Abstract
Mechanical load plays a significant role in bone and growth-plate development. Chondrocytes sense and respond to mechanical stimulation; however, the mechanisms by which those signals exert their effects are not fully understood. The primary cilium has been identified as a mechano-sensor in several cell types, including renal epithelial cells and endothelium, and accumulating evidence connects it to mechano-transduction in chondrocytes. In the growth plate, the primary cilium is involved in several regulatory pathways, such as the non-canonical Wnt and Indian Hedgehog. Moreover, it mediates cell shape, orientation, growth, and differentiation in the growth plate. In this work, we show that mechanical load enhances ciliogenesis in the growth plate. This leads to alterations in the expression and localization of key members of the Ihh-PTHrP loop resulting in decreased proliferation and an abnormal switch from proliferation to differentiation, together with abnormal chondrocyte morphology and organization. Moreover, we use the chondrogenic cell line ATDC5, a model for growth-plate chondrocytes, to understand the mechanisms mediating the participation of the primary cilium, and in particular KIF3A, in the cell’s response to mechanical stimulation. We show that this key component of the cilium mediates gene expression in response to mechanical stimulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It has been long known that mechanical stimulation plays a critical role in the development and maintenance of the skeleton [46], including the growth plate [44]. Chondrocytes possess mechano-sensing abilities [63], but how these mechano-signals are received by the cells and what mechanisms mediate their conversion into biological responses are only partially understood [62].
One possible candidate for this mechanical processing is the primary cilium. The primary cilium is critical to skeletal development; the embryonic cilium plays a role in the earliest cellular determinative events establishing left–right axis asymmetry [35] and primary cilia in the early mesenchyme is necessary for proper anterior-posterior limb patterning (reviewed by [15, 16]). Primary cilium is also vital for proper endochondral ossification, primarily due its regulatory role in Ihh signaling [32]. Several pathologies associated with ciliary dysfunction (ciliopathies) exhibit bone deformities. For example, Jeune asphyxiating thoracic dystrophy (ATD), an autosomal-recessive chondrodysplasia characterized by short ribs and a narrow thorax, short long bones, inconstant polydactyly, and trident acetabular roof [7]. This organelle functions as a complex signaling center [2] and has been identified as a mechano-sensor in several cell types, including renal epithelial cells and endothelium [42, 55]. In the skeletal system, the primary cilium has also been shown as an important mechano-sensor: in osteoblasts and osteocytes, the primary cilium is required for bone cell response (increase in the expression of osteopontin) to dynamic fluid flow [34], for formation of new bone in response to cyclic mechanical loading [76] and in mediating the osteogenic differentiation of human mesenchymal stem cells to osteoblasts in response to fluid flow [18]. Moreover, it was recently shown to be essential for the transduction of mechanically regulated signals in response to compression in sterna chondrocytes [83] and to be required for modulation of Indian hedgehog (Ihh) signal transduction in response to hydrostatic compression loading of growth-plate chondrocytes [67].
The primary cilium is present on almost all eukaryotic cell types, including chondrocytes [52, 84]. It consists of a membrane-coated axoneme that projects from the cell surface into the extracellular microenvironment. The axonemes consist of nine microtubule doublets in a radially symmetric arrangement which is commonly referred to as 9 + 0 arrangement (in apposed to 9 + 2 arrangement which characterize the motile cilia). In addition, the primary cilium contains an intracellular basal body [53] which originates from the mother centriole of the cell. The basal body functions as both an anchor for the primary cilium and a template for cilium extension [21], [53].
Axoneme growth occurs by a process called intraflagellar transport (IFT), wherein cargo is transported up and down the axoneme via complexes of IFT adaptor proteins by the motor proteins kinesin-II and cytoplasmic dynein 1b, respectively [64]. Disruption of either the IFT or kinesin-II proteins leads to impaired cilium assembly and function [47].
The link between cilium function and growth-plate development has been described in a number of studies. Primary cilium has been shown to be involved in several signaling pathways essential for regulation of the growth plate, such as the non-canonical Wnt and Ihh pathways [15, 37, 49, 71, 72]. Chondrocytes express a single primary cilium, which projects from the cell surface into the extracellular matrix (ECM) and interacts with molecules such as collagen and glycoprotein [22, 36, 39, 52, 53, 85]. It is found in all zones of the growth plate, oriented parallel to the longitudinal axis of the bone, and it represents the axis of chondrocyte polarity inside the growth plate [8]. The primary cilium mediates cell shape, orientation, growth, and differentiation in the growth plate as deletion of KIF3A, a subunit of the motor protein kinesin-II, results in defects in the columnar organization of the growth plate together with reduced cell division, accelerated hypertrophic differentiation, and disruption of cell shape and orientation relative to the long axis of the bone [72].
In this work, we use a previously reported protocol in which bags are harnessed to the back of young chickens, without surgical procedure or dramatic alterations in their environmental conditions [56–58], to study the cilium’s involvement in transducing mechanical load in chondrocytes. In addition, we use the chondrogenic cell line ATDC5, a model for growth-plate chondrocytes, to understand the mechanisms mediating the participation of the cilium, and in particular KIF3A, in the cell’s response to mechanical stimulation. We show that growth-plate chondrocytes respond to mechanical load by increasing the number of primary cilium in vivo and by activating characteristic signaling pathways both in vivo and in vitro. We further establish the role of the primary cilium as a mechano-sensor and regulator of mechano-signaling in the growth plate.
Materials and methods
In vivo experiment
Animals and the loading model
As reported previously [56–58], 2-day-old broiler chickens [48] were divided into two groups: control group (n = 30) and the “BAG” group (n = 30) in which the chickens were harnessed for 4 days with small bags (2.5 × 4 cm) filled with sand, weighing 10 % of their BW. This protocol applies a moderate supraphysiological load by using a physiological loading pattern. Thus, the loading in this experiment mimics but nevertheless enhances the loads applied to bones under physiological conditions. At the end of the loading period (when they were 6 days old), chickens were sacrificed and proximal tibia samples were processed for histology or for RNA. The experiment was approved by the committee of ethics in animal experiments.
RNA isolation, reverse transcription (RT), and real-time PCR of chick’s growth-plates
RNA was extracted from proximal tibia growth-plates with RNeasy Maxi kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The RNA samples were prepared from pools of five growth plates (from each group). Three identical, independent experiments were performed. Total RNA (1 µg) was reverse-transcribed with Reverse-RT kit (ABgene, Epsom, UK).
Real-time PCR was performed using the fluorescent dye SYBR Green I (ABgene) with specific primers (Table 1) and Gallus gallus ribosomal RNA as a normalizing control.
In-situ hybridization, immunohistochemistry, and immunofluorescence
Tibial growth plates were fixed overnight in 4 % (v/v) Para formaldehyde (Sigma Chemical Co., St. Louis, MO, USA) at 4 °C, dehydrated in graded ethanol solutions, cleared in chloroform, and embedded in Paraplast.
Sections (5 µm) were hybridized with either digoxigenin-labeled antisense probes for collagen type II (Col II) and collagen type X (Col X) or (35S)-labeled probes for Ihh and ptc1 as described in our previous reports [56, 70, 79], and photographed using dark and bright-field microscopy. The radioactive signal of the (35S)-labeled probes was intensified using emulsion (Eastman Kodak Company, Rochester, NY, USA) for 1 month, in the dark at room temperature. No signal was observed in the sense-probe hybridizations, used as controls.
Probes for ISH were prepared by PCR amplification of cDNA from both chicken growth plates and primary cultured chondrocytes, with the following primers:
Col II (F): ATATCCACGCCAAACTCCTG Col II (R): GCTCCCAGAACGTCACCTAC
Col X (F): CCACCTGGATTCTCCACTGT Col X (R): TTCAAATCCTGGAAGACCTG
Ihh (F): CATCATCTTCAAGGACGAGGAGAACIhh (R): TACTTGTTGCGGTCCCTGTCTGAC
ptc (F): CTCCTTTGGACTGCTTCTGG ptc (R): AGGCAGAACCTGAGTTGTGG
Proliferating cell nuclear antigen (PCNA) immunohistochemistry was preformed as described by Hasky-Negev et al. [14]. Sections were deparaffinized with xylene, washed twice in 100 % ethanol, and treated with 3 % H2O2 in distilled water for 30 min to block endoge-nous peroxidase activity. Next, the sections were rehydrated through a graded series (95, 80, 70 %) of ethanol solutions, rinsed in 0.05 % PBS-Tween, and incubated for 1 h in 3 % normal goat serum in PBS-Tween. Sections were then incubated for 1 h with biotinylated mouse anti-PCNA primary antibody (Zymed) and washed 3 times for 10 min each in 0.05 % PBS-Tween. After washing, sections were incubated with streptavidine peroxidase for 10 min and washed again with PBS-Tween. Next, sections were incubated with substrate solution for PCNA (5 mg 3,3-diaminobenzidine, 5 microL of H2O2 in 10 ml H2O of double-distilled water) for 15 min and washed with double-distilled water.
For the detection of primary cilia, growth-plate sections were incubated in hyaluronidase (Sigma-Aldrich, Rehovot, Israel) for 2 h at 37 °C and permeabilized using 0.5 % (v/v) Triton X-100 in PBS for 5 min, followed by 5 % (v/v) goat serum in PBS for 30 min at RT. Sections were labeled overnight at 4 °C with a primary antibody raised against acetylated α-tubulin followed by goat anti-mouse FITC second antibody (Jackson ImmunoResearch, West Grove, PA, USA) labeling for 2 h at RT. Nuclei were stained with DAPI staining (Jackson ImmunoResearch).
Histomorphometric analysis
Histomorphometric measurements of the cells in the proliferative and hypertrophic zones (PZ and HZ, respectively) were performed by imaging the section under a 100X objective, and acquiring at least three fields of view from three different sections in each group. Using Olympus Cell software, the number of cells was counted within a defined region of 2.28 mm2. The cells’ area was measured for each field of view and expressed as mean cell area/µm2 (±s.e.m).
For assessment of the flattened morphology in the PZ and cell morphology in the HZ, the dimensions of 60 cells in three fields of view from three different sections in each group were measured on the X (horizontal) and Y (vertical) axes. The X/Y ratio was calculated and used as an indicator of flattened morphology, expressed as a mean (±s.e.m).
In vitro experiments
Mechanical stimulation of the cells
For all in vitro experiments, ATDC5 cell line was used [1]. Cells were cultured as described previously [5, 20, 69]. For mechanical stimulation, cells were seeded at 2.5 × 104 cells/well into 4 cm2 silicon Stretch chambers ST-CH-04 (B-bridge, Cupertino, CA, USA) coated with 0.1 % gelatin. Mechanical stimulation was applied with the STREX cell device (B-Bridge). The applied protocol was stimulation for short time points (30, 60, 240, or 360 min), at 1 HZ and 20 % elongation.
Isolation of human articular chondrocytes
292gG human articular chondrocytes were isolate from healthy cartilage taken from patients undergoing total knee replacement. All procedures were done according to the ethics regulations of Georg-August-University, Goettingen, Germany.
Scanning electron microscopy imaging
ATDC5 chondrocytes cells were seed on coverslips coated with 0.1 % gelatin and serum starved for 24 h. Cells were fixed in 2.5 % glutaraldehyde/DMEM for 40 min, washed with 0.1 M sodium phosphate buffer, and fixed in Karnovsky’s fixative (2 % paraformaldehyde, 2.5 % glutaraldehyde, and 0.1 M sodium phosphate buffer) for 1.5 h. Following fixation, cells were washed with 0.1 M PBS dehydrated through a graded ethanol series, and processed for critical point drying and coating. Samples were imaged with a JCM6000 bench top scanning electron microscope (Jeol).
Immunocytochemistry staining
For immunocytochemistry staining cells were seeded on cover slip or in silicon chambers. Cells were fixed with 3 % PFA +0.5 % Triton × 100 at 37 °C for 3 min and than with 3 % PFA for another 20 min. Block was preformed with 1 % BSA + 5 % Goat or Donkey serum in TBST, followed by overnight incubation with primary antibodies (see list). Next day, cells were incubated with secondary antibodies (see list) for 1.5 h. Cells were viewed under the light/florescence microscopy eclipse E400 Nikon using light or the florescence filters for FITC, TRITC or DAPI. Confocal images were taken with the DMI4000B Confocal Microscopy (Leica, Wetzlar, Germany).
Quantification of the prevalence of ciliated cells and measurements of the cilia length in unstimulated cells compared to mechanical stimulated cells for short time points (30–120 min) were performed using the “cell A” software of the Olympus DP71 camera.
Antibodies for immunocytochemistry
Primary antibodies: mouse monoclonal anti-Acetylated α Tubulin (6-11B-1), pericentrin (ab448, Zotal) mouse monoclonal anti-p-p38 (D-8)(SC-7973), rabbit polyclonal anti-patched (H-267)(SC-9016), rabbit polyclonal anti-STAT1 (E-23)(sc-346) (Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit polyclonal anti-GFP (A11122) (Invitrogen, Grand Island, NY, USA).
Secondary antibodies: anti-mouse Alexa 488, anti-mouse Alexa 594, anti-rabbit Alexa 488, anti-rabbit Alexa 594 (Invitrogen), anti-mouse Dylight 594 (Pierce, Rockford, IL, USA), DAPI (Santa Cruz), Phalloidin-TRICT (Sigma-Aldrich).
RNA isolation, reverse transcription (RT), and real-time PCR
RNA was isolated from the cells using TRI reagent (Sigma-Aldrich). cDNA was synthesized from 1 µg of RNA using the high-capacity cDNA reverse-transcription kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocols.
Real-time PCR was performed using platinum SYBR Green (Invitrogen) with 2 µl of cDNA template, and Mus musculus gene specific primer sets (Table 1). Relative quantification of the specific gene was normalized to GAPDH housekeeping gene.
Protein isolation and western blot
Total protein was extracted from ATDC5 cells. Protein concentration was measured using a bicinchoninic acid (BCA) protein assay reagent kit (Pierce Biotechnology) according to the manufacturer’s protocols. Lysates (30 µg protein) were separated by 10 % SDS-PAGE, transferred to nitrocellulose membranes, incubated overnight with primary antibody (listed bellow) at 4 °C, followed by incubation with peroxidase-conjugated secondary antibody, and detected with ECL [69].
Antibodies for western blot
Primary antibodies: rabbit polyclonal anti-p38 (c-20) (sc-535), mouse monoclonal anti-p-p38 (D-8)(SC-7973), rabbit polyclonal anti-STAT1(E-23)(sc-346), mouse monoclonal anti-GAPDH (A-3)(sc-137175) (Santa Cruz Biotechnology), rabbit polyclonal anti- KIF3A (ab11259) (abcam, Zotal), rabbit polyclonal anti-GFP (A11122) (Invitrogen).
Secondary antibodies: goat anti mouse conjugated Peroxidase (115-035-003) (Jackson ImmunoResearch), Goat anti rabbit conjugated HRP (sc-2004) (Santa Cruz Biotechnology).
Flow cytometry (FACS) analysis for cell cycle
ATDC5 cells were detached from the culture wells, centrifuged (600 g, 5 min) and resuspended in 1 % paraformaldehyde for 30 min, then centrifuged again. The pellet was incubates for 1 h, in dark conditions at 4 °C in a solution containing 50 μg/mL of propidium iodide, 0.1 % sodium citrate, and 0.1 % Triton X-100. Cell cycle was analyzed by fluorescence-activated cell sorting using a linear scale, FL2 channel, excitation at 488 nm, and emission at 575 nm. Data were collected from 50,000 cells. Analyses of the FACS data were done with the WniMDI 2.9 free software [69].
Constructs and transient transfection of ATDC5
The EGFP protein was sub cloned to the multiple cloning site of the pCDNA3.1 with EcoRI and NotI restriction enzymes. This pCDNA-GFP vector was used as the backbone for the TRC-GFP vectors. The shRNA target sequence for KIF3A, with the U6 promoter, was amplified from the pLKO.1 vectors (TRCN0000090404- Mature Antisense: ATAGTAATTGTAAAGATGGCG, TRCN0000090405 Mature Antisense: TTTGCAGAACGCTTTCTTCGC) and pLKO.1 Empty vector for the control (Open biosystems, Epsom, UK) with specific primers and sub cloned to the BglII site of the pCDNA-GFP plasmid.
Transient transfection of ATDC5 cells was carried out with TransIT-LTI reagent (Mirus Bio LLC, Madison, WI, USA) according to the manufacturer protocol. 48 h post transfection, cells were taken for mechanical stimulation tests, genes and protein analysis etc.
Statistical analysis
The results for the in vivo experiment are expressed as mean ± s.e.m and for all in vitro experiments as mean ± SD Differences between groups were tested by analysis of variance using Student’s t test or one-way analysis of variance and Tukey–Kramer HSD test, using JMP 8.0 Statistical Discovery Software (SAS Institute 2000). Differences were considered significant at P < 0.05.
Results
Mechanical load alters the Ihh–PTHrP loop in the growth plate
Ihh, directly through its receptor Patched-1 (ptc1), increases chondrocyte proliferation and inhibits its hypertrophic differentiation through induction of Parathyroid hormone-related protein (PTHrP) expression [81, 82].
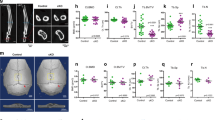
We examined the effect of mechanical load on the expression of Ihh (Fig. 1a, a′, c) and its receptor, ptc1 (Fig. 1b, b′, c), in the growth plates of supraphysiologically loaded vs. physiologically loaded chickens (for convenience, we will regard them from now on as loaded vs. non-loaded chickens). In the control growth plates, Ihh was expressed in the pre-hypertrophic zone (Fig. 1a, a′) and ptc1 appeared as a typical “double band” [80] in the resting and pre-hypertrophic zones (Fig. 1b, b′). Upon mechanical loading, quantitative analysis of gene expression (RNA extracted from a pool of growth plates) showed a significant decrease in the expression of Ihh and ptc1 (Fig. 1c). Moreover, the pattern of ptc1 expression became narrower and restricted to the resting zone and upper proliferative zone (Fig. 1b′). Interestingly, the relative localization of these two genes in the growth plate was changed by mechanical loading; overlaying the expression zone of Ihh (in green) on that of its receptor ptc1 (in purple) demonstrated a reduction in the gap between them, creating a zone in which both are expressed simultaneously (Fig. 1d). This suggested that mechanical loading alters the Ihh pathway in the growth plate.
Mechanical load modifies the expression and localization of Ihh-PTHrP loop members. Growth-plates from loaded (BAG) or control (CTRL) chicks were subjected to in situ hybridization with Ihh (a, a′), ptc (b, b′), PTHrP (e), and PPR (f, f′) (35S)-labeled antisense probes shown at 15X (a, b, e, f) and ×40 (a′, b′, f′) magnification. (c, g) Ihh, ptc, PTHrP, and PPR expression was quantified using real-time PCR on growth-plate RNA. Values are expressed as mean ± s.e.m of three replicates, normalized to 18S. Significantly different at P < 0.05. (d) Ihh expression overlaying on ptc expression in growth plates from BAG and CTRL chicks, showing the relative localization of these genes at ×15 magnification. (g) AC-articular cartilage, PZ proliferative zone, HZ hypertrophic zone
PTHrP is regulated by Ihh [82], hence we assumed that disrupted Ihh signaling might alter its gene expression. Indeed, PTHrP expression, which was limited to the reserve zone in the control growth plates (Fig. 1e), almost vanished in the loaded growth plates (Fig. 1e). This reduction was confirmed by real-time PCR analysis of PTHrP expression (Fig. 1g). The expression of PTHrP receptor was also modified by the load; in the control group it was expressed in the pre-hypertrophic zone, and upon loading, its expression zone became narrower and more intense (Fig. 1f, f′). Quantification of this gene’s expression also indicated an increase upon loading (Fig. 1g).
Taken together, these results proved that mechanical loading alters the expression and localization of the major components of the Ihh–PTHrP loop in the growth plate, and thus likely modifies the functionality of these key regulators.
Mechanical load affects growth plate and chondrocyte organization and morphology
We studied different growth-plate parameters that might be connected to the dysfunctional Ihh–PTHrP loop. We previously showed that growth plates respond to 4 days of mechanical loading by exhibiting reduced thickness, narrowed expression zones of collagen types II and X, and reduced cell numbers in those zones [56]. Here, using PCNA immunohistochemistry, we found that the reduction in cell number is due to reduced proliferation (Fig. 2a). While observing the plates, an interesting phenomenon was detected: the transition between the proliferative zone (positive for collagen II) and hypertrophic zone (positive for collagen X) was more homogeneous in the growth plates that were subjected to loading (Fig. 2b), suggesting that not only proliferation is altered by the load, but also the switch between proliferation and differentiation is altered. This strengthened our hypothesis that the Ihh–PTHrP loop is involved in mediating the growth plate’s response to load and maintaining its normal functionality.
Mechanical load inhibits chondrocyte proliferation, sharpens the transition between the zones and affects chondrocytes morphology and organization. a PCNA immunohistochemistry was performed on growth-plates from loaded (BAG) and control (CTRL) chicks: PCNA-positive cells were counted, showing a reduction in the number of proliferating cells after 4 days of loading. Values expressed as mean ± SD of four different samples. b Sections were subjected to in situ hybridization with digoxygenin-labeled riboprobes of antisense chicken collagen type II located in the proliferative zone [54] and collagen type X located in the hypertrophic zone (HZ). Red line indicating transition between the zones. c Sections were stained with H&E and are presented at ×1,000 magnification. Lines extending from the center of the first cell in a column are drawn to demonstrate the deviation of the chondrocytes from the columns in the proliferative zone in these groups. Figures represent a typical field used for the morphometric measurements of cells in the proliferative and hypertrophic zones. Morphometric analyses were performed on three different fields from three different animals d the number of cells per defined area. e The mean cell area f the ratio of X/Y cell dimensions as an indication of cell flatness. g The vertical (Y) and horizontal (X) axis length of cells. Significantly different at P < 0.05
Next, we examined the morphology and organization of the chondrocytes in the different zones of the growth plate. The loaded growth plates showed higher organization, as cells in the proliferative zone deviated less from the center of the column (marked by a line extending from the first cell in each column in Fig. 2c, PZ) compared with the control growth plates in which more cells deviated from the column line (Fig. 2c). Morphometric analysis showed a significant increase in the number of cells per defined area (Fig. 2d) and a decrease in the average cell area (Fig. 2e) of chondrocytes in both zones as a result of the load. Calculating the relative areas in the growth plates occupied by cells or matrix showed no significant differences between the control and ‘load’ groups in either zone. These results indicated that mechanical load increases the number of cells but decreases the size of each cell in a defined area compared to the control.
The X/Y ratio of cell dimensions, reflecting cell flatness, was significantly increased in the proliferative zone and decreased in the hypertrophic zone as a result of loading (Fig. 2f). The differences in the ratio resulted from a reduction in the vertical (Y) axis length in the proliferative zone with no change in the horizontal (X) axis, while in the hypertrophic zone a reduction was observed in the horizontal (X) axis length with no change in the vertical (Y) axis (Fig. 2g). Thus, the cells in the proliferative zone were more spread out, whereas those in the hypertrophic zone were more spherical, suggesting that mechanical load affects chondrocyte morphology and organization within the growth plate.
Taken together, our results showed that growth-plate chondrocytes are sensitive to mechanical load. This response involves alteration in the function of the Ihh–PTHrP loop, affecting cell proliferation and the switch between proliferation and differentiation, as well as chondrocyte shape and gross growth-plate morphology.
Mechanical load affects ciliogenesis in chondrocytes
A plausible candidate for the transduction of these load-induced effects in the growth plate is the primary cilium. Up-to-date information on the primary cilium as a mechano-sensor in chondrocytes is reviewed in [40, 62]. To examine this possibility, we studied the expression of genes that have been shown to be associated with cilium assembly and function [26, 28, 37, 72] such as kinesin II (KIF3A), IFT88/Polaris, and the two polycystic kidney disease genes (PKD1 and 2). We found significant up-regulation of all of these genes in the growth plate after 4 days of mechanical loading (Fig. 3a). Accordingly, the number of cells presenting primary cilium was increased in both the proliferative and hypertrophic zones of the loaded plates (Fig. 3b, c). These results show that loading induces ciliogenesis in growth-plate chondrocytes in vivo, and supported our suggestion that the primary cilium is involved in the response to mechanical load. The results of the in vivo model are summarized in a diagram (Fig. 4).
Mechanical load increase ciliogenesis in the growth plate. a Expression of genes involved with primary cilium: KIF3A, IFT88, PKD1 and PKD2 was examined by real-time PCR on growth-plate RNA isolated from mechanical load (BAG) compared to non-loaded (CTRL) chicks. Values are expressed as mean ± s.e.m of three replicates, normalized to 18S. Significantly different at P < 0.05. b Acetylated α-tubulin stained the primary cilia (arrows) in strong fluorescent green, while the cell cytoplasm stained light green. Nuclei are stained with DAPI (blue) in the proliferative [54] and hypertrophic (HZ) zones. Scale bar 20μm. c Cells presenting primary cilium were counted
Diagram describing the ultrastructure of the primary cilium in growth plate (GP) chondrocytes and a possible connection of the primary cilium to mechanical stimulation effects on the cell. The primary cilium consists of a membrane-coated axoneme that project from the cell surface into the extracellular microenvironment. a Unstimulated GP chondrocyte: Ihh, directly through its receptor Patched-1 (ptc1), located in the cilium, increases chondrocyte proliferation and inhibits its hypertrophic differentiation through induction of Parathyroid hormone-related protein (PTHrP) expression. b Mechanical stimulated GP chondrocyte: morphological change of the cell together with up-regulation of cilia related genes (IFT88, KIF3A, PKD1 and PKD2) and formation of stress fibers. Decrease in the expression of Ihh and ptc1 results in major decrease of PTHrP expression following reduced proliferation and switch for differentiation
Response of ATDC5 cells to mechanical stimulation by activation of typical pathways and changes in primary cilium-related genes
To better understand the role of the primary cilium in the growth plate’s response to load, we continued our work with the chondrogenic cell line ATDC5 which serves as an excellent model for chondrogenesis in vitro [1, 5, 20, 69]. Mechanical stimulation was applied to the cells using the STREX device. The applied protocol in all experiments was stimulation for short times, over a period of up to 6 h, at 1 Hz and 20 % elongation. This strain magnitude lies within the lower range of magnitudes required for effective bone healing in vivo, and normal cartilage maintenance [10, 45, 51].
First, we examined whether the cells’ response to mechanical stimulation was as reported in the literature. Changes in the cytoskeleton in response to mechanical stimulation have been reported in articular chondrocytes [4] and intervertebrate disc cells [29]. Here we show that mechanical stimulation of chondrocytes induces modifications in the cellular arrangement of the cytoskeleton (changes in F-actin) and formation of stress fibers along the strain axis (Fig. 5a, arrowhead). In addition, we showed that chondrocytes respond to mechanical stimulation by activation of p38 MAPK (Mitogen-activated protein kinases) (Fig. 5b, d), as previously reported in mesenchymal stem cells [30], and showed for the first time that mechanical stimulation activates the Stat1 (signal transducers and activators of transcription) pathway (Fig. 5c, d). The mechanical load slightly induced the expression of the cilia related genes PKD1, KD2, IFT88 and IFT172 (Fig. 5e). While dramatically and time dependent affecting FOS and Early growth response protein (EGR1), two genes which have been identified as immediate-early response genes in osteoblasts’ response to load and stretch [45, 74], as well as that of osteopontin (OPN) and aggrecan (AGC1) (Fig. 5f), two chondrogenic genes known to respond to mechanical load [30, 56, 77].
ATDC5 chondrocytes respond to mechanical stimulation. ATDC5 cells were subjected to a 2D dynamic strain of 1 Hz and 20 % elongation for different time periods. a Actin arrangement was detected by immunocytochemistry staining with phalloidin (red). Stress fibers accumulation oriented with the strain direction is pointed with arrow. Scale bar 20μm. b Immunocytochemistry with anti phospho-p38 (green) antibody demonstrates p38 phosphorylation in respond to the mechanical stimulation. Nuclei stained with DAPI. Scale bars for DAPI and p-p38 50μm. Scale bar for the overlay 100μm c Immunocytochemistry with anti Stat1 (green) antibody showing changes in the Stat1 levels and localization with time. Scale bar 50μm. d Protein levels of p-p38, p-38, and Stat1 were quantified using western blot analysis. Cell lysates were separated on SDS-PAGE followed by bloting with antibodies against p-p38, p38, and Stat1. e Expression levels of PKD1, PKD2, IFT88, and IFT172 (F) FOS, EGR1, osteopontin (OPN), aggrecan (AGC1), were examined by real-time PCR. Values are expressed as mean ± SD of three replicates. Significantly different at P < 0.05. g SEM presentation of primary cilia on ATDC5 chondrocyte (H) Immunocytochemistry with acetylated α-tubulin (A-tb, green) and pericentrin (red) antibodies for the detection of the primary cilium, and DAPI for the nuclei (DAPI, blue)
Moreover, ATDC5 chondrocytes exhibited primary cilium, as demonstrated by double staining with α acetylated tubulin and pericentrin antibodies as well as using scanning electron microscopy (Fig. 5g, h). In order to understand whether and how load application affects primary cilium distribution in chondrocytes in vitro. For this purpose, quantification of the cilia presenting cells were performed in cultured ATDC5 chondrocytes subjected to mechanical stimulation experiment (for 0, 30, 60 and 120 min). Measurements were calculated for more than 500 cells from 3 different experiments, and showed no significant differences between the unstimulated cells and the stimulated cells for the different time points. In parallel to the ciliated cells counting, we also measured cilia length of more than 150 ciliated cells per time point, and compared between the stimulated vs. unstimulated cells. Again, we did not observe any significant difference in the cilia length caused by the mechanical stimulation. In all checked samples, around 80 % of the cells presented cilia (counted according to acetylated-tubulin staining in comparison to nucleus DAPI staining), and cilia length was 2.4 μM, as demonstrated also by others [78]. These conflicting results between our in vivo and in vitro model can be attributed to the different time scale of the mechanical stimulation that the two models were subjected to. While the in vitro model examine the immediate response of the cells to mechanical stimulation on ciliogenesis, the in vivo model tested a more prolong stimulation period of 4 days.
Contrary to our data previous research [38] has shown that cyclic compression (0–15 %; 1 Hz) for 24 and 48 h reduce cilia incidence in articular chondrocytes grown in 3D agarose culture. We suspect that the cause for these inconsistencies is once again, the time scale that was investigated. However, our results are compatible with a recent study that subjected primary articular chondrocytes to cyclic tensile strain up to 20 % for 1 h at 0.33 Hz and conclude that the primary cilia prevalence was not altered in response to this stimulation [78]. But affected cilia length of cells subjected to the highest strain by reduction of approximately 15.1 %. In conclusion, our findings suggest an influence of mechanical stimulation on the process of ciliogenesis in vivo, presumably due to the long duration of stimulation.
IFT88 and ptc1 receptor localize to the chondrocytic primary cilium
The IFT complex is essential for cilium formation. IFT88 is part of this complex and has been shown to localize to the primary cilium [41]. Here we identified IFT88 in the primary cilium in ATDC5 murine chondrocytes (Fig. 6a) and in 292gG human articular chondrocytes (Fig. 6b). Second, we found that activation of the Ihh pathway by SAG [6] results not only in increased ptc1 (which is known to be downstream of Ihh) [9] mRNA expression (Fig. 7d), but also in accumulation of ptc1 at the chondrocytic cilium (Fig. 6c).
IFT88 and ptc1 receptor localized at the chondrocytic primary cilium. a, b ATDC5 cell line and 292gG primary human articular chondrocytes were subjected to immunocytochemistry with anti acetylated α-tubulin antibody for the detection of primary cilium (tb, green), anti IFT88 antibody (IFT88, red) and DAPI for nuclei staining (DAPI, blue). Shifted overlay (right panel) demonstrating the localization of IFT88 to the primary cilium. c ATDC5 cells were treated with SAG for the activation of the Hh pathway. The localization of ptc1 to the primary cilium was detected by immunocytochemistry with anti acetylated α-tubulin antibody for the detection of primary cilium (tb, red) and anti ptc1 antibody (Ptc1, green). Results showing accumulation of ptc1 in the primary cilia following SAG stimuli
Knocking down KIF3A modifies cilium morphology and functionality. KIF3A was silenced by transient tranfection. a KIF3A protein levels were determined by western blot analysis. (b, c) Control (empty) and KIF3A knock-down cells were immuno-stained with acetylated α-tubulin Abs (tb, red) to detect the primary cilium and DAPI for nuclei staining (DAPI, blue). b The number of cells presenting primary cilium on transfected cells (GFP) was counted and presented as percent of total green cells. c Staining showing the changed cilium morphology in the KIF3A knock-down cells compared with control. The shKIF3A-1 cells showed a thinner staining of the tubulin structure while the shKIF3A-2 cells demonstrate a truncated cilium structure. d ATDC5 cells were treated with SAG for the activation of the Hh pathway. Ptc1 mRNA was quantified by real-time PCR. KIF3A knock-down attenuates the increase in ptc1 expression following SAG stimulation
Knocking down KIF3A modifies cilium morphology and functionality
Inactivation of KIF3A, one of the primary cilium’s building blocks [13, 17], results in deletion [31] or significant reduction [25] in the number of primary cilium. In growth-plate chondrocytes, inactivation of KIF3A results in reduced Ihh expression [28].
To understand the role of the primary cilium in the chondrocytes’ response to mechanical load, we partially knocked-down KIF3A using transient transfection of shRNAi for Kif3A (shKIF3A). We used a construct that knocked-down KIF3A while simultaneously expressing GFP reporter gene, so that cells in which KIF3A was silenced appeared green. 48 h post transfection we show in Fig. 6a depletion of KIF3A protein in cells transfected with two shKIF3A vectors. Although protein levels were dramatically reduced, the presence of primary cilium was not abolished from the transfected cells (Fig. 7b, c), but only moderately (albeit significantly) reduced in cells transfected with the shKIF3A-2 vector (Fig. 7b).
However, functionality of the primary cilium was indeed damaged by KIF3A silencing. This was demonstrated by the altered response of the cells to SAG stimulation: while the control cells (nontransfected and empty vector) showed an increase in ptc1 expression following SAG treatment (Fig. 7d), the response of the shKIF3A cells to SAG stimulation was markedly inhibited (Fig. 7d).
KIF3A is involved in the chondrocytic response to mechanical stimulation
Finally, we wanted to characterize the role of the primary cilium in the ATDC5 chondrocyte’s response to mechanical stimulation. We transfected cells with either shKIF3A or control vectors; 48 h post-transfection, mechanical stimulation was applied. KIF3A reduction was validated by the significantly lower protein-expression levels compared to control cells (Fig. 8a). Next, a panel of mechano-sensitive genes was examined by extensive real-time PCR analysis, to: (1) characterize their response to mechanical stimulation in chondrocytes, (2) differentiate these responses in cells with normal KIF3A expression vs. silenced KIF3A. The examined genes included chondrocyte-related genes, mechanical force-related genes and primary cilium-related genes. Based on our results, we clustered genes according to their pattern of KIF3A dependence (Fig. 8b). Four different response clusters were characterized:
KIF3A is involved in the chondrocytic response to mechanical stimulation. a KIF3A protein levels were determined by western blot analysis. b Mechanical stimulation was applied on control (empty) and KIF3A knock-down cells. mRNA expression of mechano-sensitive genes was quantified using real-time PCR. The genes were divided to 4 clusters depending on their dependence on KIF3A
-
1.
the KIF3A-independent mechano-sensing cluster the response of EGR1 to mechanical stimulation was unaffected by KIF3A suppression;
-
2.
the partially KIF3A-dependent mechano-sensing cluster the response pattern of FOS, RUNX2, collagen type II, and ATF3 to mechanical stimulation was not affected by KIF3A knockdown. However the magnitude of the response changed;
-
3.
the KIF3A-dependent mechano-sensing cluster this cluster included AGC1, Ihh, collagen type X and p21 genes, whose response to mechanical stimulation was abolished following KIF3A suppression;
-
4.
the KIF3A ‘switching’ mechano-sensing cluster The response pattern of OPN, PKD1, PKD2, IFT88, IFT172, and SOX9 to mechanical stimulation switched to the opposite direction following KIF3A suppression.
Taken together, these results suggested that normal KIF3A levels are necessary for both maintenance of primary cilium function and facilitation of mechanical-load signaling in chondrocytes. Moreover, disrupted KIF3A levels and subsequently damaged cilium functionality resulted in altered response of the chondrocytes to mechanical load.
Discussion
In this study, we show for the first time that mechanical load up-regulates primary cilium number in the chicken growth plate concomitant with altered chondrogenesis. Moreover, we demonstrate that KIF3A is necessary for primary cilium function and its ablation hampers the chondrocyte’s response to mechanical load by altering the expression of mechano-sensitive as well as chondrocyte-specific genes. Chondrocytes, osteoblasts and osteoclasts are constantly exposed to physical forces that modulate their cellular phenotype and gene expression during development and postnatal growth. During normal physical activities, the cartilage, as a tissue, experience compression as the primary mechanical load. At the cellular level, the compression converts to various mechanical loads including direct compression, hydrostatic pressure, shear and tensile loading, interstitial fluid flow, electrokinetic effects, and osmotic pressure [12, 65]. At the cellular level, these biomechanical strains, together with biological factors (e.g., growth factors, cytokines etc.), play important roles in modulating chondrocytes physiology, metabolism, and response to external loads. Numerous studies have investigated the effects of increased or decreased load on the structure of mature bones [11, 48, 61]. However, only a few studies have described the effect of mechanical load on bone and growth-plate development at a young age, during the rapid growth phase. In recent years, this has been a major focus of our group [56–58]. We used a unique loading protocol which mimics physiological loading pattern to apply a moderate supraphysiological load, consisting of 10 % of the young chickens’ body weight. We have shown that even at low magnitude, addition of load reduces the length and diameter of the long bones (femur and tibia). This load also results in a narrower growth plate, while mineralization and ossification are increased. Here we show that mechanical load increases ciliogenesis, which leads to alteration and probably malfunction of the Ihh–PTHrP loop in the growth plate. This is followed by decreased chondrocyte proliferation and irregularities in the switch between proliferation and differentiation. Furthermore, the increased number of primary cilium results in higher organization of the chondrocyte columns. In vitro, we mimicked the cellular micro-environment experiencing shear as well as tensile and compression stresses: tensile load is induced during stretching while compression load is induced during release from stretch. Both acts induce fluid flow, thus creating shear load. Taken together, the combination of these three types of load represents the nature of loads experienced by growth-plate chondrocytes. Using this platform, we demonstrate the requirement of primary cilium in the chondrocyte’s response to mechanical stimulation, and show the role of KIF3A in mediating gene expression in response to load.
The process of mechano-transduction in chondrocytes is not fully understood. The primary cilium’s unique characteristics make this organelle a potential player in receiving and mediating a variety of signals into the cell. Moreover, different mechano-receptors, such as integrins and calcium channels, have been found to localize in the primary cilium [27, 36, 54, 66]. Here we show that mechanical load increases ciliogenesis in the growth plates in vivo and the expression of cilium-related genes in chondrocytes in vitro. Cell organization in cartilage elements is directly linked to primary cilium [8, 72], and its disruption results in impaired ciliogenesis accompanied by misoriented cells in the growth plate [28, 37]. We show that mechanical loading results in the reciprocal phenotype: increased ciliogenesis, increased expression of cilium-related genes and increased cell organization. This provides further evidence for the primary cilium’s involvement in the growth plate’s response to mechanical load.
The connection of the primary cilium to the cell cycle and proliferation is well established [50]. In normal proliferating cells, the presence of cilia on the cell surface is most commonly observed in the G1 phase [59]. Moreover, cell-cycle regulators have been found to control the formation or loss of cilia [50] and vice versa, some of the IFT proteins, long thought to be relevant solely to protein trafficking within the cilium, have recently been found to function in cell-cycle control. For example, over-expression of IFT88 prevents the G1–S transition, whereas depletion of IFT88 causes cilium disappearance and promotes cell-cycle progression [60]. Therefore, it is reasonable to suggest that the observed decrease in chondrocyte proliferation as a result of mechanical loading is also connected to the primary cilium. However, it is unclear whether the reduced proliferation enables the appearance of primary cilium, or conversely, the increased ciliogenesis affects the cell cycle.
One possible explanation is that the changes in cell proliferation are also associated with the alterations in the Ihh–PTHrP loop observed in the loaded growth plates. This loop is a master regulator of bone development, coordinating chondrocyte proliferation and differentiation [23, 73]. Ihh is considered an essential mediator of mechanotransduction in cartilage; its expression is highly induced by cyclic mechanical stress in proliferative sternal chondrocytes [87], as well as in mandibular condylar cartilage where it was shown to convert mechanical strain into cellular proliferation and cartilage formation [43], [75]. Recently, the primary cilium was shown to modulate Ihh signal transduction in response to hydrostatic loading of growth plate chondrocytes [67]. In vivo, the mechanical load altered the expression and localization of Ihh and its receptor ptc1 as well as PTHrP, which is downstream of Ihh, thereby modifying the functionality of the Ihh–PTHrP loop in the growth plate. This is probably the main reason for the decreased proliferation rate and abnormal switch between proliferation and differentiation induced by the load. Again, this also suggests involvement of the primary cilium, given that Ihh signaling is mediated by this organelle [24].
In our in vitro model, we used this fact to prove that knockdown of KIF3A damages the functionality of the cilium: in control cells, mechanical stimulation induced Ihh expression, and activation of the pathway by SAG stimulation increased the expression of ptc1 and its accumulation in the cilium. Knockdown of KIF3A abolished these responses. This gave us the opportunity to study the role of KIF3A (and consequently of the primary cilium) in mechano-signaling by comparing the response of wild-type and damaged cilium cells to mechanical stimulation. By examining a panel of genes, we were able to establish four clusters which differ in their dependence on the cilium (i.e. KIF3A) for a normal response to load. It is important to note that although we focused on the cells’ immediate response in 2D culture conditions. This approach is not common in mechanical stimulation experiments, for two main reasons: first most of these experiments are aimed at discovering a phenotype for this manipulation, such as cell differentiation; second, the commonly used systems, such as 3D approaches, are too complex for an examination of the short-term kinetics of mechanical stimulation [30]. However, the response to mechanical stimulation is significantly affected by cell shape which is in turn dictated by culture conditions i.e. 3D. Despite these limitation we still believe that investigation of the immediate response to mechanical stimulation in 2D culture can shed light on the role of the primary cilium in mechano-transduction pathways.The four clusters described the extent of KIF3A’s involvement in chondrocytes’ response to mechanical stimulation. Clusters 1 and 2 represent genes which respond to mechanical stimulation but are KIF3A-independent. These genes are probably regulated through other mechanisms, such as the MEK1/2 signaling pathway [45]. Alternatively, these changes might be related to the changes observed in the cytoskeleton following mechanical stimulation. The cytoskeleton is vastly involved in mechanotransduction; forces can be applied directly through the ECM or transmitted through the cytoskeleton to mechanosensitive components to mediate cellular response which can result in either reinforcement or fluidization of the cytoskeleton (Reviewed by Hoffman et al. [19]. Here we have shown that mechanical stimulation of chondrocytes resulted in changes in F-actin and formation of stress fibers along the strain axis. Interestingly, primary cilium length is also regulated through changes in either the actin or microtubule network of the cytoskeleton [68], and loss of ciliary genes such as IFT88 and KIF3A leads to hyper-acetylation of cytosolic microtubules [3]. Clusters 3 and 4 also respond to mechanical stimulation, but their response is KIF3A-dependent. Upon KIF3A knockdown, genes in cluster 3 abolish the response whereas those in cluster 4 reverse the response. Interestingly, cluster 3 includes mostly “chondrocytic” genes. AGC1 and collagen type X are two major ECM proteins in the growth plate. They are both highly up-regulated in response to mechanical stimulation in control cells, probably to strengthen the cell environment [86]. Ihh is a key component in chondrogenesis [82], and is highly up-regulated following mechanical stimulation [87]. Here we show that this regulation is KIF3A-dependent. In addition, we show that p21’s response to load is also KIF3A-dependent. Cell-cycle arrest has been shown to be one of the phenotypes of mechanical stimulation [33]. Moreover, it is well known that the primary cilium is connected to cell-cycle status [50]. Hence, it is not surprising that the response of this cell-cycle inhibitor to mechanical loading is KIF3A-dependent. Interestingly, we show that regulation of the primary cilium-related genes PKD1, PKD2, IFT88, and IFT172 is KIF3A-dependent. This proves that once a key protein such as KIF3A is disrupted, other key ciliary genes are down-regulated. The reduction in cilium number after KIF3A knockdown might be mediated by this mechanism. However further studies are required to confirm this.
To conclude, this work proves that the primary cilium is involved in growth-plate chondrocytes’ response to mechanical load both in vivo and in vitro. Key components of this organelle mediate gene expression, which ultimately alters cell proliferation and differentiation, as well as cell morphology and organization in the growth plate.
References
Atsumi T, Miwa Y, Kimata K, Ikawa Y (1990) A chondrogenic cell line derived from a differentiating culture of AT805 teratocarcinoma cells. Cell Differ Dev 30(2):109–116
Berbari NF, O’Connor AK, Haycraft CJ, Yoder BK (2009) The primary cilium as a complex signaling center. Curr Biol 19(13):R526–R535
Berbari NF, Sharma N, Malarkey EB, Pieczynski JN, Boddu R, Gaertig J, Guay-Woodford L, Yoder BK (2013) Microtubule modifications and stability are altered by cilia perturbation and in cystic kidney disease. Cytoskeleton (Hoboken) 70(1):24–31
Blain EJ (2009) Involvement of the cytoskeletal elements in articular cartilage homeostasis and pathology. Int J Exp Pathol 90(1):1–15
Challa TD, Rais Y, Ornan EM (2010) Effect of adiponectin on ATDC5 proliferation, differentiation and signaling pathways. Mol Cell Endocrinol 323(2):282–291
Chen JK, Taipale J, Young KE, Beachy PA (2002) Small molecule modulation of smoothened activity. Proc Natl Acad Sci USA 99(22):14071–14076
Dagoneau N, Goulet M, Genevieve D, Sznajer Y, Martinovic J, Smithson S, Huber C, Baujat G, Flori E, Tecco L, Cavalcanti D, Delezoide AL, Serre V, Le Merrer M, Munnich A, Cormier-Daire V (2009) DYNC2H1 mutations cause asphyxiating thoracic dystrophy and short rib-polydactyly syndrome, type III. Am J Hum Genet 84(5):706–711
de Andrea CE, Wiweger M, Prins F, Bovee JV, Romeo S, Hogendoorn PC (2010) Primary cilia organization reflects polarity in the growth plate and implies loss of polarity and mosaicism in osteochondroma. Lab Invest 90(7):1091–1101
Ehlen HW, Buelens LA, Vortkamp A (2006) Hedgehog signaling in skeletal development. Birth Defects Res C Embryo Today 78(3):267–279
Epari DR, Taylor WR, Heller MO, Duda GN (2006) Mechanical conditions in the initial phase of bone healing. Clin Biomech (Bristol, Avon) 21(6):646–655
Goodship AE, Lanyon LE, McFie H (1979) Functional adaptation of bone to increased stress: an experimental study. J Bone Joint Surg Am 61(4):539–546
Guilak F, Alexopoulos LG, Upton ML, Youn I, Choi JB, Cao L, Setton LA, Haider MA (2006) The pericellular matrix as a transducer of biomechanical and biochemical signals in articular cartilage. Ann N Y Acad Sci 1068:498–512
Haraguchi K, Hayashi T, Jimbo T, Yamamoto T, Akiyama T (2006) Role of the kinesin-2 family protein, KIF3, during mitosis. J Biol Chem 281(7):4094–4099
Hasky-Negev M, Simsa S, Tong A, Genina O, Monsonego Ornan E (2008) Expression of matrix metalloproteinases during vascularization and ossification of normal and impaired avian growth plate. J Anim Sci 86(6):1306–1315
Haycraft CJ, Zhang Q, Song B, Jackson WS, Detloff PJ, Serra R, Yoder BK (2007) Intraflagellar transport is essential for endochondral bone formation. Development 134(2):307–316
Haycraft CJ, Serra R (2008) Cilia involvement in patterning and maintenance of the skeleton. Curr Top Dev Biol 85:303–332
Hirokawa N (2000) Determination of left-right asymmetry: role of Cilia and KIF3 motor proteins. News Physiol Sci 15:56
Hoey DA, Tormey S, Ramcharan S, O'Brien FJ, Jacobs CR (2012) Primary cilia-mediated mechanotransduction in human mesenchymal stem cells. Stem Cells 30(11):2561–2570
Hoffman BD, Grashoff C, Schwartz MA (2011) Dynamic molecular processes mediate cellular mechanotransduction. Nature 475(7356):316–323
Idelevich A, Rais Y, Monsonego-Ornan E (2012) Bone Gla protein increases HIF-1alpha-dependent glucose metabolism and induces cartilage and vascular calcification. Arterioscler Thromb Vasc Biol 31(9):e55–e71
Jacobs CR, Temiyasathit S, Castillo AB (2010) Osteocyte mechanobiology and pericellular mechanics. Annu Rev Biomed Eng 12:369–400
Jensen CG, Poole CA, McGlashan SR, Marko M, Issa ZI, Vujcich KV, Bowser SS (2004) Ultrastructural, tomographic and confocal imaging of the chondrocyte primary cilium in situ. Cell Biol Int 28(2):101–110
Karp SJ, Schipani E, St-Jacques B, Hunzelman J, Kronenberg H, McMahon AP (2000) Indian hedgehog coordinates endochondral bone growth and morphogenesis via parathyroid hormone related-protein-dependent and -independent pathways. Development 127(3):543–548
Kim J, Kato M, Beachy PA (2009) Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci USA 106(51):21666–21671
Kinumatsu T, Shibukawa Y, Yasuda T, Nagayama M, Yamada S, Serra R, Pacifici M, Koyama E (2011) TMJ development and growth require primary cilia function. J Dent Res 90(8):988–994
Kolpakova-Hart E, Nicolae C, Zhou J, Olsen BR (2008) Col2-Cre recombinase is co-expressed with endogenous type II collagen in embryonic renal epithelium and drives development of polycystic kidney disease following inactivation of ciliary genes. Matrix Biol 27(6):505–512
Kottgen M, Buchholz B, Garcia-Gonzalez MA, Kotsis F, Fu X, Doerken M, Boehlke C, Steffl D, Tauber R, Wegierski T, Nitschke R, Suzuki M, Kramer-Zucker A, Germino GG, Watnick T, Prenen J, Nilius B, Kuehn EW, Walz G (2008) TRPP2 and TRPV4 form a polymodal sensory channel complex. J Cell Biol 182(3):437–447
Koyama E, Young B, Nagayama M, Shibukawa Y, Enomoto-Iwamoto M, Iwamoto M, Maeda Y, Lanske B, Song B, Serra R, Pacifici M (2007) Conditional Kif3a ablation causes abnormal hedgehog signaling topography, growth plate dysfunction, and excessive bone and cartilage formation during mouse skeletogenesis. Development 134(11):2159–2169
Li S, Duance VC, Blain EJ (2008) Zonal variations in cytoskeletal element organization, mRNA and protein expression in the intervertebral disc. J Anat 213(6):725–732
Li J, Zhao Z, Yang J, Liu J, Wang J, Li X, Liu Y (2009) p38 MAPK mediated in compressive stressinduced chondrogenesis of rat bone marrow MSCs in 3D alginate scaffolds. J Cell Physiol 221(3):609–617
Lin F, Hiesberger T, Cordes K, Sinclair AM, Goldstein LS, Somlo S, Igarashi P (2003) Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc Natl Acad Sci USA 100(9):5286–5291
Liu A, Wang B, Niswander LA (2005) Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development 132(13):3103–3111
Luo W, Xiong W, Zhou J, Fang Z, Chen W, Fan Y, Li F (2011) Laminar shear stress delivers cell cycle arrest and anti-apoptosis to mesenchymal stem cells. Acta Biochim Biophys Sin (Shanghai) 43(3):210–216
Malone AM, Anderson CT, Tummala P, Kwon RY, Johnston TR, Stearns T, Jacobs CR (2007) Primary cilia mediate mechanosensing in bone cells by a calcium-independent mechanism. Proc Natl Acad Sci USA 104 (33):13325–13330
Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS (1999) Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci USA 96(9):5043–5048
McGlashan SR, Jensen CG, Poole CA (2006) Localization of extracellular matrix receptors on the chondrocyte primary cilium. J Histochem Cytochem 54(9):1005–1014
McGlashan SR, Haycraft CJ, Jensen CG, Yoder BK, Poole CA (2007) Articular cartilage and growth plate defects are associated with chondrocyte cytoskeletal abnormalities in Tg737orpk mice lacking the primary cilia protein polaris. Matrix Biol 26(4):234–246
McGlashan SR, Knight MM, Chowdhury TT, Joshi P, Jensen CG, Kennedy S, Poole CA (2010) Mechanical loading modulates chondrocyte primary cilia incidence and length. Cell Biol Inter 34(5):441–446
Meier-Vismara E, Walker N, Vogel A (1979) Single cilia in the articular cartilage of the cat. Exp Cell Biol 47(3):161–171
Muhammad H, Rais Y, Miosge N, Ornan EM (2012) The primary cilium as a dual sensor of mechanochemical signals in chondrocytes. Cell Mol Life Sci 69(13):2101–2107
Murcia NS, Richards WG, Yoder BK, Mucenski ML, Dunlap JR, Woychik RP (2000) The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development 127(11):2347–2355
Nauli SM, Zhou J (2004) Polycystins and mechanosensation in renal and nodal cilia. BioEssays 26(8):844–856
Ng TC, Chiu KW, Rabie AB, Hagg U (2006) Repeated mechanical loading enhances the expression of Indian hedgehog in condylar cartilage. Front Biosci 11:943–948
Olsen B (2005) Nearly all cells in vertebrates and many cells in invertebrates contain primary cilia. Matrix Biol 24(7):449–450
Ott CE, Bauer S, Manke T, Ahrens S, Rodelsperger C, Grunhagen J, Kornak U, Duda G, Mundlos S, Robinson PN (2009) Promiscuous and depolarization-induced immediate-early response genes are induced by mechanical strain of osteoblasts. J Bone Miner Res 24(7):1247–1262
Papachristou DJ, Papachroni KK, Basdra EK, Papavassiliou AG (2009) Signaling networks and transcription factors regulating mechanotransduction in bone. BioEssays 31(7):794–804
Pazour GJ, Witman GB (2003) The vertebrate primary cilium is a sensory organelle. Curr Opin Cell Biol 15(1):105–110
Pearson SJ, Cobbold M, Harridge SD (2004) Power output of the lower limb during variable inertial loading: a comparison between methods using single and repeated contractions. Eur J Appl Physiol 92(1-2):176–181
Pedersen LB, Veland IR, Schroder JM, Christensen ST (2008) Assembly of primary cilia. Dev Dyn 237(8):1993–2006
Plotnikova OV, Golemis EA, Pugacheva EN (2008) Cell cycle-dependent ciliogenesis and cancer. Cancer Res 68(7):2058–2061
Poole CA, Flint MH, Beaumont BW (1984) Morphological and functional interrelationships of articular cartilage matrices. J Anat 138(Pt 1):113–138
Poole CA, Flint MH, Beaumont BW (1985) Analysis of the morphology and function of primary cilia in connective tissues: a cellular cybernetic probe? Cell Motil 5(3):175–193
Poole CA, Zhang ZJ, Ross JM (2001) The differential distribution of acetylated and detyrosinated alpha-tubulin in the microtubular cytoskeleton and primary cilia of hyaline cartilage chondrocytes. J Anat 199 (Pt4):393–405
Praetorius HA, Leipziger J (2012) Primary cilium-dependent sensing of urinary flow and paracrine purinergic signaling. Semin Cell Dev Biol 24(1):3–10
Praetorius HA, Spring KR (2003) The renal cell primary cilium functions as a flow sensor. Curr Opin Nephrol Hypertens 12(5):517–520
Reich A, Jaffe N, Tong A, Lavelin I, Genina O, Pines M, Sklan D, Nussinovitch A, Monsonego-Ornan E (2005) Weight loading young chicks inhibits bone elongation and promotes growth plate ossification and vascularization. J Appl Physiol 98(6):2381–2389
Reich A, Sharir A, Zelzer E, Hacker L, Monsonego-Ornan E, Shahar R (2008) The effect of weight loading and subsequent release from loading on the postnatal skeleton. Bone 43(4):766–774
Reich A, Maziel SS, Ashkenazi Z, Ornan EM (2010) Involvement of matrix metalloproteinases in the growth plate response to physiological mechanical load. J Appl Physiol 108(1):172–180
Rieder CL, Jensen CG, Jensen LC (1979) The resorption of primary cilia during mitosis in a vertebrate (PtK1) cell line. J Ultrastruct Res 68(2):173–185
Robert A, Margall-Ducos G, Guidotti JE, Bregerie O, Celati C, Brechot C, Desdouets C (2007) The intraflagellar transport component IFT88/polaris is a centrosomal protein regulating G1-S transition in non-ciliated cells. J Cell Sci 120(Pt 4):628–637
Rubin CT, Lanyon LE (1984) Regulation of bone formation by applied dynamic loads. J Bone Joint Surg Am 66(3):397–402
Ruhlen R, Marberry K (2014) The chondrocyte primary cilium. Osteoarthr Cartil S1063–4584(14):01090–01095
Sanz-Ramos P, Mora G, Ripalda P, Vicente-Pascual M, Izal-Azcarate I (2012) Identification of signalling pathways triggered by changes in the mechanical environment in rat chondrocytes. Osteoarthr Cartil 20(8):931–939
Scholey JM, Anderson KV (2006) Intraflagellar transport and cilium-based signaling. Cell 125(3):439–442
Schulz RM, Bader A (2007) Cartilage tissue engineering and bioreactor systems for the cultivation and stimulation of chondrocytes. Eur Biophys J 36(4-5):539–568
Seeger-Nukpezah T, Golemis EA (2012) The extracellular matrix and ciliary signaling. Curr Opin Cell Biol 24(5):652–661
Shao YY, Wang L, Welter JF, Ballock RT (2012) Primary cilia modulate Ihh signal transduction in response to hydrostatic loading of growth plate chondrocytes. Bone 50(1):79–84
Sharma N, Kosan ZA, Stallworth JE, Berbari NF, Yoder BK (2011) Soluble levels of cytosolic tubulin regulate ciliary length control. Mol Biol Cell 22(6):806–816
Simsa-Maziel S, Monsonego-Ornan E (2012) Interleukin-1beta promotes proliferation and inhibits differentiation of chondrocytes through a mechanism involving down-regulation of FGFR-3 and p21. Endocrinology 153(5):2296–2310
Simsa S, Ornan EM (2007) Endochondral ossification process of the turkey (Meleagris gallopavo) during embryonic and juvenile development. Poult Sci 86(3):565–571
Sloboda RD, Rosenbaum JL (2007) Making sense of cilia and flagella. J Cell Biol 179(4):575–582
Song B, Haycraft CJ, Seo HS, Yoder BK, Serra R (2007) Development of the post-natal growth plate requires intraflagellar transport proteins. Dev Biol 305(1):202–216
St-Jacques B, Hammerschmidt M, McMahon AP (1999) Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev 13(16):2072–2086
Tanaka SM, Sun HB, Roeder RK, Burr DB, Turner CH, Yokota H (2005) Osteoblast responses one hour after load-induced fluid flow in a three-dimensional porous matrix. Calcif Tissue Int 76(4):261–271
Tang GH, Rabie AB, Hagg U (2004) Indian hedgehog: a mechanotransduction mediator in condylar cartilage. J Dent Res 83(5):434–438
Temiyasathit S, Tang WJ, Leucht P, Anderson CT, Monica SD, Castillo AB, Helms JA, Stearns T, Jacobs CR (2012) Mechanosensing by the primary cilium: deletion of Kif3A reduces bone formation due to loading. PLoS One 7(3):e33368
Terai K, Takano-Yamamoto T, Ohba Y, Hiura K, Sugimoto M, Sato M, Kawahata H, Inaguma N, Kitamura Y, Nomura S (1999) Role of osteopontin in bone remodeling caused by mechanical stress. J Bone Miner Res 14(6):839–849
Thompson CL, Chapple JP, Knight MM (2014) Primary cilia disassembly down-regulates mechanosensitive hedgehog signalling: a feedback mechanism controlling ADAMTS-5 expression in chondrocytes. Osteoarthr Cartil 22(3):490–498
Tong A, Reich A, Genin O, Pines M, Monsonego-Ornan E (2003) Expression of chicken 75-kDa gelatinase B-like enzyme in perivascular chondrocytes suggests its role in vascularization of the growth plate. J Bone Miner Res 18(8):1443–1452
van der Eerden BC, Karperien M, Gevers EF, Lowik CW, Wit JM (2000) Expression of Indian hedgehog, parathyroid hormone-related protein, and their receptors in the postnatal growth plate of the rat: evidence for a locally acting growth restraining feedback loop after birth. J Bone Miner Res 15(6):1045–1055
Vortkamp A (2000) The Indian hedgehog–PTHrP system in bone development. Ernst Schering Res Found Workshop 29:191–209
Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ (1996) Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 273(5275):613–622
Wann AK, Zuo N, Haycraft CJ, Jensen CG, Poole CA, McGlashan SR, Knight MM (2012) Primary cilia mediate mechanotransduction through control of ATP-induced Ca2+ signaling in compressed chondrocytes. Faseb J 26(4):1663–1671
Wheatley DN, Bowser SS (2000) Length control of primary cilia: analysis of monociliate and multiciliate PtK1 cells. Biol Cell 92(8-9):573–582
Wilsman NJ, Farnum CE, Reed-Aksamit DK (1980) Incidence and morphology of equine and murine chondrocytic cilia. Anat Rec 197(3):355–361
Wu Q, Zhang Y, Chen Q (2001) Indian hedgehog is an essential component of mechanotransduction complex to stimulate chondrocyte proliferation. J Biol Chem 276(38):35290–35296
Wu QQ, Chen Q (2000) Mechanoregulation of chondrocyte proliferation, maturation, and hypertrophy: ion-channel dependent transduction of matrix deformation signals. Exp Cell Res 256(2):383–391
Acknowledgments
We would like to thank our technician Svetlana Penn for technical assistance. This research was supported by the Israel Science Foundation (Grant No. 292/07) and by the United States–Israel Binational Science Foundation (BSF) (Grant No. 2011393).
Author information
Authors and Affiliations
Corresponding author
Additional information
Y. Rais and A. Reich are these authors contributed equally to the paper.
Rights and permissions
About this article
Cite this article
Rais, Y., Reich, A., Simsa-Maziel, S. et al. The growth plate’s response to load is partially mediated by mechano-sensing via the chondrocytic primary cilium. Cell. Mol. Life Sci. 72, 597–615 (2015). https://doi.org/10.1007/s00018-014-1690-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-014-1690-4