGata, SPola, Hondo, and EbroD. The best GLM for heterotrophic ery-S production included salinity and virus abundance as predictors (Table 4). Once the site effect was controlled, we observed a significant negative relationship between heterotrophic ery-S production and salinity (Fig. 6a), as well as viral abundance (Fig. 6b). On the other hand, the best GLM for heterotrophic ery-R production was coherent with the first set of GLMs, including all the saline wetlands; the best GLM included TDN and site as predictors (Table supplementary S5). This result confirms the robust relationship between TDN and heterotrophic ery-R production in the study wetlands.
Patterns between heterotrophic erythromycin-sensitive production and their chemical and biological predictors. Partial effects for relationships between salinity (a) and virus abundance (b) and the residuals of heterotrophic erythromycin-sensitive production, based on the best models shown in Table 4. In each case the Y variable represents the residuals taken from the selected model after fitting the other predictor variables
Discussion
The best-generalized linear model (GLM), including all the coastal wetlands, indicated that the concentration of TDN positively affected the abundance of prokaryotes and heterotrophic ery-R production. TDN was consistently associated with higher ery-R heterotrophic production, even considering all the alternative GLMs with other predictor variables, such as DOC concentration and viral abundance (VA). In contrast, TDN negatively affected heterotrophic ery-S production, but this result changed when DOC and VA were also considered as predictor variables in alternative GLMs. The negative relationship between TDN and heterotrophic ery-S production appeared to be mediated by salinity and VA. Heterotrophic ery-S production declined as salinity and viruses increased; therefore, we observed a switch from heterotrophic ery-S production (predominantly bacterial production) to heterotrophic ery-R production (predominantly achaeal production) as salinity and VA increased. Archaea could be the main microorganisms processing dissolved nitrogen in the most saline and eutrophic wetlands.
In this across-wetlands study, salinity did not directly affect heterotrophic ery-R production (predominantly achaeal production), whereas total dissolved nitrogen was consistently the best predictor, regardless of the variables considered, in all the alternative generalized linear models. Historically, archaea were associated with extreme environments, such as salterns, where salt concentrations are close to saturation (Oren 1994, 2011; Antón et al. 1999). Now, we know that archaea occur in a broader variety of environments and perform several functions in N cycling, including nitrification and denitrification (Francis et al. 2007, Offre et al. 2013). On the other hand, our results emphasize that heterotrophic ery-R production, measured as the uptake of 3H-leucine, was coupled to total dissolved nitrogen concentration (Fig. 4c). Denitrification is primarily a heterotrophic, microaerophilic or anoxic process widespread along salinity gradients, which can be carried out by archaea (Zumft 1997). Coastal wetlands provide optimal conditions for archaeal denitrification due to their low oxygen concentrations as a result of high salinities, and high concentrations of organic matter and nitrate (Seitzinger 1988; Seitzinger et al. 2007). Furthermore, Orsi et al. (2016) demonstrated the organo-heterotrophic nature of some archaeal groups, which take up dissolved proteins. Therefore, heterotrophic archaeal metabolism via denitrification and protein assimilation affects the nitrogen cycle, ameliorating coastal eutrophication. However, a more detailed analysis of N functional genes, beyond leucine incorporation, is needed to assess the actual contribution of archaea to nitrogen cycling in coastal wetlands.
Early works emphasized the importance of organic carbon derived from phytoplankton as a critical driver of heterotrophic prokaryotic (bacterial + archaeal) production in diverse aquatic ecosystems (e.g., Cole et al. 1988; Baines and Pace 1991; Ortega-Retuerta et al. 2008). However, in this study, we did not find a difference between the best GLMs for heterotrophic ery-R production with and without the concentration of dissolved organic carbon (DOC) (Table S3). Indeed, the total pool of DOC concentration was not a predictive variable, at least for heterotrophic ery-R production. In saline wetlands, extreme evapoconcentration (Batanero et al. 2017) and high loads of organic matter from freshwater inflows (Walton et al. 2015) result in high concentrations of DOC in oligo- to eusaline wetlands, likely ensuring plenty of DOC for heterotrophic metabolism. Similarly, TDP was not included as a predictor variable in the best GLMs for the different microbial variables, except for the abundance of cyanobacteria. Bacterioplankton growth is assumed to be limited by the availability of phosphorus in many aquatic ecosystems (Cotner et al. 1997; Smith and Prairie 2004; Ortega-Retuerta et al. 2007). However, more recently, Cotner et al. (2010) demonstrated that bacteria exhibit high flexibility in their P content and stoichiometry, questioning the ranges of P-limitation. Our results suggest that neither phosphorus nor dissolved organic carbon appears to significantly affect the prokaryotic production in the studied saline wetlands.
In the study wetlands, heterotrophic ery-S production appeared to be controlled by viral abundance (VA) and salinity (Fig. 6), providing an indirect explanation for the negative correlation between total dissolved nitrogen and heterotrophic ery-S production (Fig. 4b). The results showed that, within a given wetland complex, VA increased significantly with salinity (Table 3, Fig. 5) and negatively affected the heterotrophic ery-S production (Table 4, Fig. 6b) but did not have a significant effect on heterotrophic heterotrophic ery-R production (Table supplementary S5). These findings suggest that most of the viruses were likely bacteriophages that caused significant bacterial mortality in these extremely saline wetlands. Thus, external factors such as evaporation would trigger an increase in VA, which would increase bacterial mortality, while archaeal communities would be more resilient to such changes. Guixa-Boixereu et al. (1996) observed the highest prokaryotic losses by viral lysis at the highest salinities in solar salterns. Another complementary explanation is that as the salinity increases, there is a coupled decline in oxygen solubility, promoting suboxic conditions. High salinity and low oxygen may be more energetically favourable conditions for some halophilic archaeal groups (Oren, 2001).
The projected scenario of climatic warming includes wetland salinization due to sea-level rise and decreases of freshwater discharges from rivers, which will allow marine waters further into the estuaries, salinizing the surrounding wetlands (Herbert et al. 2015; Schuerch et al. 2018). On the other hand, human population growth and crop production lead to more nitrogen inputs (Mulholland et al. 2008; Mekonnen and Hoekstra, 2015). These two environmental risks can interact, affecting the potential services of coastal wetlands. Eutrophication can boost the mineralization of organic carbon, reducing the carbon storage capacity of coastal marshes (Deegan et al. 2012), and salinization can affect the bacterial capacity to remove nitrogen (Franklin et al. 2017). Our results imply that microbial activity will change from bacterial-dominated processes to archaeal-dominated processes along with the increases of nitrogen and salinity. More studies linking the processing rates of dissolved nitrogen and organic carbon with specific archaeal and bacterial taxa will allow more accurate predictions in under future scenarios of global change.
Conclusions
The concentration of total dissolved nitrogen appeared to determine the abundance of heterotrophic prokaryotes and, mainly, the heterotrophic ery-R (predominantly archaea) production in the suite of coastal wetlands studied. In contrast, total dissolved nitrogen negatively affected the heterotrophic ery-S (predominantly bacterial) production. This negative relationship between total dissolved nitrogen and heterotrophic ery-S production appeared to be mediated by salinity and virus abundance. Heterotrophic ery-S production declined as salinity and viruses increased; therefore, there was a switch from heterotrophic ery-S production in wetlands with lower nitrogen content and salinity to heterotrophic ery-R production in wetlands with higher nitrogen and salinity. Archaeal taxa could be the main prokaryotes processing nitrogen in the most saline wetlands. Therefore, changes in nitrogen inputs and salinities associated with global change can alter prokaryotic functional groups and, consequently, the ecosystem services provided by coastal wetlands.
Data availability
Additional figures and tables of data can be found in the supplementary information.
References
Álvarez-Salgado XA, Miller AEJ (1998) Simultaneous determination of dissolved organic carbon and total dissolved nitrogen in seawater by high temperature catalytic oxidation: conditions for precise shipboard measurements. Mar Chem 62:325–333. https://doi.org/10.1016/S0304-4203(98)00037-1
American Public Health Association (1992) Standard methods for the examination of water and wastewater. APHA, 18th edn
Antón J, Llobet-Brossa E, Rodriguez-Valera F, Amann R (1999) Fluorescence in situ hybridization analysis of the prokaryotic community inhabiting crystallizer ponds. Environ Microbiol 1:517–523. https://doi.org/10.1046/j.1462-2920.1999.00065.x
Ardón M, Morse JL, Colman BP, Bernhardt ES (2013) Drought-induced saltwater incursion leads to increased wetland nitrogen export. Global Change Biol 19(10):2976–2985. https://doi.org/10.1111/gcb.12287
Athearn ND, Takekawa JY, Shinn JM (2009) Avian response to early tidal salt marsh restoration at former commercial salt evaporation ponds in San Francisco Bay, California, USA. Nat Resour Environ 15(1):14
Baines SB, Pace ML (1991) The production of dissolved organic matter by phytoplankton and its importance to bacteria: patterns across marine and freshwater systems. Limnol Oceanogr 36(6):1078–1090. https://doi.org/10.4319/lo.1991.36.6.1078
Bass-Becking LGM (1931) Historical notes on salt and salt-manufacture. Sci Mon 32(5):434–446
Batanero GL, León-Palmero E, Li L, Green AJ, Rendón-Martos M, Suttle CA, Reche I (2017) Flamingos and drought as drivers of nutrients and microbial dynamics in a saline lake. Sci Rep 7(1):12173. https://doi.org/10.1038/s41598-017-12462-9
Battye W, Aneja VP, Schlesinger WH (2017) Is nitrogen the next carbon? Earth’s Future 5:894–904. https://doi.org/10.1002/2017EF000592
Britton RH, Johnson AR (1987) An ecological account of a Mediterranean salina: the Salin de Giraud, Camargue (S. France). Biol Conserv 42:185–230. https://doi.org/10.1016/0006-3207(87)90133-9
Brunet RC, Garcia-Gil LJ (1996) Sulfide-induced dissimilatory nitrate reduction to ammonia in anaerobic freshwater sediments. FEMS Microbiol Ecol 21(2):131–138. https://doi.org/10.1111/j.1574-6941.1996.tb00340.x
Brussaard CPD, Payet JP, Winter C, Weinbauer MG (2010) Quantification of aquatic viruses by flow cytometry. Man Aquatic Viral Ecol 11:102–109. https://doi.org/10.4319/mave.2010.978-0-9845591-0-7.102
Buesing N, Gessner M (2003) Incorporation of radiolabeled leucine into protein to estimate bacterial production in plant litter, sediment, epiphytic biofilms, and water samples. Microb Ecol 45:291–301. https://doi.org/10.1007/s00248-002-2036-6
Canfield DE, Glazer AN, Falkowski PG (2010) The evolution and future of earth’s nitrogen cycle. Science 330:192–196. https://doi.org/10.1126/science.1186120
Casamayor EO, Massana R, Benlloch S, Ovreas L, Diez B, Goddard VJ, Gasol JM, Joint I, Rodriguez-Valera F, Pedros-Alio C (2002) Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ Microbiol 4:338–348. https://doi.org/10.1046/j.1462-2920.2002.00297.x
Chmura G, Anisfeld S, Donald C, Lynch JC (2003) Global carbon sequestration in tidal, saline wetland soils. Global Biogeochem Cycles 17(4):1111. https://doi.org/10.1029/2002GB001917
Cole JJ, Findlay S, Pace ML (1988) Bacterial production in fresh and saltwater ecosystems: a cross system overview. Mar Ecol Prog Ser 43:1–10
Cotner JB, Ammerman JW, Peele ER, Bentzen E (1997) Phosphorus-limited bacterioplankton growth in the Sargasso Sea. Aquatic Microb Ecol 13:141–149. https://doi.org/10.3354/ame013141
Cotner JB, Hall EK, Scott JT, Heldal M (2010) Freshwater bacteria are stoichiometrically flexible with a nutrient composition similar to seston. Front Microbiol 1:132. https://doi.org/10.3389/fmicb.2010.00132
Deegan L, Johnson D, Warren R et al (2012) Coastal eutrophication as a driver of salt marsh loss. Nature 490:388–392. https://doi.org/10.1038/nature11533
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci USA 89:5685–5689. https://doi.org/10.1073/pnas.89.12.5685
Du J, Qi W, Niu Q, Hu Y, Zhang Y, Yang M, Li YY (2016) Inhibition and acclimation of nitrifiers exposed to erythromycin. Ecol Eng 94:337–343. https://doi.org/10.1016/j.ecoleng.2016.06.006
Francis CA, Beman JM, Kuypers MM (2007) New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J 1:19–27. https://doi.org/10.1038/ismej.2007.8
Frank AH, Pontiller B, Herndl GJ, Reinthaler T (2016) Erythromycin and GC7 fail as domain specific inhibitors for bacterial and archaeal activity in the open ocean. Aquat Microb Ecol 77:99–110. https://doi.org/10.3354/ame01792
Franklin RB, Morrissey EM, Morina JC (2017) Changes in abundance and community structure of nitrate-reducing bacteria along a salinity gradient in tidal wetlands. Pedobiologia 60:21–26. https://doi.org/10.1016/j.pedobi.2016.12.002
Fuhrman JA, McCallum K, Davis AA (1992) Novel major archae-bacterial group from marine Plankton. Nature 356:148–149. https://doi.org/10.1038/356148a0
García Vargas, E. y Martínez Maganto, J (2006) “La sal de la Bética: Algunas notas sobre su producción y comercio”. Habis 37: 253–274. https://doi.org/10.12795/Habis.2006.i37.19
Gasol J, Casamayor E, Join I, Garde K, Gustavson K, Benlloch S, Diez B, Schauer M, Massana R, Pedrós-Alio C (2004) Control of heterotrophic prokaryotic abundance and growth rate in hypersaline planktonic environments. Aquat Microb Ecol 34:193–206. https://doi.org/10.3354/ame034193
Gasol J, del Giorgio P (2000) Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci Mar 64:197–224. https://doi.org/10.3989/scimar.2000.64n2197
Geertz-Hansen O, Montes C, Duarte C, Sand-Jesen K, Marba N, Grillas P (2011) Ecosystem metabolism in a temporary Mediterranean marsh (Doñana National Park, SW Spain). Biogeosciences. https://doi.org/10.5194/bg-8-963-2011
Ghai R, Pašić L, Fernández AB, Martin-Cuadrado AB, Megumi Mizuno C, McMahon KD, Papke RT, Stepanauskas R, Rodriguez-Brito B, Rohwer F, Sánchez-Porro C, Ventosa A, Rodríguez-Valera F (2011) New abundant microbial groups in aquatic hypersaline environments. Sci Rep 1:135. https://doi.org/10.1038/srep00135
Gruber N, Galloway J (2008) An Earth-system perspective of the global nitrogen cycle. Nature 451:293–296. https://doi.org/10.1038/nature06592
Guixa-Boixereu N, Calderon-Paz JI, Heldal M, Bratbak G, Pedrós-Alió C (1996) Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat Microb Ecol 11:215–227. https://doi.org/10.3354/ame011215
Hahn MW (2006) The microbial diversity of inland waters. Curr Opin Biotechnol 17:256–261. https://doi.org/10.1016/j.copbio.2006.05.006
Herbert ER, Boon P, Burgin AJ, Neubauer SC, Franklin RB, Ardon M, Hopfensperger KN, Lamers LPM, Gell P (2015) A global perspective on wetland salinization: Ecological consequences of a growing threat to freshwater wetlands. Ecosphere 6(10):206. https://doi.org/10.1890/ES14-00534
Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A, Pernthaler J (2005) Contribution of archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol 71:2303–2309. https://doi.org/10.1128/AEM.71.5.2303-2309
Ingalls AE, Shah SR, Hansman RL, Aluwihare LI, Santos GM, Druffel ERM, Perason A (2006) Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA 103:6442–6447. https://doi.org/10.1073/pnas.0510157103
Jeppesen E, Brucet S, Naselli-Flores L, Papastergiadou E, Stefanidis K, Noges T, Noges P, Attayde JL, Zohary T, Coppens J, Bucak T, Menezes RF, Freitas FRS, Kernan M, Sondergaard M, Beklioglu M (2015) Ecological impacts of global warming and water abstraction on lakes and reservoirs due to changes in water level and related changes in salinity. Hydrobiologia 750:201–227. https://doi.org/10.1007/s10750-014-2169-x
Justice NB, Pan C, Mueller R, Spaulding SE, Shah V, Sun CL, Yelton AP, Miller CS, Thomas BC, Shah M, VerBerkmoes N, Hettich R, Banfield JF (2012) Heterotrophic archaea contribute to carbon cycling in low-pH, suboxic biofilm communities. Appl Environ Microbiol 78:8321–8330. https://doi.org/10.1128/AEM.01938-12
Karner M, DeLong EF, Karl DM (2001) Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507–509. https://doi.org/10.1038/35054051
Lamers LPM, Tomassen HBM, Roelofs JGM (1998) Sulfate-induced entrophication and phytotoxicity in freshwater wetlands. Environ Sci Technol 32:199–205. https://doi.org/10.1021/es970362f
Laverman AM, Canavan RW, Slomp CP, Van Cappellen P (2007) Potential nitrate removal in a coastal freshwater sediment (Haringvliet Lake, The Netherlands) and response to salinization. Water Res 41(14):3061–3068. https://doi.org/10.1016/j.watres.2007.04.002
Lazar CS, Baker BJ, Seitz K, Hyde AS, Dick GJ, Hinrichs KU, Teske AP (2016) Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ Microbiol 18:1200–1211. https://doi.org/10.1111/1462-2920.13142
Luo M, Huang JF, Zhu WF, Tong C (2019) Impacts of increasing salinity and inundation on rates and pathways of organic carbon mineralization in tidal wetlands: a review. Hydrobiologia 827(1):31–49. https://doi.org/10.1007/s10750-017-3416-8
Mekonnen MM, Hoekstra AY (2015) Global gray water footprint and water pollution levels related to anthropogenic nitrogen loads to fresh water. Environ Sci Technol 49:12860–12868. https://doi.org/10.1021/acs.est.5b03191
Miranda MR, Guimarães JRD, Coelho-Souza AS (2007) [3H] Leucine incorporation method as a tool to measure secondary production by periphytic bacteria associated to the roots of floating aquatic macrophyte. J Microbiol Methods 71(1):23–31. https://doi.org/10.1016/j.mimet.2007.06.020
Mitsch W, Bernal B, Hernández M (2015) Ecosystem services of wetlands. Int J Biodivers Sci Ecosyst Serv Manage 11(1):1–4. https://doi.org/10.1080/21513732.2015.1006250
Mitsch W, Gosselink J (2000) The value of wetlands: importance of scale and landscape setting. Ecol Econ 35:25–33. https://doi.org/10.1016/S0921-8009(00)00165-8
Morrissey EM, Gillespie JL, Morina JC, Franklin RB (2014) Salinity affects microbial activity and soil organic matter content in tidal wetlands. Glob Change Biol 20(4):1351–1362
Mulholland PJ, Helton AM, Poole GC, Hall RO, Hamilton SK, Peterson BJ, Dodds WK (2008) Stream denitrification across biomes and its response to anthropogenic nitrate loading. Nature 452(7184):202–205. https://doi.org/10.1038/nature06686
Murphy J, Riley JP (1962) A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta 27:31–36. https://doi.org/10.1016/S0003-2670(00)88444-5
Offre A, Spang SC (2013) Archaea in biogeochemical cycles. Annu Rev Microbiol 67:437–457. https://doi.org/10.1146/annurev-micro-092412-155614
Oren A (1990) The use of protein synthesis inhibitors in the estimation of the contribution of halophilic archaebacteria to bacterial activity in hypersaline environments. FEMS Microbiol Ecol 6:187–192. https://doi.org/10.1016/0378-1097(90)90729-A
Oren A (1994) The ecology of the extremely halophilic archaea. FEMS Microbiol Rev 13:415–440. https://doi.org/10.1016/0168-6445(94)90063-9
Oren A (2001) The bioenergetic basis for the decrease in metabolic diversity at increasing salt concentrations: implications for the functioning of salt lake ecosystems. Hydrobiologia 466:61–72. https://doi.org/10.1023/A:101455711
Oren A (2011) Ecology of halophiles. In: Horikoshi K (ed) Extremophile handbook. Springer, Tokyo, pp 343–361. https://doi.org/10.1007/978-4-431-53898-1_16
Orsi RH, Wiedmann M (2016) Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl Microbiol Biotechnol 100:5273–5287. https://doi.org/10.1007/s00253-016-7552-2
Ortega-Retuerta E, Pulido-Villena E, Reche I (2007) Effects of dissolved organic matter photoproducts and mineral nutrient supply on bacterial growth in mediterranean inland waters. Micro Ecol 54(1):161–169. https://doi.org/10.1007/s00248-006-9186-x
Ortega-Retuerta E, Reche I, Pulido-Villena E, Agustí S, Duarte CM (2008) Exploring the relationship between active bacterioplankton and phytoplankton in the Southern Ocean. Aquat Micro Ecol 52(1):99–106. https://doi.org/10.3354/ame01216
Pedrós-Alió C, Calderón-Paz JI, MacLean MH, Medina G, Marrasé C, Gasol JM, Guixa-Boixereu N (2000) The microbial food web along salinity gradients. FEMS Microbiol 32(2):143–155. https://doi.org/10.1111/j.1574-6941.2000.tb00708.x
Razinkovas-Baziukas A, Povilanskas, R (2012) Introducing transitional waters. In: Nilsson, Henrik; Povilanskas, Ramūnas; and Stybel, Nardine (2012) Trans boundary management of Transitional Waters - Code of Conduct and Good Practice examples 8–14. http://commons.wmu.se/artwei/1
Roehm CL (2005) Respiration in wetland ecosystems. In: del Giorgio AP, Williams PJ (eds) Respiration in aquatic ecosystems. Oxford University, New York, pp 83–102
Sánchez MI, Green AJ, Castellanos EM (2006) Temporal and spatial variation of an aquatic invertebrate community subjected to avian predation at the Odiel salt pans (SW Spain). Arch Hydrobiol 166:199–223. https://doi.org/10.1127/0003-9136/2006/0166-0199
Saunders DL, Kalff J (2001) Nitrogen retention in wetlands, lakes and rivers. Hydrobiology 443:205–212. https://doi.org/10.1023/A:1017506914063
Schlesinger WH (2009) On the fate of anthropogenic nitrogen. Proc Natl Acad Sci USA 106:203–208. https://doi.org/10.1073/pnas.0810193105
Schuerch M, Spencer T, Temmerman S, Kirwan ML, Wolff C, Lincke D, McOwen CJ, Pickering MD, Reef R, Vafeidis AT, Hinkel J, Nicholls RJ, Brown S (2018) Future response of global coastal wetlands to sea-level rise. Nature 561:231–234. https://doi.org/10.1038/s41586-018-0476-5
Seitzinger SP (1988) Denitrification in freshwater and coastal marine ecosystems Ecological and geochemical significance. Limnol Oceanogr 33:702–724. https://doi.org/10.4319/lo.1988.33.4part2.0702
Seitzinger SP, Harrison J, Bohlke J, Bouwman A, Lowrance R, Peterson B, Tobias C, Drecht G (2007) Denitrification across landscapes and waterscapes: A synthesis. Ecol Appl 16:2064–2090. https://doi.org/10.1890/10510761(2006)016[2064:DALAWA]2.0.CO;2
Smith DC, Azam F (1992) A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Microb Food Webs 6:107–114
Smith EM, Prairie YT (2004) Bacterial metabolism and growth efficiency in lakes: the importance of phosphorus availability. Limnol Oceanogr 49(1):137–147. https://doi.org/10.4319/lo.2004.49.1.0137
Ventosa A, Fernández AB, León MJ, Sánchez-Porro C, Rodriguez-Valera F (2014) The Santa Pola saltern as a model for studying the microbiota of hypersaline environments. Extremophiles 18(5):811–824
Verhoeven JTA, Arheimer B, Yin CQ, Hefting MM (2006) Regional and global concerns over wetlands and water quality. Trends Ecol Evol 21:96–103. https://doi.org/10.1016/j.tree.2005.11.015
Walton MEM, Vilas C, Canavate P, Gonzalez-Ortegon E, Prieto A, van Bergeijk SA, Green AJ, Librero M, Mazuelos N, Le Vay L (2015) A model for the future: Ecosystem services provided by the aquaculture activities of Veta la Palma, Southern Spain. Aquaculture 448:382–390. https://doi.org/10.1016/j.aquaculture.2015.06.017
Weston NB, Dixon RE, Joye SB (2006) Ramifications of increased salinity in tidal freshwater sediments: geochemistry and microbial pathways of organic matter mineralization. J Geophys Res-Biogeo 111(G1):1. https://doi.org/10.1029/2005JG000071
Weston NB, Vile MA, Neubauer DC, Velinsky DJ (2011) Accelerated microbial organic matter mineralization following salt-water intrusion into tidal freshwater marsh soils. Biogeochemistry 102:135–151. https://doi.org/10.1007/s10533-010-9427-4
Yokokawa T, Sintes E, De Corte D, Olbrich K, Herndl GJ (2012) Differentiating leucine incorporation of Archaea and Bacteria throughout the water column of the eastern Atlantic using metabolic inhibitors. Aquat Microb Ecol 66:247–256. https://doi.org/10.3354/ame01575
You J, Das A, Dolan EM, Hu Z (2009) Ammonia-oxidizing archaea involved in nitrogen removal. Water Res 43(7):1801–1809. https://doi.org/10.1016/j.watres.2009.01.016
Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61(533–665):616
Acknowledgements
We especially thank Alejandra Fernández, Francisco Perfectti, and Eulogio Corral for their help in the field and laboratory. We also thank the facilities and technical staff of Odiel Marshes, Veta la Palma (Doñana National Park), El Hondo, Santa Pola, Ebro Delta, Tour du Valat (Camargue), Parco Naturale Molentargius Saline (Sardinia), and Université de Gabês (Tunisia). We also thank the logistic support of Josefa Antón at the University of Alicante, Arnaud Béchet at Tour du Valat, and M.A. Chokri and S. Selmi in the Tunisian site. We thank Ignacio Peralta-Maraver for his help with the Figures.
Funding
Funding for open access charge: Universidad de Granada / CBUA. This research was funded by the projects FLAMENCO (CGL2010-15812) and CRONOS (RTI2018-098849-B-I00) of the Spanish Ministry of Economy and Competitiveness and Ministry of Science and Innovation, the Modeling Nature Scientific Unit (UCE.PP2017.03), European Regional Development Fund (ERDF) and a PhD fellowship FPI (Formación del Personal Investigador: BES-2011–043658) to GLB.
Author information
Authors and Affiliations
Contributions
I.R., A.J.G., J.A.A., M.V. and C.A.S. designed this study; G.L.B., A.J.G., J.A.A., M.V., and I.R. performed the samplings and most of biological and chemical analysis. G.L.B. and C.A.S. analyzed the virus abundance. G.L.B., A.J.G., and I.R. analyzed the data and wrote the manuscript with input from all the authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Batanero, G.L., Green, A.J., Amat, J.A. et al. Patterns of microbial abundance and heterotrophic activity along nitrogen and salinity gradients in coastal wetlands. Aquat Sci 84, 22 (2022). https://doi.org/10.1007/s00027-022-00855-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00027-022-00855-6