Abstract
In the mammalian retina, light signals generated in photoreceptors are passed to bipolar and horizontal cells via synaptic contacts. In various pathological conditions, these second-order neurons extend neurites into the outer nuclear layer (ONL). However, the molecular events associated with this neurite outgrowth are not known. Here, we characterized the morphological synaptic changes in the CNGA3/CNGB1 double-knockout (A3B1) mouse, a model of retinitis pigmentosa. In these mice, horizontal cells looked normal until postnatal day (p) 11, but started growing neurites into the ONL 1 day later. At p28, the number of sprouting processes decreased, but the remaining sprouts developed synapse-like contacts at rod cell bodies, with an ultrastructural appearance reminiscent of ribbon synapses. Hence, neurite outgrowth and ectopic synaptogenesis in the A3B1 retina were precisely timed events starting at p12 and p28, respectively. We therefore performed microarray analysis of retinal gene expression in A3B1 and wild-type mice at those ages to evaluate the genomic response underlying these two events. This analysis identified 163 differentially regulated genes in the A3B1 retina related to neurite outgrowth or plasticity of synapses. The global changes in gene expression in the A3B1 retina were consistent with activation of signaling pathways related to Tp53, Smad, and Stat3. Moreover, key molecules of these signaling pathways could be localized at or in close proximity to outgrowing neurites. We therefore propose that Tp53, Smad, and Stat3 signaling pathways contribute to the synaptic plasticity in the A3B1 retina.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mammalian retinal neurons are arranged in three distinct nuclear layers interconnected by two synaptic (plexiform) layers. Multiple chemical and electrical synaptic contacts within the plexiform layers establish a complex neural signaling network that processes and transfers light information from the photoreceptors to the retinal ganglion cells. In the first plexiform layer (outer plexiform layer, OPL), rod and cone photoreceptors connect with bipolar cells (BCs) and horizontal cells (HCs) in a highly precise manner. Rods synapse onto rod bipolar cell (RBC) dendrites and HC axons, whereas cones form synaptic contacts with cone bipolar cells and HC dendrites [1]. Although synaptogenesis in the OPL was described in detail in several anatomical studies [2, 3], the mechanisms that allow for the emergence of this specific synaptic network are still unknown. Light-driven activity seems to be of minor relevance for the establishment of OPL synapses, since they are formed prior to development of photoreceptor outer segments [2, 3]. Furthermore, mice lacking rod and cone function develop normal synaptic contacts in the OPL [4]. Interestingly, both BCs and HCs respond to a number of pathological conditions by extending sprouting processes into the outer nuclear layer (ONL). This behavior can be caused by a number of stimuli, ranging from retinal detachment [5] to aging [6]. Neurite outgrowth in the ONL is also common in mutant mice lacking functional photoreceptors [4, 7] or bassoon, a protein known to be important for ribbon synapse formation [8]. Similarly, mouse mutants with reduced neurotransmission at the photoreceptor synapse, caused by either loss or functional defects of the L-type voltage-gated calcium channel, show extensive outgrowth of RBC and HC neurites and ectopic synaptogenesis [9–13].
Here, we set out to characterize neurite outgrowth in the pathologic retina and to identify genes that could underlie this behavior. We combined neuroanatomical methods with microarray-based gene expression analysis and bioinformatics analysis to study the neurite outgrowth of retinal neurons in response to a lack of photoreceptor function in the CNGA3/CNGB1 double-knockout (A3B1) mouse model of retinitis pigmentosa (RP). These mice lack the CNGA3 and CNGB1 proteins, essential subunits of the cone and rod cyclic nucleotide-gated channels, respectively [7, 14]. We identified 163 differentially regulated genes in the A3B1 retina related to neurite outgrowth or synapse plasticity. Among those genes, we found 64 that map to the “axonal guidance signaling pathway”. In addition, our analysis identified significant up-regulation of Tp53, as well as Smad and Stat3-related signaling in the A3B1 retina at key time points for neurites outgrowth and ectopic synapse formation.
Materials and methods
Animals
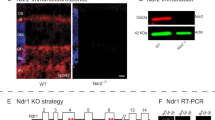
CNGA3/CNGB1 double-knockout (A3B1) mice were generated by cross-breeding CNGA3−/− mice [14] with CNGB1−/− mice [7] on a mixed C57BL/6 N × 129S2/SvHsd genetic background. The resulting A3B1s in the F2 generation were identified by PCR. Age-matched wild-type mice were used for comparison. All experiments were approved by the local animal care committee and were in accordance with the law for animal experiments issued by the German government (Tierschutzgesetz). For the light microscopic studies, in total 64 mice with an age of 9–44 days were examined under various lighting conditions (in general, at least two animals each age and genotype). In the dark-rearing experiments, mice were born and weaned under constant darkness, mice exposed to a normal light/dark (LD) cycle of 12:12 h served as controls (lights on at 6 a.m. and off at 6 p.m.; 100 lux at the bottom of the cages). Mice were kept under constant laboratory conditions with free access to food and water. For removal of tissue, they were anesthetized with isoflurane and killed by decapitation, during dark conditions under dim red light. For the electron microscopical evaluation of the ectopic synapses in the outer retina, in total 12 mice (wild type and A3B1) with an age of 29–44 days were examined.
Immunohistochemistry
Eyes were enucleated after decapitation, the anterior segments were removed and the posterior eyecups immersion-fixed for 15–60 min in 4 % paraformaldehyde in 0.1 M phosphate buffer, pH 7.4 (PB) at room temperature. Fixed eyecups were rinsed in PB and cryoprotected in graded sucrose solutions (10, 20, 30 %). Retinas were dissected from the eyecups, and sectioned vertically at 14 μm using a cryostat. Sections were collected on Super-Frost glass slides and stored at −20 °C until use.
Immunolabeling was carried out using the indirect fluorescence method. Horizontal cells were labeled with antibodies against calbindin (either rabbit anti-calbindin; 1:2000; Swant, Bellinzona, Switzerland, or mouse anti-calbindin; 1:1000; Sigma, St. Louis, MO, USA) [15]. Synaptic ribbons were visualized with an antibody against the C-terminal binding protein 2 (mouse anti-CtBP2; 1:5000; BD Transduction, Heidelberg, Germany) [16]. Rod bipolar cells were immunostained with an antibody against protein kinase C alpha (biotinylated mouse anti-PKCα; 1:20; Leinco Technologies, St. Louis, MO, USA) and visualized with Dy-555-Avidin (1:3000; Dyomics, Jena, Germany). Other antibodies used were: guinea pig anti-mGluR6 (1:200, GP13105, Neuromics, Edina, MN, USA), goat anti-Lgals3 (1:300, sc-19283, Santa Cruz Biotechnologies, Heidelberg, Germany), rat anti-Plxnc1 (1:200, M652, Amgen, Seattle, WA, USA) [17], rabbit anti-phospho-Smad1/5/8 (1:300, #9511, Cell Signaling Technology), rabbit anti-phospho-Src (Tyr416) (1:20, #2101, Cell Signaling Technology, Danvers, MA, USA), rabbit anti-phospho-Stat3 (Tyr705) (1:100, #9145, Cell Signaling Technology) and rabbit anti-TGFβ1 (1:200, G122A, Promega, Mannheim, Germany).
Sections were incubated overnight with primary antibodies in 5 % Chemiblocker (Chemicon, Hofheim, Germany) or 3 % normal goat serum, 0.5 % Triton X-100 and 0.05 % sodium azide in PB. After washing in PB, secondary antibodies were applied for 1 h. These were conjugated either to Cy3 (Dianova, Hamburg, Germany) or Alexa TM 488 (Molecular Probes, Eugene, OR, USA).
Samples were imaged on a Zeiss LSM5 Pascal or LSM510Meta confocal microscope (Zeiss, Oberkochen, Germany) equipped with argon and helium–neon lasers. Optical sections were acquired with a 40×/1.3 Plan-Neofluar objective, and projections of stacks of 3–6 μm were used for figures. Brightness and contrast of the final images were adjusted using Adobe Photoshop. The imaging conditions (laser intensity, pinhole diameter, and detector gain) and digital processing were identical for all images within a series/experiment.
Density (average number of aberrant sprouts ± SEM) and total length of sprouts (in μm ± SEM) were quantified per confocal stack volume (230.34 × 89.98 × 9 μm). An unpaired Student’s t test was performed for the comparison between groups.
To estimate the co-localization of phospho-Src and calbindin signals within growing neurites in the ONL, we evaluated the overlap of the two signals with Pearson’s correlation coefficients [18] from confocal images using the Histology Tool of the LSM 510 v4.0SP1 Software (Zeiss).
Western blot
For protein isolation from mouse retinas, the retinas were homogenized using a mortar and suspended in homogenization buffer (2 % SDS, 50 mM Tris, and proteinase inhibitor cocktail mix). After heating at 95 °C for 15 min followed by centrifugation at 1,000 × g for 10 min at 4 °C, the resulting supernatant was used in Western-blot analysis. The proteins were separated by 10 % SDS-PAGE followed by Western-blot analysis according to standard procedures. The following antibodies were used: goat anti-Lgals3 (1:25, sc-19283, Santa Cruz Biotechnologies), rabbit anti-phospho-Smad1/5/8 (1:1000, #9511, Cell Signaling Technology), rabbit anti-phospho-Src (Tyr416) (1:1000, #2101, Cell Signaling Technology), rabbit anti-pwildthospho-Stat3 (Tyr705) (1:500, #9145, Cell Signaling Technology) and rabbit anti-TGFβ1 (1:1000, G122A, Promega) and mouse anti-Tubulin (1:2000, Dianova, Hamburg, Germany). All antibodies detected distinct bands at the predicted molecular weights in wild-type and/or A3B1 retinal lysates. The Western blots were reproduced in 2–3 independent experiments. All Western blots shown were from the same batch of retinal lysates.
Transmission electron microscopy
For optimal tissue preparation, posterior eyecups or isolated retinas were immediately fixed in 2 % paraformaldehyde and 2.5 % glutaraldehyde in phosphate-buffered saline (PBS) for 15 h. Subsequently, tissue was rinsed in PBS containing 6.8 % (w/v) sucrose, postfixed in osmium tetroxide (2 % (w/v) in PBS) for 90 min, washed three times in PBS, and dehydrated in a graded series of acetone. Tissue was flat-embedded in Epon (Serva, Heidelberg, Germany). Transverse sections (50 nm thick) were mounted onto one-hole Formvar-coated copper grids (Serva), stained with 8 % (w/v) uranyl acetate (10 min), and contrasted with lead citrate for 5 min. In randomly selected sections, the retinas were qualitatively evaluated using a LEO TEM 906E (Zeiss, Oberkochen, Germany), usually at a primary magnification of ×27800.
RNA extraction
Retinas of 12- and 28-day-old wild-type and A3B1 mice were placed in 350 μl of RLT buffer (Qiagen, Hilden, Germany) containing 1 % β-mercaptoethanol (Sigma-Aldrich Chemie, Steinheim, Germany). Extraction of total RNA was performed using the RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions for tissues. QIAshredder mini-spin columns (Qiagen) as well as needle and syringe homogenization were applied. RNA quality was verified with an Agilent 2100 Bioanalyzer using the RNA 6000 Nano LabChip Kit (Agilent Technologies, Boeblingen, Germany) following the manufacturer’s instructions.
cDNA synthesis and qRT-PCR
One microgram of RNA was applied to the cDNA synthesis using the QuantiTect® Reverse Transcription Kit (Qiagen), which includes digestion of genomic DNA. qRT-PCR was performed with the LightCycler480 System (Roche, Mannheim, Germany) using the QuantiTect® SYBR® Green PCR Kit (Qiagen) according to the manufacturer’s instructions.
Standard curves for each amplified transcript were generated to obtain the PCR efficiency. CP values were determined by the LightCycler® Software 480 (Roche). Expression levels of each sample were detected in triplicate reactions. Pyruvate dehydrogenase β-subunit (PDH) was used as reference gene to calculate the relative expression of each target gene applying the efficiency-corrected equation by Pfaffl et al. [19]. Oligonucleotides for the qRT-PCR were designed using Primer3 Software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The sequences of the oligonucleotides will be made available upon request.
Microarray analysis
Microarray experiments of retinal tissue were performed at two different ages (p12 and p28) comparing gene expression of A3B1 and wild-type animals using an Affymetrix platform according to the manufacturer’s instructions. Fragmented and labeled cRNA of three wild-type and three A3B1 retinas were hybridized on Affymetrix Mouse Genome 430 2.0 Arrays, respectively. A probe-level summary was determined using the Affymetrix GeneChip Operating Software using the MAS5 algorithm. Normalization of raw data was performed by the Array Assist Software 4.0 (Stratagene, La Jolla, CA, USA), applying the GC-robust multichip average (RMA) algorithm. Significance was calculated using a t test without multiple testing correction (Array Assist software), selecting all transcripts with a minimum change in expression level of 1.5-fold together with a p value <0.05. The heatmaps were generated using Multi Experiment Viewer Software (MeV v4.8.1.).
Gene regulation networks
Gene regulation networks were generated by Ingenuity Pathways Analysis (IPA) 8.8 (http://www.ingenuity.com). For that purpose, data sets containing gene identifiers and the corresponding expression and significance values were uploaded into the application. These genes, called Focus Genes, were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. Networks of these Focus Genes were then algorithmically generated based on their connectivity.
Functional analyses
Ingenuity functional analysis identified the biological functions and/or diseases that were most significant to the data set. Genes from the data set that met the negative logarithmic significance cut-off of five or higher, and were associated with biological functions and/or diseases in the Ingenuity Pathways knowledge base were considered for the analysis. Fisher’s exact test was used to calculate a p value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone.
Genomatix-based promoter analysis
We extracted the mouse promoter sequences from the ElDorado database (Genomatix Suite-ElDorado, release 4.8, Mouse Genome build 37, Genomatix, Munich, Germany). The GEMS launcher task “FrameWorker” using the available weight matrix library (GEMS launcher version 4.4; Genomatix; http://www.genomatix.de) was used to generate the model of the promoter framework. The FastM task of GEMS Launcher was used to optimize models. ModelInspector (a GEMS launcher task) was used to search the mouse promoter database (Genomatix promoter database, GPD; Genomatix) with the optimized model. Default parameters were used for the initial analyses in all programs, if not indicated otherwise.
Results
Retinal morphology in A3B1 mice
We used immunofluorescence labeling against calbindin to reveal horizontal cell morphology in adult A3B1 (Fig. 1b) and wild-type (Fig. 1c) mice. Calbindin immunofluorescence also labels the perikarya of many amacrine cells and ganglion cells, and three strata of dense processes within the inner plexiform layer (IPL): the two cholinergic bands and one band in the center of the IPL [15]. While all these structures appeared normal in the A3B1 mouse (Fig. 1b), we observed some horizontal cell processes growing deep into the ONL (arrows, Fig. 1b) at P36 that were not observed in the wild type (Fig. 1c).
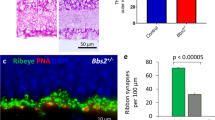
Altered horizontal cell morphology in A3B1 mice. a Nomarski micrograph of a vertical retinal section through the A3B1 retina at P36. OS outer segments, IS inner segments, ONL outer nuclear layer, OPL outer plexiform layer, INL inner nuclear layer, IPL inner plexiform layer, GCL ganglion cell layer. b Confocal images of the same section as in a and of age-matched wild-type (wt) retina (c) immunolabeled for calbindin. Calbindin is present in horizontal cells, amacrine cells, and ganglion cells in both retinas. Calbindin also identified outgrowing horizontal cell processes in the A3B1 retina but not in the wild type. d–f Confocal images of calbindin-labeled horizontal cells in the A3B1 mouse at p11 (d) and p12 (e) and wt control retina at p12 (f). Scale bar 25 μm
Onset of neurite outgrowth in the A3B1 retina
To determine the time point at which horizontal cells started sprouting and analyze their evolution, we stained retinal slices of P9–P43 mice with calbindin. Until p11, only sporadic outgrowth of neurites could be observed in A3B1 mice (Fig. 1d). However, only 1 day later, the density and total length of horizontal cell neurites growing into the ONL increased threefold (p11: 11.1 ± 1.1, n = 4, p12: 33.7 ± 3.3, n = 5; p = 0.0007) and 4.8 fold (p11: 130.4 ± 21.1 μm, n = 4, p12: 625.9 ± 69.5 μm, n = 5; p = 0.0005), respectively (Fig. 1e). This phenomenon was completely absent in age-matched wild-type mice (Fig. 1f).
Since p12 corresponds to the age of eye opening in the mouse, this outgrowth of neurites in the double-knockout might represent a response of HCs to missing light-dependent inputs. To clarify the involvement of light-dependent stimuli, we analyzed dark-reared A3B1 and wild-type mice. However, dark rearing did not cause sprouting in wild-type mice, nor it did cause any visible change in the behavior of HCs in A3B1 mice (data not shown).
Formation of ectopic synapses in the outer nuclear layer
We examined the temporal evolution of ectopic photoreceptor synapses in A3B1 mice using immunofluorescence. Presynaptic ribbons were labeled with antibodies against the C-terminal binding protein 2 (CtBP2), and horizontal cells with antibodies against calbindin (Fig. 2). At p18, calbindin-positive processes had no CtBP2 labeling at their tips (Fig. 2a). The sprouting neurites extended across the full depth of the ONL and only a few CtBP2 puncta, indicative of the existence of synapses, could be observed within the deepest 5 μm of the ONL. At p28, HC sprouting was less evident but ectopic ribbons began to appear in more superficial parts of the ONL (arrows, Fig. 2b). After an additional week and at later stages, all aberrant horizontal cell processes were associated with ectopic CtBP2-positive ribbons at their tips (Fig. 2c, arrows in d). Using high-resolution confocal microscopy, we analyzed the structure of synaptic contacts in detail and observed that the shape of the ectopic synapses in the ONL was comparable to those in the OPL (Fig. 2c, inset). In addition to CtBP2 associated with tips of the HC sprouts, there were several processes with ectopic synapses evident along their shafts (arrowheads in Fig. 2d).
Outgrowing horizontal cells and the formation of ectopic synapses. Confocal images of the outer retina of A3B1 mice at p18 (a), p28 (b), P36 (c), and P43 (d) double labeled for calbindin (green) and the presynaptic ribbon marker CtBP2 (magenta). Arrows in b mark the first ectopic appearing synapses in the ONL. Magnification view of a horizontal cell process tip in the ONL directly surrounded by a horseshoe-shaped ribbon is shown in the inset in c. d At P43, all aberrant horizontal cell processes were associated with ectopic synapses at their tips (arrows), some processes were flanked by ectopic synapses along the shaft (arrowheads). e–g Rod bipolar cell (RBC) outgrowth in the A3B1 mouse at different time points. e RBC staining (PKCα, magenta) reveals no sprouting at P18. f, g Double labeling of HCs (Calbindin, green) and RBCs (PKCα, magenta) reveals that the aberrant processes from these two cell types frequently exit the OPL at the same points and navigate together through the ONL. The arrow in f marks a RBC dendrite on the way along the same path as the HC process. h Double labeling of PKCα (magenta) and mGluR6 (green) reveals ectopic mGluR6 expression on sprouting RBC dendrites. Scale bars 10 μm in b (for a–d), in e (for e–g) and h
In summary, our data demonstrate that horizontal cells in the A3B1 mouse produce extensive outgrowths, with no apparent target by the time of eye opening. However, 2–3 weeks later, the processes that formed new normal-looking ectopic synaptic contacts with rod photoreceptors in the ONL survived while all others disappeared.
Immunolabeling of A3B1 retinas with antibodies against PKCα revealed that rod bipolar cell (RBC) dendrites also extended aberrantly into the ONL (Fig. 2f–h), and labeling both RBCs and HCs indicated that the aberrant processes from these two cell types often exit the OPL at the same points (Fig. 2f, g). However, RBCs extended their processes at a later time. RBC sprouting was not evident at P18 (Fig. 2e). At P36, RBC sprouting was less evident than HC sprouting. Some of the HC processes were flanked by RBC dendrites, and a few of the RBC dendrites were on their way along the same path as the HC processes (Fig. 2f, arrow). At later time points, most of the aberrant HC processes co-fasciculated with RBC dendrites (Fig. 2g). To analyze the functional relevance of the ectopic synapses in the A3B1 mouse, we studied the localization of the metabotropic glutamate receptor mGluR6, which is postsynaptically expressed on the dendritic tips of all ON BCs [20]. We found clear examples of mGluR6 puncta at the tips of sprouting RBCs at P43 (Fig. 2h). To examine the ultrastructure of the ectopic synapses, we performed transmission electron microscopy and searched for ectopic ribbon synapses in the ONL of A3B1 retinas. Figure 3 shows examples of ectopic synapses within the ONL. An electron-dense ribbon in the presynaptic cytoplasm and several postsynaptic elements characterize the ribbon synapses. The synaptic elements that lie deeper and lateral to the synaptic ribbons are most likely processes from axon terminals of the horizontal cells while the central elements are rod bipolar cell dendrites. We observed no structural differences between the ectopic ribbon synapses in the ONL and the ribbon synapses in the OPL.
Ultrastructure of ectopic ribbon synapses. a, b ONL of a 2-month-old A3B1 retina with an ectopic synapse (frame). The higher magnification in b shows a typical triad containing a presynaptic ribbon, two lateral horizontal cell processes (h), and a central element (star), which is most likely a rod bipolar cell dendrite. c–d ONL of A3B1 at P44 with two examples of ectopic synapses (frame and arrow). The framed ectopic synapse is magnified in d and contains two electron-dense ribbons in the presynaptic cytoplasm and some postsynaptic processes. The lateral elements are most likely horizontal cell processes (h). e High magnification of an ectopic synapse in the ONL at P36 with a presynaptic ribbon, two lateral horizontal cell processes (h), and a central element (star). f Example of a ribbon synapse at the ONL/OPL border in a 2-month-old A3B1 retina. Scale bars 2 μm in a and c; 0.5 μm in b and d–f
Microarray-based gene expression analysis in the A3B1 retina
The molecular events associated with neurite outgrowth and ectopic synaptogenesis in the outer retina are not well understood. The A3B1 retina shows precisely timed and very pronounced HC and RBC neurite outgrowth starting at p12 and ectopic synaptogenesis beginning around p28. To evaluate the genomic response underlying neurite outgrowth and synapse formation, we determined the global retinal gene expression profiles in p12 and p28 A3B1 mice using an Affymetrix MOE 430 2.0 microarray platform and compared it with the gene expression profile of the respective wild-type control retinas. At p12, we identified 911 transcripts (listed in Supplementary Table 1) with a minimum change in expression level of 1.5 fold in combination with a p value less than 0.05. A total of 794 of these transcripts are from known genes (546 up-regulated and 248 down-regulated). At p28, we found 1,842 differently regulated (≥1.5-fold, p value <0.05) transcripts (specified in Supplementary Table 2), 1,625 of those being from known genes (1,076 up-regulated and 549 down-regulated). There was an 8 % overlap between the two age groups. Categorization of the differentially regulated genes using the Ingenuity Pathway Analysis (IPA) software showed that the top functional classes were “enzyme”, “transcription regulation”, “transporter”, and “kinase” (Supplementary Fig. 1).
Validation of the microarray data
To test the validity of the microarray data, we performed additional in silico analyses using the IPA software. Our previous results on the retinal phenotype of the single knockout models [7, 14, 21] suggested that genes involved in the phototransduction pathway of rods and cones should be down-regulated in the A3B1 retina. Confirming the validity of the microarray data, we found that “ophthalmic disease” was among the top three IPA biological functions in the dataset comparing the A3B1 with the wild-type retina at p28 (67 genes, p = 1.7 × 10−4–5.7 × 10−17). Moreover, in the same dataset “phototransduction pathway” was the top IPA canonical pathway (p = 2.4 × 10−8). Fifteen out of 65 genes that map to this canonical pathway were down-regulated in the p28 A3B1 retina when compared to the wild-type control (Fig. 4a). Importantly, six of the down-regulated genes were cone-specific genes not expressed in other retinal cells (Fig. 4a). Thus, although the microarray was performed with cRNA from whole retina, it was sensitive enough to detect changes in gene expression in cone photoreceptors that make up <5 % of the total retinal cell population in mice.
Validation of microarray analysis. a Top panel Heat map showing the relative expression of photoreceptor-specific genes in the wild-type and A3B1 replicates at p12 and p28. Rod-specific genes are labeled in black and cone-specific in red. Bottom panel Centroid graph view of the heat map data. The up-regulation of photoreceptor-specific genes in the wild-type retina at p28 is missing in the A3B1 retina. b Quantitative RT-PCR-based validation of selected transcripts at p12 and p28 (left and right from dotted vertical line, respectively). Red bars show fold-change values from the qPCR (n = 3) and grey bars fold-change values from the microarray experiment. The upper and lower horizontal lines denote the +1 and −1 fold-change levels, respectively
We next selected ten genes for an experimental, qPCR-based validation. Three genes were tested at p12, four genes at p28, and another three genes at both ages resulting in 13 analyses. We took care to include genes with large as well as small changes in expression between wild-type and A3B1 retinas. In 12 out of 13 analyses we obtained changes in the predicted direction (one down, 11 up); of these, ten were significant at the level of p < 0.05 (see Fig. 4b). For Gsn and Mmp9, the observed up-regulation at p12 was not significant. Based on the microarray data Src was expected to be up-regulated at p12 and p28, but the qPCR could confirm the up-regulation only at p28 (Fig. 4b).
Identification of genes involved in neurite outgrowth and synaptogenesis
Following the validation, we performed a bioinformatics analysis of the p12 and p28 datasets of differentially expressed genes in the A3B1 retinas, focusing on genes linked to neurite and synapse function. The Ingenuity analysis identified a significant enrichment of 17 biological functions and one canonical pathway related to axon or neurite growth and guidance or synaptic transmission and plasticity (Table 1). At p12, 67 transcripts (52 up- and 15 down-regulated) were associated with at least one biological function related to neurite outgrowth and synaptogenesis (Supplementary Table 3). Among others, this analysis identified “guidance of neurites” (25 up-, ten down-regulated; p = 0.0027) and “growth of neurites” (21 up, eight down; p = 0.0022) as significantly enriched functions at p12. At p28, the enrichment of biological functions related to neurite outgrowth and synapse formation was even stronger (Table 1). For instance, the number of genes involved in the functions “guidance of neurites” or “growth of neurites” increased to 85 (68 up-, 17 down-regulated; p = 2.5 × 10−9) and 68 (55 up, 13 down; p = 2.15 × 10−8), respectively. The overall number of differentially regulated genes with related annotations increased to 115 (96 up- and 19 down-regulated genes) at this age (Supplementary Table 4). Nineteen of those genes were also differentially regulated at p12 and 16 of those showed a regulation in the same direction at both time points. In summary, this analysis identified 163 differentially expressed genes in the A3B1 retinas linked to “axon/neurite growth and guidance” or “synaptic transmission and plasticity”. A heatmap with a hierarchical clustering of these 163 genes is shown in Supplementary Fig. 2. In general, most of the 163 genes showed higher expression levels in A3B1 than in wild-type retinas. The hierarchical clustering also identified a group of 19 genes specifically up-regulated in the p12 A3B1 retinas and another cluster of seven genes up-regulated in the A3B1 retinas at both ages. There were only three genes specifically down-regulated in the p12 A3B1 retinas and five at both p12 and p28.
Transcription factor analysis using Ingenuity software
The second largest functional class among the differentially regulated genes in the A3B1 retinas was “transcription regulators” (Supplementary Fig. 1). We therefore performed a transcription factor analysis using Ingenuity software to identify transcriptional regulators contributing to the observed changes in gene expression. This analysis identified 14 transcriptional regulators with a regulation z-score above 2, which were predicted to be activated and another ten transcriptional regulators with a regulation z-score below 2 predicted to be inhibited (Table 2). Most of the phototransduction genes shown in Fig. 4a are known as Crx target genes [20]. In good agreement with this, Crx was the transcription factor with the highest predicted inhibition in A3B1 retinas (Table 2). The transcription factor with the highest predicted activation in the A3B1 retina as judged by the highest regulation z-score was the tumor suppressor Tp53 (Table 2). In addition to its well established functions (e.g., in cell cycle progression and apoptosis), Tp53 was recently shown to play a role in neurite outgrowth and axon regeneration [22, 23]. In line with this, 25 of the 163 genes related to axon and neurite function have expression changes (22 up-regulated and three down-regulated) consistent with Tp53 activation (see Fig. 5a). One of those genes was Src, encoding a kinase also implicated in neurite outgrowth [24, 25]. We stained wild-type and A3B1 retinas with an antibody against the phosphorylated, and thus active, form of Src (pSrc) that detects a single 60-kDa band in retinal lysates from wild-type or A3B1 mice. In wild-type retinas, we observed low levels of pSrc (Fig. 5c), mainly in HCs (Fig. 5e); however the signal appeared up-regulated in A3B1 retinas (Fig. 5d). Fine immunopositive puncta or fibers were evident in the ONL and the OPL of the A3B1 that partly co-localized with calbindin-immunoreactive HC neurites (Fig. 5f, f’) (Pearson’s co-localization coefficient 0.45 ± 0.02, n = 5).
a Ingenuity network showing the Tp53-regulated differentially expressed genes from the subset of 163 genes related to neurite outgrowth and synaptogenesis. Up-regulated genes are shown in red and down-regulated genes are green. The fold-change values from the microarray are given below the molecules. b Western blot on retinal lysates from wt and A3B1 mice probed with anti-phospho-Src (Y416) (top) and anti-tubulin (bottom). c–f Confocal images from wild-type (c, e) and A3B1 (d, f) retina labeled with antibodies directed against Y416-phosphorylated Src (magenta). e, f Double labeling for pSrc (magenta) and calbindin (green). d’, f’ High magnification view of the framed area in d and f, respectively. Scale bars mark 20 μm
The transcription factor analysis also predicted an activation of Smad signaling (Table 2). Moreover, transcription factor binding site analysis using the Genomatix promoter database revealed that more than 40 % (43 and 45 % at p12 and p28, respectively) of the genes with strong changes in expression (fold change ≥2.0, p value <0.05) had Smad binding sites in their promotor regions. The subset of 163 genes related to axon and neurite function also included 15 genes with changes in gene expression consistent with activation of Smad2, Smad3, or Smad4 and inhibition of Smad7 (Fig. 6a). Together, these findings suggested activation of Smad-dependent signaling in the A3B1 retina. To test this experimentally, we performed Western-blot and immunohistochemical experiments using an antibody against phospho-Smad1, phospho-Smad5 and phospho-Smad8 (anti-phospho-Smad 1/5/8). These Smads form dimers with Smad4 upon phosphorylation and translocate as a complex to the nucleus to drive expression of target genes [26]. The antibody detected a weak band at approx. 60 kDa in wild-type and A3B1 retinal lysates (Fig. 6b). The signal appeared stronger in the A3B1 retina. In the wild-type immunostainings, anti-phospho-Smad 1/5/8 strongly labeled the two cholinergic strata in the inner plexiform layer, weakly stained Müller glia cells (MGCs) and the nuclei of HCs (Fig. 6c, e). The signal in the cholinergic strata did not significantly change in the A3B1 retina; however, the labeling intensity of HCs and other inner retinal cells increased substantially (Fig. 6d, f). Activation of Smad-dependent signaling is also consistent with increased activity of TGF-beta. We therefore stained wild-type and A3B1 retina with a TGFβ1-specific antibody that detects a single approx. 25-kDa band in Western blots from wild-type and A3B1 retinal lysates (Fig. 7a). Interestingly, although TGFβ1 was not among the differentially regulated genes, we observed a clear and defined up-regulation of the signal in the ONL and the OPL of A3B1 mice (Fig. 7d–e). A large number of fine TGFβ1-positive puncta appeared in the A3B1 retina and co-localized with calbindin at the tips of outgrowing neurites (Fig. 7e’).
a Ingenuity network showing the Smad-regulated differentially expressed genes from the subset of 163 genes related to neurite outgrowth and synaptogenesis. Up-regulated genes are shown in red, down-regulated genes are green. The fold-change values from the microarray are given below the molecules. b Western blot on retinal lysates from wt and A3B1 mice probed with anti-phospho-SMAD1/5/8 (S463/465 in Smad1 or 5 and S426/428 in Smad 8) (top) and anti-tubulin (bottom). c–f Confocal images from wild-type (c, e) and A3B1 (d, f) retina labeled with an antibody directed against S463/465 (Smad1 or 5) or S426/428 (Smad8) phosphorylated Smads (magenta). e, f Double labeling for pSmad1/5/8 and calbindin (green). e The phosphorylated Smad signal in horizontal (arrows) and other retinal cells is increased in the A3B1 retina. Scale bar marks 20 μm
Immunolocalization of TGFβ1 and Lgals3 in the A3B1 retina. a–c Western blot on retinal lysates from wt and A3B1 mice probed with anti-TGFβ1 (a) and Lgals3 (b). c Tubulin loading control for Western blots shown in b. Loading control for a is shown in Fig. 5 (bottom panel B). d–g Confocal images from wild-type (d, f) and A3B1 (e, g) retina labeled with TGFβ1-specific (d–e) or Lgals3-specific (f–g) antibodies (magenta). (e’) Magnification view from e with overlaid calbindin signal (green). g’ Magnification view from g with overlaid calbindin signal (green). Scale bars mark 20 μm in d–e and f–g and 10 μm in e’ and g’)
Lgals3, also termed galectin-3, is known to regulate TGFβ and the pathogenesis of lung fibrosis [27]. Interestingly, Lgals 3 was one of the highly up-regulated genes in the A3B1 retina (see Fig. 4b). We could also confirm this up-regulation at the protein level (Fig. 7b, f–g). The anti-Lgals3 antibody detected a faint 33-kDa band in wild-type retinal lysates, which was strongly up-regulated in the A3B1 retina. The Lgals3-specific antibody faintly stained MGCs in wild-type mouse retina (Fig. 7f), which was in line with previous reports showing expression of Lgals3 in rat and pig retinal MGCs [28, 29]. The signal in MGCs was strongly increased in the A3B1 retina (Fig. 7g). Double labeling experiments showed that calbindin-positive HC neurites grow along Lgals3-positive MGCs (Fig. 7g’) suggesting an interaction of MGCs with outgrowing HC neurites.
Stat3 was another transcription factor implicated in neurite outgrowth [23] and predicted to be activated (Table 2) by the dataset. Moreover, the microarray detected increased Stat3 transcript levels in the A3B1 retina. In total, 20 of the 163 genes from our dataset implicated in axon and neurite function showed changes in gene expression in the A3B1 retina were consistent with an activation of Stat3 (Fig. 8a). To confirm Stat3 activation and to localize the activated protein, we stained A3B1 and wild-type retinas with an antibody that specifically detects the active form of Stat3 (92 kDa) (Fig. 8b–f). We observed an up-regulation of the immunosignal in the A3B1 retina (Fig. 8b, d, f) with punctuate signal in the OPL and the ONL that localized close to calbindin-positive outgrowing HC neurites.
a Ingenuity network showing the Stat3-regulated differentially expressed genes from the subset of 163 genes related to neurite outgrowth and synaptogenesis. Up-regulated genes are shown in red, down-regulated genes are green. The fold-change values from the microarray are given below the molecules. b Western blot on retinal lysates from wt and A3B1 mice probed with anti-phospho-Stat3 (Y705) (top) and anti-tubulin (bottom). c–f Confocal images from wild-type (c, e) and A3B1 (d, f) retina labeled with an antibody directed against Y705-phosphorylated Stat3 (magenta). e, f Double labeling for pStat3 and calbindin (green). Scale bar marks 20 μm
Canonical pathway analysis
To further analyze which signaling pathways may underlie the differential regulation of the subset of 163 genes related to axon and neurite outgrowth and synapse formation, we performed a canonical pathway analysis using Ingenuity software. In line with the idea that the list of 163 genes contains only genes related to neurite and synapse function, neurite outgrowth, or synaptogenesis, the analysis identified “axonal guidance signaling” as the most significant canonical pathway (p = 6.8 × 10−55) (Fig. 9a). Sixty-four of the 430 molecules (ratio: 0.149) that map to this canonical pathway were among the subset of 163 differentially expressed genes (52 up- and 12 down-regulated) (Supplementary Fig. 3). Plexin C1 (Plxnc1) and its ligand Sema7a, were both among the up-regulated “axonal guidance signaling” genes (Fig. 9b). We therefore analyzed the localization of Plxnc1 in A3B1 and wild-type retinas. This protein was not detectable in the wild-type retina (Fig. 9c and e). However, in the A3B1 retina the Plxnc1 antibody strongly labeled puncta in the ONL and the OPL (Fig. 9d and f). Some Plxnc1-positive puncta were also calbindin-positive (Fig. 9F).
a Bar graph showing the top 3 canonical pathways and ten additional relevant canonical pathways significantly regulated in the A3B1 retina. The left and right y-axis show the log p value and the ratio of genes within the subset of 163 genes relative to all known genes that map to the canonical pathway. b Ingenuity network with PlexinC1/Sema7a-related signaling with overlaid expression levels in the A3B1 retina. Up-regulated genes are shown in red, down-regulated genes are green. The fold-change values from the microarray are given below the molecules. c–f Confocal images from wild-type (c, e) and A3B1 (d and f) retina labeled with a Plxnc1-specific antibody (magenta). d, f Double labeling for Plxcn1 and calbindin (green). Scale bar marks 20 μm
The second and third most significant canonical pathways were “ephrin receptor signaling” (p = 3.7 × 10−23, ratio 0.141) and “Cxcr4 signaling” (p = 2.2 × 10−18, ratio 0.137) with 28 (21 up and seven down) and 23 (17 up and six down) differentially regulated genes, respectively. Both signaling pathways are also related to axon and neurite guidance and outgrowth [30, 31] and partially overlap with the “axonal guidance signaling” pathway. Moreover, 21 of the 163 differentially expressed genes mapped to the “actin cytoskeleton signaling” pathway, which was also among the significant canonical pathways (p = 5.3 × 10−13, ratio 0.087).
Discussion
At the first chemical synapse of the mammalian retina, rod and cone photoreceptors transfer light signals to bipolar and horizontal cells. In the present study, we aimed at analyzing the response of these postsynaptic partners (e.g., the horizontal cells) to the loss of photoreceptor function. We used a mouse model with targeted deletions of the genes CNGA3 and CNGB1. These genes encode essential subunits of the cone and rod cyclic nucleotide-gated channels, respectively [17]. Knockout of CNGA3 renders cone photoreceptors non-functional [14], while the deletion of CNGB1 results in dramatically reduced rod photoresponses [7]. Accordingly, the double-knockout mice lack normal rod and cone function. We show that horizontal cells in the double-knockout mice react to the missing input by extending sprouting neurites into the ONL. By analyzing the developmental time-course of horizontal cell sprouting, we show for the first time that neurite outgrowth in the diseased retina is a precisely timed and very fast-developing phenomenon. In the A3B1 retina, the number and total length of outgrowing neurites increased significantly from p11 to p12. Thus, neurite outgrowths several micrometers in length, fully develop within a few hours in the A3B1 retina. The trigger for this synaptic plasticity is not known. Since the onset of neurite outgrowth coincides with eye opening at p12, the sprouting response might depend on light activity. However, we observed the same amount of horizontal cell sprouting in A3B1 mice kept under constant darkness throughout retinal development, and could not induce neurite sprouting in wild-type mice that were dark-reared during development (data not shown). Thus, although the sprouting phenomenon coincides with eye opening at p12, it does not depend on light activity. At the time of eye opening the horizontal cell outgrowths seemed to have no apparent target. However, at p28, most horizontal cell sprouts in the outer nuclear layer were apposed to synaptic ribbons at their tips. Electron microscopy confirmed the presence of synaptic ribbons at rod cell bodies with characteristic postsynaptic elements undistinguishable from normal synaptic contacts at rod cell bodies located close to the outer plexiform layer. Moreover, the expression of mGluR6 on sprouting RBC dendrites approved the functional relevance of these ectopic synapses. Horizontal cell neurite outgrowth can also be observed in the CNGA3−/− (single knockout) mouse [21]. However, compared to the A3B1 mouse, this HC sprouting in the CNGA3−/− mouse can be considered a rare phenomenon. We cannot exclude that outgrowing HC neurites in the A3B1 retina also grow towards cones and eventually establish synaptic contacts to cone cell bodies within the ONL, but since such events would be rather rare we were not able to detect them.
The molecular mechanisms mediating neurite outgrowth and formation of ectopic synapses are largely unknown. To shed some light on the molecular mechanisms underlying these phenomena, we determined the genomic response in 12- and 28 day-old A3B1 mice using microarray analysis of global retinal gene expression. We identified 163 differentially regulated genes in the A3B1 retina related to neurite outgrowth or synaptogenesis. Sixty-four of those genes mapped to the “axonal guidance signaling pathway”. Thus, although the microarray was performed with cRNA from whole retina, we were able to detect changes in gene expression corresponding to the synaptic changes in the A3B1 retina. The pattern of regulation of gene expression pointed towards activation of Tp53, Smad, and Stat3 signaling. Most of the differentially regulated genes were up-regulated. Importantly, selected key molecules from these signaling pathways could be localized to or at close proximity to outgrowing horizontal cell neurites. Many of the proteins (e.g., TGF-beta, phosphorylated Stat3, and PlexinC1) showed a spotty pattern of up-regulation in the OPL and the ONL of A3B1 mice. Other proteins, e.g., Lgals3 were up-regulated in Müller glia cells, suggesting an involvement of glial cells in neurite outgrowth and synaptogenesis in the A3B1 retina. Interestingly, Lgals3 was recently shown to regulate TGF-beta function in lung fibrosis [27]. This might suggest that TGFbeta/Lgals3 signaling in HCs and MGCs and neuron-glia interactions could play a role in neurite outgrowth in the A3B1 retina.
Our microarray analysis identified up-regulation of several paracrine factors and related receptors or signaling molecules including bone morphogenic proteins (BMPs), wingless (Wnt) family members, and TGF-beta. Some of the genes were already up-regulated at p12, suggesting that early intraretinal cell–cell communication in the diseased retina might drive this synaptic plasticity.
This study sheds new light on the mechanisms involved in neurite outgrowth and synaptic plasticity by identifying differentially regulated gene networks in the A3B1 retina related to these phenomena.
Aberrant connectivity of neurons has been described in several human genetic diseases [32] and is currently discussed in many other neurological diseases [32, 33]. Rewiring of retinal neurons was described in several mouse models of retinal degeneration [18, 34]. The new information from the present study on induced/repressed molecular signaling pathways in the A3B1 retina might help establishing ways to manipulate/stimulate neurite outgrowth for the normalization of synaptic networks after treatment of retinal diseases or neuronal diseases in general.
References
Wässle H (2004) Parallel processing in the mammalian retina. Nat Rev Neurosci 5(10):747–757
Blanks JC, Adinolfi AM, Lolley RN (1974) Synaptogenesis in the photoreceptor terminal of the mouse retina. J Comp Neurol 156(1):81–93
Olney JW (1968) An electron microscopic study of synapse formation, receptor outer segment development, and other aspects of developing mouse retina. Invest Ophthalmol 7(3):250–268
Claes E, Seeliger M, Michalakis S, Biel M, Humphries P, Haverkamp S (2004) Morphological characterization of the retina of the CNGA3(-/-)Rho(-/-) mutant mouse lacking functional cones and rods. Invest Ophthalmol Vis Sci 45(6):2039–2048
Lewis GP, Linberg KA, Fisher SK (1998) Neurite outgrowth from bipolar and horizontal cells after experimental retinal detachment. Invest Ophthalmol Vis Sci 39(2):424–434
Liets LC, Eliasieh K, van der List DA, Chalupa LM (2006) Dendrites of rod bipolar cells sprout in normal aging retina. Proc Natl Acad Sci USA 103(32):12156–12160
Hüttl S, Michalakis S, Seeliger M, Luo DG, Acar N, Geiger H, Hudl K, Mader R, Haverkamp S, Moser M, Pfeifer A, Gerstner A, Yau KW, Biel M (2005) Impaired channel targeting and retinal degeneration in mice lacking the cyclic nucleotide-gated channel subunit CNGB1. J Neurosci 25(1):130–138
Dick O, Dieck S, Altrock WD, Ammermuller J, Weiler R, Garner CC, Gundelfinger ED, Brandstatter JH (2003) The presynaptic active zone protein bassoon is essential for photoreceptor ribbon synapse formation in the retina. Neuron 37(5):775–786
Ball SL, Pardue MT, McCall MA, Gregg RG, Peachey NS (2003) Immunohistochemical analysis of the outer plexiform layer in the nob mouse shows no abnormalities. Vis Neurosci 20(3):267–272
Bayley PR, Morgans CW (2007) Rod bipolar cells and horizontal cells form displaced synaptic contacts with rods in the outer nuclear layer of the nob2 retina. J Comp Neurol 500(2):286–298
Chang B, Heckenlively JR, Bayley PR, Brecha NC, Davisson MT, Hawes NL, Hirano AA, Hurd RE, Ikeda A, Johnson BA, McCall MA, Morgans CW, Nusinowitz S, Peachey NS, Rice DS, Vessey KA, Gregg RG (2006) The nob2 mouse, a null mutation in Cacna1f: anatomical and functional abnormalities in the outer retina and their consequences on ganglion cell visual responses. Vis Neurosci 23(1):11–24
Haeseleer F, Imanishi Y, Maeda T, Possin DE, Maeda A, Lee A, Rieke F, Palczewski K (2004) Essential role of Ca2+-binding protein 4, a Cav1.4 channel regulator, in photoreceptor synaptic function. Nat Neurosci 7(10):1079–1087
Mansergh F, Orton NC, Vessey JP, Lalonde MR, Stell WK, Tremblay F, Barnes S, Rancourt DE, Bech-Hansen NT (2005) Mutation of the calcium channel gene Cacna1f disrupts calcium signaling, synaptic transmission and cellular organization in mouse retina. Hum Mol Genet 14(20):3035–3046
Biel M, Seeliger M, Pfeifer A, Kohler K, Gerstner A, Ludwig A, Jaissle G, Fauser S, Zrenner E, Hofmann F (1999) Selective loss of cone function in mice lacking the cyclic nucleotide-gated channel CNG3. Proc Natl Acad Sci USA 96(13):7553–7557
Haverkamp S, Wassle H (2000) Immunocytochemical analysis of the mouse retina. J Comp Neurol 424(1):1–23
Dieck S, Altrock WD, Kessels MM, Qualmann B, Regus H, Brauner D, Fejtova A, Bracko O, Gundelfinger ED, Brandstatter JH (2005) Molecular dissection of the photoreceptor ribbon synapse: physical interaction of Bassoon and RIBEYE is essential for the assembly of the ribbon complex. J Cell Biol 168(5):825–836
Walzer T, Galibert L, Comeau MR, De Smedt T (2005) Plexin C1 engagement on mouse dendritic cells by viral semaphorin A39R induces actin cytoskeleton rearrangement and inhibits integrin-mediated adhesion and chemokine-induced migration. J Immunol 174(1):51–59
Zinchuk V, Zinchuk O, Okada T (2007) Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta histochemica et cytochemica 40(4):101–111
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30(9):e36
Vardi N, Morigiwa K (1997) ON cone bipolar cells in rat express the metabotropic receptor mGluR6. Vis Neurosci 14(4):789–794
Michalakis S, Geiger H, Haverkamp S, Hofmann F, Gerstner A, Biel M (2005) Impaired opsin targeting and cone photoreceptor migration in the retina of mice lacking the cyclic nucleotide-gated channel CNGA3. Invest Ophthalmol Vis Sci 46(4):1516–1524
Di Giovanni S, Rathore K (2012) p53-dependent pathways in neurite outgrowth and axonal regeneration. Cell Tissue Res 349(1):87–89
Floriddia EM, Rathore KI, Tedeschi A, Quadrato G, Wuttke A, Lueckmann JM, Kigerl KA, Popovich PG, Di Giovanni S (2012) p53 regulates the neuronal intrinsic and extrinsic responses affecting the recovery of motor function following spinal cord injury. J Neurosci 32(40):13956–13970
He JC, Gomes I, Nguyen T, Jayaram G, Ram PT, Devi LA, Iyengar R (2005) The G alpha(o/i)-coupled cannabinoid receptor-mediated neurite outgrowth involves Rap regulation of Src and Stat3. J Biol Chem 280(39):33426–33434
Yang LT, Alexandropoulos K, Sap J (2002) c-SRC mediates neurite outgrowth through recruitment of Crk to the scaffolding protein Sin/Efs without altering the kinetics of ERK activation. J Biol Chem 277(20):17406–17414
Whitman M (1998) Smads and early developmental signaling by the TGFbeta superfamily. Genes Dev 12(16):2445–2462
Mackinnon AC, Gibbons MA, Farnworth SL, Leffler H, Nilsson UJ, Delaine T, Simpson AJ, Forbes SJ, Hirani N, Gauldie J, Sethi T (2012) Regulation of TGF-beta1 driven lung fibrosis by galectin-3. Am J Respir Crit Care Med 185(5):537–546
Kim J, Moon C, Ahn M, Joo HG, Jin JK, Shin T (2009) Immunohistochemical localization of galectin-3 in the pig retina during postnatal development. Mol Vis 15:1971–1976
Uehara F, Ohba N, Ozawa M (2001) Isolation and characterization of galectins in the mammalian retina. Invest Ophthalmol Vis Sci 42(10):2164–2172
Klein R (2009) Bidirectional modulation of synaptic functions by Eph/ephrin signaling. Nat Neurosci 12(1):15–20
Stumm R, Hollt V (2007) CXC chemokine receptor 4 regulates neuronal migration and axonal pathfinding in the developing nervous system: implications for neuronal regeneration in the adult brain. J Mol Endocrinol 38(3):377–382
Engle EC (2010) Human genetic disorders of axon guidance. Cold Spring Harbor Perspect Biol 2(3):a001784
Schmitt A, Hasan A, Gruber O, Falkai P (2011) Schizophrenia as a disorder of disconnectivity. Eur Arch Psychiatry Clin Neurosci 261(Suppl 2):S150–S154
Haverkamp S, Michalakis S, Claes E, Seeliger MW, Humphries P, Biel M, Feigenspan A (2006) Synaptic plasticity in CNGA3(−/−) mice: cone bipolar cells react on the missing cone input and form ectopic synapses with rods. J Neurosci 26(19):5248–5255
Acknowledgments
We thank G.-S. Nam, Ilse von Graevenitz and Jennifer Schmidt for technical assistance, Jeroen Pasterkamp (University Medical Center Utrecht) for helpful discussion on PlexinC1, and Heinz Wässle for financial support. Grant sponsor: Deutsche Forschungsgemeinschaft (DFG).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Michalakis, S., Schäferhoff, K., Spiwoks-Becker, I. et al. Characterization of neurite outgrowth and ectopic synaptogenesis in response to photoreceptor dysfunction. Cell. Mol. Life Sci. 70, 1831–1847 (2013). https://doi.org/10.1007/s00018-012-1230-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-012-1230-z