Abstract
Stem cell senescence is considered deleterious because it may impair tissue renewal and function. On the other hand, senescence may arrest the uncontrolled growth of transformed stem cells and protect organisms from cancer. This double function of senescence is strictly linked to the activity of genes that the control cell cycle such as the retinoblastoma proteins RB1, RB2/P130, and P107. We took advantage of the RNA interference technique to analyze the role of these proteins in the biology of mesenchymal stem cells (MSC). Cells lacking RB1 were prone to DNA damage. They showed elevated levels of p53 and p21cip1 and increased regulation of RB2/P130 and P107 expression. These cells gradually adopted a senescent phenotype with impairment of self-renewal properties. No significant modification of cell growth was observed as it occurs in other cell types or systems. In cells with silenced RB2/P130, we detected a reduction of DNA damage along with a higher proliferation rate, an increase in clonogenic ability, and the diminution of apoptosis and senescence. Cells with silenced RB2/P130 were cultivated for extended periods of time without adopting a transformed phenotype. Of note, acute lowering of P107 did not induce relevant changes in the in vitro behavior of MSC. We also analyzed cell commitment and the osteo-chondro-adipogenic differentiation process of clones derived by MSC cultures. In all clones obtained from cells with silenced retinoblastoma genes, we observed a reduction in the ability to differentiate compared with the control clones. In summary, our data show evidence that the silencing of the expression of RB1 or RB2/P130 is not compensated by other gene family members, and this profoundly affects MSC functions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alterations in stem cell function have been extensively reported in a variety of tissues and experimental systems. On the one hand, impaired stem cell functionality may induce defective tissue regeneration and aging, while on the other hand, uncontrolled self-renewal and proliferation can trigger tumorigenesis. The anti-proliferative effect of the senescence clearly indicates that this process acts as a tumor-suppression mechanism. Early on, senescence was found to be mediated by the two main tumor suppressor pathways of the cell, the ARF/p53 and the INK4a/RB pathways [1]. For these reasons, studies on senescence mechanisms in stem cells are of great interest to dissect the pathways that may control both aberrant cell proliferation and aging phenomena.
Mitotic quiescence is a typical characteristic of stem cells and involves a temporary exit from the cell cycle. This event must be carefully controlled since it is distinct from permanent cell cycle exit. The latter represents the fundamental step to induce a novel program of gene expression, which leads to the elaboration of a specialized phenotype (differentiation) or, alternatively, triggers senescence. It is then clear that components of cell cycle machinery can play a key role in the regulation of stem cell self-renewal, proliferation, differentiation, and senescence.
The analysis of which molecules participate in cell cycle regulation of several stem cell lineages is not yet exhaustive. Moreover, the strict relation between cell cycle regulation, cell self-renewal, cell commitment, and induction of differentiation has not been fully understood and seems to depend on the cell type [2, 3].
The retinoblastoma family genes RB1, RB2/P130, and P107 play a major role in controlling the cell cycle G1/S transition through the negative regulation of the E2F family of transcription factors. In addition, this protein family plays an important role in regulating other cellular processes, such as terminal differentiation and senescence. Initial observations suggested that retinoblastoma family proteins showed overlapping functions, and initial knock-out studies suggested that Rb2/p130 and p107 had an ancillary role. However, several studies have evidenced functional differences among these proteins [2–4].
Recently, it has become clear that the role of Rb1, Rb2/p130, and p107 depends on several parameters such as animal species under investigation, cell type, and cell status (stem cell, progenitor, differentiated cell) [5–8].
Data on the function of retinoblastoma family members in the control of stem cell biology have been biased by the fact that the majority of authors have focused their attention mainly on the Rb1 protein. Data on the role of p107 and Rb2/p130 in stem cell biology are very scant.
Moreover, the functions of Rb1, p107, and Rb2/p130 in the biology of mesenchymal stem cells (MSCs) remain largely uncharacterized. These cells contribute significantly to the maintenance of tissue homeostasis in the body [9–11]. Consequently, any loss in numbers or the functionality of MSCs would have profound consequences for the maintenance of tissue viability [12–14]. In-depth studies on the biology of MSCs also have a great therapeutic value since they are being tested in cell and gene therapy for a number of human diseases [15].
For these reasons, we analyzed mechanisms that regulate the senescence of MSCs and their association with self-renewal and cell commitment. In particular, we focused our attention on the contribution of the retinoblastoma gene family to these decisions.
Materials and methods
All detailed procedures can be found in Supplemental file 1.
MSC cultures
Bone marrow was obtained from healthy donors after informed consent. We separated cells on a Ficoll density gradient (GE Healthcare, Italy), and the mononuclear cell fraction was collected and washed in PBS. We seeded 1–2.5 × 105 cells/cm2 in an alpha-MEM containing 10 % FBS and bFGF. After 72 h, non-adherent cells were discarded, and adherent cells were further cultivated to confluency. We carried out silencing experiments on these cells (passage 0) and then harvested cells for all the assays as reported below.
We verified that in our experimental conditions, MSC cultures fulfilled the three proposed criteria to define MSC [16]. All cell culture reagents were obtained from Euroclone Life Sciences (Italy) and Hyclone (UT, USA).
Silencing
shRNAs targeting the human RB1, RB2/P130, and P107 mRNA, as well as scrambled control shRNAs, were designed following the procedure described by the RNAi consortium of Broad Institute (Cambridge, MA, USA), we used the following pLKO.1 vectors to express the shRNAs, available at the RNAi consortium: clone TRCN0000010418 to silence RB1 (NCBI Reference Sequence: NM_000321.2); clone TRCN0000039923 to silence RB2/P130 (NCBI Reference Sequence: NM_005611.3); clone TRCN0000040022 to silence P107 (NCBI Reference Sequence: NM_183404.1).
To generate knock down cells, lentiviral particles were produced as described (http://www.broadinstitute.org/genome_bio/trc/publicProtocols.html); see also Supplemental file 1.
Cell cycle analysis and cell proliferation assay
For each assay, cells were collected and re-suspended in a hypotonic buffer containing propidium iodide. Samples were acquired on a FACSCalibur flow cytometer using the Cell Quest software (BD, NJ, USA). They were then analyzed with a standard procedure using Cell Quest and ModFitLT software (BD, NJ, USA).
Cell proliferation was evaluated by the Quick Cell Proliferation Assay Kit II (Biovision, CA, USA). Following silencing, 1,000 cells were seeded in 96-well culture plates. Cells were collected and counted at 1, 2, 15, 16, 30, and 31 days post-plating. The ratio of the total number of cells at day “n” to the number of cells at day “n−1” was regarded as the cell proliferation rate [17].
BrdU assay
For BrdU, immunostaining was performed according to manufacturer’s protocol (Supplemental file 1).
Colony-forming unit (CFU) assay
MSCs were plated as described above. Cultures were expanded to 70–80 % confluency. We carried out silencing experiments on these cells (passage 0), and then we harvested cells for CFU assay as reported [18].
Clonal differentiation
MSCs were plated as described above. Cultures were expanded to 70–80 % confluency. We carried out silencing experiments on these cells (passage 0), and then we harvested cells for clonal differentiation assay as reported [19].
Adipogenic, osteogenic, and chondrogenic differentiation
Cells were stimulated for 2 weeks with specific differentiation media (Lonza, MD, USA). Differentiated cells were evidenced as reported in the Supplemental file 1.
Annexin V assay
Apoptotic cells were detected using fluorescein-conjugated Annexin V (Roche, Italy) following the manufacturer’s instructions.
In situ senescence-associated beta-galactosidase assay
The percentage of senescent cells was calculated by the number of blue, beta-galactosidase-positive cells out of at least 500 cells in different microscope fields as already reported [20].
Quantitative senescence-associated beta-galactosidase assay
4-Methylumbelliferyl-β-d-galactopyranoside (4-MUG) is a substrate of beta-galactosidase that does not fluoresce until cleaved by the enzyme to generate the fluorophore 4-methylumbelliferone. An assay was carried out on lysates obtained from cells that were grown in 96-well plates as reported [21]. The production of the fluorophore was monitored at an emission/excitation wavelength of 365/460 nm.
Immunocytochemistry for detection of 8-oxoguanine, H2AX
8oxodG, H2AX, and HP1 were detected according to the manufacturer’s protocol. Hoechst 33342 staining was performed, and then cells were observed through a fluorescence microscope (Leica Italia, Italy). The percentage of 8oxodG, H2AX-positive cells was calculated by counting at least 500 cells in different microscope fields. Further details can be found in Supplemental file 1.
Treatment with DNA-damaging agents
MSC cultures were treated for 1 h with 300 μM H2O2. Following treatment, the medium was removed, and the complete medium was added. Cells were then collected for data analysis 24 h later.
For UV irradiation, cell plates with lids removed were irradiated with UV light by exposure to a germicidal lamp (peak sensitivity was approximately 254 nm) in a tissue culture hood (15 mJ/cm2). Following treatment, the medium was removed, and the complete medium was added. Cells were collected 24 h later.
For the induction of DNA double-strand breaks, MSCs were incubated for 6 h with 1 μM doxorubicin. Following treatment, the medium was removed, and the complete medium was added. Cells were then collected for data analysis 24 h later.
Comet assay
The single-cell gel electrophoresis assay (Comet assay) was used for the detection of DNA damage at the level of the individual cell. Further details can be found in Supplemental file 1.
RNA extraction, RT-PCR, and real-time PCR
The total RNA was extracted from cell cultures using Omnizol (Euroclone, Italy) according to the manufacturer’s protocol. mRNA levels were measured by RT-PCR amplification as previously reported [22].
Microarray analysis
Total RNA was converted to cRNA and hybridized to Affymetrix human gene ST 1.0 arrays (Affymetrix, Germany) according to the manufacturer’s instructions. We used the David web-based software (http://david.abcc.ncifcrf.gov/) to carry out a comprehensive set of functional annotation tools to understand the biological meaning behind the large list of the up-regulated and down-regulated genes in the different silencing experiments. We used a Venn diagram (http://genevenn.sourceforge.net/) to display the number of common characteristics among the RB1, RB2/P130, and P107 gene lists. Fold change values for genes were calculated as the ratio of the signal values of the experimental (silenced samples) group compared with the control group. Gene expression changes with >1.6-fold alterations were considered significant. For validation of microarray data we performed quantitative real-time RT-PCR on some of the genes that showed significant expression changes (see Supplemental file 10). Further details on the microarray data analysis are provided in Supplemental file 1.
Western blotting
Cells were lysed in a buffer containing 0.1 % Triton for 30 min at 4 °C. A total of 10–40 μg of each lysate was l-electrophoresed in a polyacrylamide gel and electroblotted onto a nitrocellulose membrane. All the primary antibodies were used according to the manufacturers’ instructions. Immunoreactive signals were detected with a horseradish peroxidase-conjugated secondary antibody (Santa Cruz, CA, USA) and reacted with ECL plus reagent (GE Healthcare, Italy).
Results
Bone marrow cells were plated as described in section “Materials and methods”. Cultures were expanded to 70–80 % confluency. We carried out silencing experiments on these cells (passage 0) and then selected the cells expressing shRNAs by a puromycin treatment. After antibiotic selection, cells were expanded at 70–80 % and, unless otherwise stated, all the experiments were carried out soon after.
Silencing of retinoblastoma family
Human MSCs were tested for RB1, RB2/P130, and P107 knockdown after lentiviral transductions and puromycin selection. Cell expressing shRNAs against RB1, RB2/P130, and P107 were indicated as shR1, shR2, and sh107, respectively. Cells expressing scrambled shRNAs were indicated as shCTRLs.
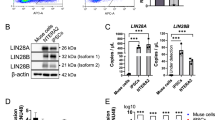
The selected shRNAs were effective in silencing and induced a decrease of their target mRNAs, as detected by RT-PCR (Fig. 1). Silencing was further verified by analyzing the protein levels of knocked-down genes. We observed a decrease in the target proteins (Fig. 1). We detected a 70 % decrease of RB1 in shR1 compared with the controls. Of interest, the levels of the P107 protein increased dramatically upon the silencing of RB1. Also, the expression of RB2/P130 increased in shR1 (Fig. 1).
Silencing of retinoblastoma family. Left: in cells expressing either shR1, or shR2, or sh107, the mRNA levels of genes under analysis were normalized with respect to HPRT, chosen as an internal control. The histogram shows the ratio of gene expression between treated and control cells (shCTRL). The mean expression values (±SD, n = 3; *p < 0.05) of each gene are presented. Right: Western-blot analysis in MSCs expressing either shR1, or shR2, sh107 or shCTRL. The arrow indicates the RB2/P130 hyperphosphorylated inactivated form. The protein levels were normalized with respect to alpha-tubulin, the loading control. The table shows the mean expression values (±SD, n = 3). The change in the protein level of treated cells was compared with that of shCTRL-transduced cells (*p < 0.05)
We observed only a 40 % decrease of RB2/P130 protein in shR2; nevertheless, the expressed protein was almost completely in its hyperphosphorylated, inactivated form. In shR2, the levels of the P107 protein also increased significantly compared with shCTRL (Fig. 1).
In cells with silenced P107, we found evidence of nearly a total decrease of the target protein and an upregulation of RB2/P130 (Fig. 1).
Silencing of RB1 but not RB2/P130 decreases proliferative activity
The cell proliferation curves showed a significant increase in the cell growth of shR2 compared with the control, whereas shR1 grew less than shCTRL (Fig. 2a). These data were in agreement with the flow cytometry carried out at the same time points of cell proliferation analysis. In several experiments, we observed a reduction of S-phase cells, along with an increase of G1 cells in samples with silenced RB1. Conversely, shR2 had an increased percentage of the S-phase cell population (p < 0.05) (Fig. 2b). Cells with silenced P107 showed no significant differences when compared with the controls (Fig. 2b).
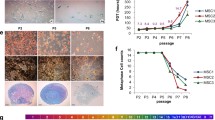
Effect of silencing on cell growth. a Cell proliferation was evaluated by a Quick Cell Proliferation Assay Kit II (Biovision, CA, USA). Following silencing cells were seeded in 96-well culture plates. At 1, 2, 15, 16, 30, 45, 60, and 80 days post-plating, cells were collected and counted. The ratio of the total number of cells at day “n” to the number of cells at day “n−1” was regarded as the cell proliferation rate. b Representative FACS analysis of MSCs transduced with lenti-siRNAs. The experiment was carried out at day 15 after plating, as reported in panel a. c BrdU assay. The micrograph shows a representative field of BrdU (brown) staining shR1, shR2, sh107 and shCTRL. The experiment was carried out at day 15 after plating, as reported in panel a. The table shows the mean expression values and standard deviations (SD) (n = 3, **p < 0.01)
It should be underscored that MSCs lacking RB1 did not show increases in the S-phase as occurs in many other biological systems [2]. This unexpected result prompted us to strengthen our data with a BrdU assay. MSCs carrying silenced RB2 displayed 12.2 % BrdU-positive cells, compared with the 5 % observed in the controls (p < 0.05) (Fig. 2c). Differences among shR1- sh107 and shCTRL were not significant (Fig. 2c).
RB1 and RB2/P130 silencing showed opposite effects on senescence and apoptosis
Annexin assays evidenced a reduced percentage of apoptotic cells in cultures from RB2/P130-silenced samples as compared to the controls, whereas no modification occurred in shR1 and sh107 (Fig. 3a and Supplemental file 4). The reduced level of apoptosis in MSCs is in agreement with our previous research, showing that overexpression of RB2/P130 in MSCs induced programmed cell death [23].
Effect of silencing on apoptosis and senescence. a Annexin assay. The micrographs show representative fields of cells stained with Annexin V (green). Nuclei were counterstained with Hoechst 33342 (blue). Arrows indicate Annexin-positive cells. The mean percentage value of apoptotic cells (±SD, n = 3) is indicated below each picture. Statistically significant variations (p < 0.05) are in bold. b Senescence assay. Representative microscopic fields of acid beta-galactosidase (blue) in cells with silenced retinoblastoma proteins are shown. The mean percentage value of senescent cells (± SD, n = 3) is indicated below each picture. Statistically significant variations (p < 0.05) are in bold. MUG indicates the results of quantitative senescence-associated beta-galactosidase assay carried out on lysates obtained from cells. Data are expressed in arbitrary units. Experiments were repeated three times. Statistically significant variations (p < 0.05) are in bold. c H2AX staining. Fluorescence photomicrographs show the merging of cells stained with anti-H2AX (green) and Hoechst 33342 (blue). A representative microscopic field for each treatment is shown. Arrows indicate double-stained cells. The degree of H2AX phosphorylation was evaluated by counting the number of H2AX foci/cell. We classified cells into three groups: H2AX-negative cells (0 foci/cell); mild H2AX activation (1–10 foci/cell); strong H2AX activation (>10 foci/cell). Mild and strong activation of H2AX indicate cells with damaged DNA. The mean percentage of H2AX-positive cells is indicated below each picture (±SD, n = 3). Statistical evaluation was carried out on the total number of H2AX-positive cells (n = 3), and significant variations (p < 0.05) are in bold. d Fluorescence photomicrographs show cells stained with anti-8-oxo-dG (green). Nuclei were counterstained with Hoechst 33342 (blue). A representative microscopic field for each treatment is shown. Arrows designate 8-oxo-dG-positive cells. The mean percentage of 8-oxo-dG-positive cells is indicated below each picture (±SD, n = 3). Statistically significant variations (p < 0.05) are in bold
We observed signs of senescence in shR1 as detected by in situ senescence-associated-beta-galactosidase assay, compared with cells transduced with controls shRNAs (Fig. 3b and Supplemental file 6). Beta-galactosidase-positive cells showed the characteristic senescent morphology (flat, enlarged cells). Moreover, we detected an increase in the percentage of bi-nucleate cells in shR1 (data not shown), which is another hallmark of senescent cells [24].
On the contrary, the percentage of in situ beta-galactosidase-positive cells was lower in shR2 compared with shCTRL (Fig. 3b and Supplemental file 5). The in situ assay is a cytochemical method based on the production of a blue-dyed precipitate that may generate false-positive results [21].
We corroborated our data with a quantitative method for measuring the beta-galactosidase activity. This was done to also detect relatively subtle changes in activity. The results of the quantitative assay confirmed data obtained with the in situ assay (Fig. 3b and Supplemental file 5).
Retinoblastoma proteins and DNA damage
Extensive research has revealed how the DNA damage-sensing and -signaling pathways, referred to as the DNA damage response network, are tied to cellular senescence and apoptosis [25, 26]. Following DNA damage, the cell responds by activating the cell cycle checkpoint and repair mechanisms. If DNA damage cannot be properly repaired, damaged cells can be eliminated either by apoptosis or senescence.
Changes in apoptosis and senescence rates in MSCs with silenced retinoblastoma proteins prompted us to investigate the degree of DNA damage in our cell system.
Indeed, in shR1, which is prone to senescence, we observed an increase in the number of cells labeled with anti phosphorylated-H2AX, which is a hallmark of damaged DNA nuclear foci (Fig. 3c and Supplemental file 6). The observed data were in agreement with the analysis of the level of DNA damage in cell nuclei by immunodetection of the mutated 8-oxo-2′-deoxyguanosine (oxo8dG) (Fig. 3d and Supplemental file 7). Conversely, in shR2, which showed minimal levels of apoptotic and senescent cells, the percentage of H2AX- and oxo8dG-positive cells was lower compared to the control (Fig. 3c, d and Supplemental files 6, 7). We did not detect any modification in sh107 in the number of H2AX- and oxo8dG-positive cells compared with the control (Fig 3c, d and Supplemental files 6, 7).
Overall, these results prompted us to hypothesize that silencing RB1 and RB2/P130 may render these cells more or less sensitive to DNA damage, respectively. To verify this hypothesis, we treated MSCs with three different DNA-damaging agents: H2O2, UV radiation, and doxorubicin [27–30].
In the control cells (shCTRL), all the treatments induced an increase in the number of cells labeled with anti-phosphorylated-H2AX (Table 1). It is noteworthy that in subsequent DNA-damaging treatments, cells lacking RB1 showed evidence of a further increase of H2AX-positive cells when compared with the controls. Alternately, MSCs with silenced RB2/P130 evidenced a minor percentage of damaged cells (Table 1).
H2O2 induces typical hydroxyl radical-induced products of DNA bases such as oxo8dG [28]. For this reason, we further evaluated its effects through the immunodetection of oxo8dG. shR1 cells treated with peroxide hydrogen showed evidence of a higher number of oxo8dG-positive cells compared with the H2O2-treated control, while in shR2 cultures incubated with H2O2, the labeled cells were significantly less than in the corresponding shCTRLs.
Doxorubicin is drug that induces mainly DNA double-strand breaks [29, 30]; we determined its effect by the detection of DNA breaks with a Comet assay. Quantitation of fragmented DNA showed a higher percentage of damaged DNA in MSCs lacking RB1 and a lower degree of DNA fragmentation in shR2 cells, compared with control cultures (Table 1).
Silencing of RB1 activates the tumor suppressor p53
TP53 is considered to be a key guardian of the genome and prevents the propagation of abnormal cells at risk of becoming cancer cells [31]. Cyclin kinase inhibitor-related pathways are strictly associated with p53, and their activation/inactivation plays a role in determining whether cells undergo senescence or apoptosis [2, 31].
We detected a significant up-regulation (p < 0.05) of p53 protein in shR1 along with an increase in the level of p27kip1 and p21cip1, whereas no modification was observed in p16Ink4a (Fig. 1). The increased expression of these genes was also observed at mRNA level (Fig. 1). This result may suggest that cell cycle arrest and senescence in cells with silenced RB1 relies on the p53–p21–p27 axis rather than on p16. Even if several research studies suggest a primary role for p16Ink4a in senescence [32], there are several reports showing that replicative senescence, as well as accelerated senescence triggered by ionizing radiation, was accompanied by the sustained nuclear accumulation of p21cip1 but did not correlate with p16Ink4a expression [33, 34].
Cells with silenced RB2/P130 showed a reduced degree of DNA damage. This was associated with a significant reduction in the mRNA expression of the P21 gene and no modification of its protein level. No significant changes in the expression of TP53 and of the other CKIs were detected (Fig. 1). This result is in good agreement with reports showing a major role for p53-p21 in cell cycle arrest following DNA damage to repair cells [2, 31]. sh107 showed only a strong up-regulation of p27kip1 compared with controls (Fig. 1).
Silencing of retinoblastoma proteins affected cell commitment and differentiation
The control of the stem cell properties (self-renewal, multipotentiality) is strictly linked to regulation of the cell cycle; hence it is reasonable to have perturbation in stem cell status in MSCs with silenced retinoblastoma proteins.
We carried out a CFU assay on these cells to test their clonogenicity, i.e., their ability to expand at a single-cell level, which is an important feature of self-renewing stem cells.
The silencing of RB2/P130 doubled the number of clones observed in the 100-mm plates seeded at low density. Conversely, the RB1 knocked-down cells showed a significant reduction of clones (Fig. 4a). This result is in good agreement with data on cell proliferation and senescence. In fact, senescence observed in cells with silenced RB1 could greatly affect the “stemness” of MSC cultures, whereas low levels of senescence and apoptosis in shR2 fits the CFU results.
Effect on stemness properties. a CFU assay. The pictures show representative crystal violet staining of clones obtained after a 14-day incubation of MSCs plated following silencing experiments. b Clonal differentiation assay. The first column of the table indicates the total number of analyzed clones of silenced retinoblastoma proteins and the control. The second column shows the clones with differentiation ability. c Clonal differentiation assay. The frequency and differentiation potential of clonal MSCs were assessed as indicated in “Materials and methods”. The table indicates clones with tri-, bi-, and unipotent ability. O Osteogenic phenotype; C Chondrogenic phenotype; A Adipogenic phenotype. The tables show the mean expression values. The change in the clones of treated cells was compared with that of shCTRL-transduced cells (*p < 0.05)
The deregulation of the cell cycle could also affect the cell commitment and differentiation process. We assessed the multipotentiality of individual MSC clones obtained by limiting dilutions. To this end, we isolated, expanded, and characterized 182 clones from control MSCs and from cultures with silenced retinoblastoma proteins. We analyzed their osteo-chondro-adipogenic (OCA) potential by in vitro assays.
Cultures of MSCs are heterogeneous in morphology and in their content of the earliest progenitor cells. For this reason, MSC cultures are composed of progenitors with tri-, bi-, and uni-potential differentiation capability [19].
In control cultures, 85 % of analyzed clones were able to differentiate (right column of table in Fig. 4b). Some clones were able to differentiate into the three lineages we analyzed, whereas other clones were bipotent or unipotent (last row of table in Fig. 4c). In contrast to other reports [19], we obtained clones able to differentiate in all the possible associations of phenotypes (OA-, OC-, CA-bipotent clones; O-, C-, A-unipotent clones) (last row of table in Fig. 4c).
We observed a significant reduction in the ability of all clones obtained from cells with silenced retinoblastoma proteins to differentiate: 50 % of RB1, 40 % of RB2/P130, and 53 % of P107 clones gave differentiated progeny, compared with 85 % of control clones (Fig. 4b). This result is of great interest: the lack of retinoblastoma proteins impaired the differentiation process of MSCs to the same extent, even if the silencing effects on the main biological phenomena we analyzed (proliferation, DNA damage, apoptosis, senescence) are different for each member. Our data showed evidence that cell differentiation is greatly affected by such effects.
The knockdown of retinoblastoma genes also influenced the multipotential properties of MSC clones (Fig. 4c). In shR1 cultures we did not find all the possible associations of phenotypes that were observed in the controls; this could be in part due to the low number of clones that were able to differentiate compared with shCTRL clones (Fig. 4c). Interestingly, we observed a bias toward adipocyte differentiation: eight out 14 clones (57 %) were A-unipotent compared with eight out 42 clones (19 %) in the control. Also, clones from shR2 cells showed a reduced number of possible phenotypes and an adipogenic differentiation bias (10 out 20). Differences in the percentage of clones between sh107 cultures and controls were not statistically significant (Fig. 4c).
Long-term cultures of cells with silenced RB2/P130
Although it has been shown that human MSCs and other stem cell populations continuously grow in vitro for 10 to 20 passages (roughly 30–90 days in vitro), it seems that MSCs, similar to all other primary cells, are subject to the Hayflick limit, i.e., to replicative senescence [35–37]. Conflicting results have been published on the ability of human MSCs to undergo spontaneous in vitro transformation [35, 37].
On this premise, we decided to analyze the silencing effect of retinoblastoma proteins on the long-term culture behavior of MSCs. This was also done because the lack of function of RB1, and to a lesser extent of RB2/P130, has been associated with neoplastic transformation [2, 38, 39]. We found evidence that shR2 cells could be grown in vitro for more than a year without evident sign of cellular transformation (Fig. 2a, Supplemental files 2, 3).
Global changes in gene expression following gene silencing
Previously, attempts to identify the impact of RB family members on gene expression have examined fibroblasts from corresponding knockout mice or from established normal human diploid fibroblast lines [40, 41]. Here, we analyzed the effect of acute inhibition of different retinoblastoma proteins on growing MSCs. Transcriptional profiling was performed on growing cells expressing shRNA, targeting RB1, RB2/P130, or P107.
Reproducible shRNA-dependent changes in gene expression were observed under all conditions. Cells with silenced RB1 underwent the most substantial changes in gene expression, as evidenced by the number of genes that were up- or down-regulated (Fig. 5). Interestingly, the three members of the RB family show either common or specific regulations of gene expression. This further strengthens the concept that retinoblastoma proteins have both specific and overlapping functions [2, 23].
Venn diagrams. Venn diagrams of up-regulated (UP) and down-regulated (DOWN) genes in MSCs treated with shRNAs against RB1, RB2/P130, and P107, respectively. Numbers refer to genes whose expression was changed following silencing experiments. Gene expression changes with >1.6-fold alterations were considered significant
We focused our attention on clusters of genes (Table 2) that may be related to the biological phenomena described above. A growing body of literature demonstrates that distinct functional characteristics of stem and progenitor cells are under redox regulation [42]. Alternately, unbalanced redox regulation may trigger cellular senescence [42–44]. Cells with silenced RB1 showed a reduction in the expression of thioredoxin, while MSCs with decreased RB2/P130 evidenced a significant up-regulation. This result is in line with the role of thioredoxin in the positive regulation of the OCT-4, a stem cell-specific transcription factor [45]. The role of oxidative stress as a major mediator of senescence appears strictly associated with changes in the intracellular glutathione status (GSH/GSSG) that is also regulated through glutathione-S-transferase. The expression of this enzyme is also modulated in RB1 and RB2/P130-silenced cells. The senescence of shR1 cells is in good agreement with modification in the expression of growth factors and changes in the extracellular matrix [32, 46]. The increased proliferation of shR2 MSCs and their resistance to DNA damage are in line with a strong up-regulation of several genes involved in DNA repair and in processes required for cell growth, including the expression of ribosomal proteins and translation initiation factors (Table 2).
Finally, it should be underscored that ribosomal RNA maturation is a phenomenon regulated collectively by all the three retinoblastoma proteins. Indeed, their silencing induced a down-regulation of several small nucleolar RNAs (snoRNAs) and of Vault RNA 1–2, a long non-coding RNA that seems involved in the regulation of ribonucleoprotein particles [47] (Table 2). It is well known that the retinoblastoma proteins may negatively regulate the synthesis of rRNA by RNA polymerases I. Suppression of these important determinants of biosynthetic capacity might provide a mechanism for restraining cell growth [48, 49]. This simplified vision is challenged by our data, showing that silencing of retinoblastoma proteins induce an up-regulation of genes involved in rRNA maturation. This may suggest that the retinoblastoma proteins regulate rRNA synthesis and maturation through a complex feedback process that has not been described until now.
Discussion
Cellular senescence is a process by which the capacity for cell division, growth and function is lost over time, ultimately leading to cell death. Cells are the fundamental structure composing our bodies, and hence cellular decline contributes to aging. According to this view, senescence recapitulates, at the cellular level, the aging of our body. In this context, cellular senescence is considered deleterious because it contributes to decrements in tissue renewal and function. On the other hand, cellular senescence has been also proposed to be an anti-cancer mechanism. This hypothesis arose from the fact that cancer cells proliferate indefinitely, and cellular senescence may arrest cancer growth. Senescence is beneficial in this respect because it protects organisms from cancer. The paradox of this double aspect of senescence has puzzled researchers for long time. Now, it is clear that this arises from the intimate connection of senescence with the cell cycle. Many cell cycle regulator genes, such as those belonging to p53- and RB-pathways, are both involved in cancer onset and also play a key role in organism aging. On this premise, we have analyzed the biology of MSCs with reduced functions of the members of the retinoblastoma family, in an effort to address the importance of these genes in senescence and the phenomena that may be impaired by senescence, cell proliferation, self-renewal, commitment, and differentiation.
The silencing of RB1 proved to be most deleterious to MSCs. It decreased proliferation, promoted the accumulation of DNA damage, and impaired stem cell properties with an increase in senescence and loss of self-renewal properties (Figs. 1b, 3b, 4a).
It is well known that RB1 is essential to regulate E2F1 activity in order to prevent premature entry into the S-phase [50]. An inactivation of RB1 disturbs the balance between the temporally programmed initiation of DNA replication and a progression through the S-phase and mitosis. These processes are a source of DNA damage [50]. Accordingly, we showed that MSC cultures with silenced RB1 exhibited an increased number of cells which were positive for gamma-H2AX and oxo8dG immunostaining (Fig. 3c, d), which are hallmarks of damaged DNA. Also, they were more sensitive to exogenous DNA damage than were control cells. This indicates that the maintenance of the RB1 function is crucial for MSCs to maintain genome stability.
Similarly, Takahashi et al. [51] found that DNA damage response cascades converge on the activation of the tumor suppressor protein TP53. As a major consequence, TP53 transactivates the promiscuous CDK inhibitor p21CIP1 and thereby blocks further passage of damaged cells through the cell cycle. The suppression of CDK activity also promotes the accumulation of hypophosphorylated p27KIP1, a CDK inhibitor that has a high affinity for cyclin E-CDK2 complexes [52], and it stabilizes RB2/P130 in its hypophosphorylated form [53]. All of these changes were noted in MSCs with silenced RB1, which provides evidence for the activation of the DNA damage response cascade downstream of gamma-H2AX.
The activation of p53 is a decisive step for the induction of cellular senescence both in human and mouse fibroblasts [54]. It coincides with the suppression of RB1 and P107 [53, 55], leaving behind RB2/P130 as the dominating retinoblastoma protein in senescent cells [56]. Our data on human MSCs show that the silencing of RB1 itself triggers the activation of p53-RB2/P130 crosstalk, which leads to an irreversible cell cycle arrest. The data indicate that neither RB2/P130 nor P107 can substitute RB1 to maintain the proliferative activity of MSCs. This is consistent with the note that RB1, p107, and RB2/P130 regulate completely different gene settings [57]. In summary, the maintenance of genomic stability in MSCs is highly dependent on a functional RB1. Impairing these control functions provokes a shift to RB2, causing premature senescence [50].
In cells with silenced RB2/P130, we observed an increase in the proliferation rate and an increase in self-renewal properties as detected by the CFU assay (Figs. 2a, c, 4a). These results are in good agreement with data showing a reduced degree of DNA damage, apoptosis, and senescence (Fig. 3a–d).
The decrease in the percentage of senescent cells in shRB2MSCs is in good agreement with the observation that cells with senescence markers show high levels of RB2/P130 [53, 58]. This suggests that Rb2/p130 is the critical retinoblastoma protein in the advanced senescent stage, participating in a growth arrest stalling cells beyond the restriction point by forming strong repressor complexes at E2F1 promoter binding sites.
Together, our data demonstrate that, at least in some cell types and in some cellular contexts, the RB1-P16 pathways may be dispensable for senescence, whereas the activation of p53 along with the accumulation of RB2/P130 may be the leading events.
Moreover, a long-term culture of shR2-MSCs further demonstrated that the accumulation of RB2/P130 is a key event for a definitive cell cycle exit and the trigger of a senescence program. On the other hand, the absence of transformation events in this long-term culture is in agreement with an ancillary role for RB2/P130 in malignant transformation. We cannot, however, exclude the possibility that cells in shR2-MSC long-term cultures had alteration in their karyotype and modification in the panel of expressed genes.
The classic role for the retinoblastoma family is the regulation of the cell cycle as repressors of the E2F family of transcription factors [2]. An equally important aspect of retinoblastoma proteins is the development and differentiation of many tissues because RB1 can activate certain differentiation transcription factors to promote cellular differentiation [59]. The retinoblastoma family has been shown to be differentially regulated during the adipogenic differentiation of preadipocyte cell lines [60, 61].
The sequestration of retinoblastoma family members by SV40 large T antigen inhibits adipocyte differentiation [62]. In addition, RB1 −/− fibroblasts are unable to differentiate without exogenous PPAR-gamma activators [63].
In contrast to the research reported above, we demonstrate that the targeted loss of RB1 or RB2 in MSCs leads to bias toward the adipocyte phenotype. To reconcile these studies, it should be emphasized that our analysis was carried out on multipotent progenitor cells, whereas other investigations were done on committed pre-adipocytes and/or fibroblasts. It is possible that retinoblastoma proteins play a complex role in adipogenesis: they may be dispensable during the adipocyte cell commitment of multipotent stem cells, whereas they could further promote differentiation process in committed cells. On the other hand, there is also a report showing that, paradoxically, RB1 can inhibit adipogenesis by attenuating the capacity of the PPAR-gamma transcription factor to drive gene expression [64]. It is evident that the role of the retinoblastoma family in adipocyte differentiation should be investigated in further depth. This was beyond the scope of the current research.
In conclusion, our study showed evidence that the retinoblastoma proteins play a key role in regulation of several aspects of MSC biology: proliferation, stemness, cell commitment, senescence, and apoptosis.
The three members of the retinoblastoma family have specific functions that only partially overlap. In fact, our data evidenced that the silencing of RB1 or RB2/P130 expression is not compensated by other gene family members, and this profoundly effected MSC functions.
References
Collado M, Blasco MA, Serrano M (2007) Cellular senescence in cancer and aging. Cell 130(2):223–233 [pii: S0092-8674(07)00890-2]
Galderisi U, Cipollaro M, Giordano A (2006) The retinoblastoma gene is involved in multiple aspects of stem cell biology. Oncogene 25(38):5250–5256
Galderisi U, Jori FP, Giordano A (2003) Cell cycle regulation and neural differentiation. Oncogene 22(33):5208–5219
Claudio PP, Howard CM, Baldi A, De Luca A, Fu Y, Condorelli G, Sun Y, Colburn N, Calabretta B, Giordano A (1994) p130/pRb2 has growth suppressive properties similar to yet distinctive from those of retinoblastoma family members pRb and p107. Cancer Res 54(21):5556–5560
Herrera RE, Sah VP, Williams BO, Makela TP, Weinberg RA, Jacks T (1996) Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol 16(5):2402–2407
MacPherson D, Sage J, Crowley D, Trumpp A, Bronson RT, Jacks T (2003) Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol Cell Biol 23(3):1044–1053
MacPherson D, Sage J, Kim T, Ho D, McLaughlin ME, Jacks T (2004) Cell type-specific effects of Rb deletion in the murine retina. Genes Dev 18(14):1681–1694
Haigis K, Sage J, Glickman J, Shafer S, Jacks T (2006) The related retinoblastoma (pRb) and p130 proteins cooperate to regulate homeostasis in the intestinal epithelium. J Biol Chem 281(1):638–647
Beyer Nardi N, da Silva Meirelles L (2006) Mesenchymal stem cells: isolation, in vitro expansion and characterization. Handb Exp Pharmacol 174:249–282
Bianco P, Riminucci M, Gronthos S, Gehron Robey P (2001) Bone marrow stromal stem cells: nature, biology and potential applications. Stem Cells 19:180–192
Muller-Sieburg CE, Deryugina E (1995) The stromal cells’ guide to the stem cell universe. Stem Cells 13(5):477–486
Beausejour C (2007) Bone marrow-derived cells: the influence of aging and cellular senescence. Handb Exp Pharmacol 180:67–88
Pelicci PG (2004) Do tumor-suppressive mechanisms contribute to organism aging by inducing stem cell senescence? J Clin Invest 113(1):4–7
Sethe S, Scutt A, Stolzing A (2006) Aging of mesenchymal stem cells. Ageing Res Rev 5(1):91–116
Giordano A, Galderisi U, Marino IR (2007) From the laboratory bench to the patient’s bedside: an update on clinical trials with mesenchymal stem cells. J Cell Physiol 211(1):27–35
Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E (2006) Minimal criteria for defining multipotent mesenchymal stromal cells. International Society for Cellular Therapy position statement. Cytotherapy 8(4):315–317
Nakamura S, Yamada Y, Baba S, Kato H, Kogami H, Takao M, Matsumoto N, Ueda M (2008) Culture medium study of human mesenchymal stem cells for practical use of tissue engineering and regenerative medicine. Biomed Mater Eng 18(3):129–136
Pochampally R (2008) Colony-forming unit assays for MSCs. Methods Mol Biol 449:83–91
Muraglia A, Cancedda R, Quarto R (2000) Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. J Cell Sci 113(Pt 7):1161–1166
Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O (2009) Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc 4(12):1798–1806
Gary RK, Kindell SM (2005) Quantitative assay of senescence-associated beta-galactosidase activity in mammalian cell extracts. Anal Biochem 343(2):329–334
Galderisi U, Di Bernardo G, Cipollaro M, Peluso G, Cascino A, Cotrufo R, Melone MA (1999) Differentiation and apoptosis of neuroblastoma cells: role of N-myc gene product. J Cell Biochem 73(1):97–105
Jori FP, Melone MA, Napolitano MA, Cipollaro M, Cascino A, Giordano A, Galderisi U (2005) RB and RB2/p130 genes demonstrate both specific and overlapping functions during the early steps of in vitro neural differentiation of marrow stromal stem cells. Cell Death Differ 12(1):65–77
Walen KH (2006) Human diploid fibroblast cells in senescence; cycling through polyploidy to mitotic cells. In Vitro Cell Dev Biol Anim 42(7):216–224
Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW (2005) DNA repair, genome stability, and aging. Cell 120(4):497–512
Roos WP, Kaina B (2006) DNA damage-induced cell death by apoptosis. Trends Mol Med 12(9):440–450
Cleaver JE, Crowley E (2002) UV damage, DNA repair and skin carcinogenesis. Front Biosci 7:d1024–d1043
D’Errico M, Parlanti E, Dogliotti E (2008) Mechanism of oxidative DNA damage repair and relevance to human pathology. Mutat Res 659(1–2):4–14
Kurz EU, Douglas P, Lees-Miller SP (2004) Doxorubicin activates ATM-dependent phosphorylation of multiple downstream targets in part through the generation of reactive oxygen species. J Biol Chem 279(51):53272–53281
Nitiss JL (2002) DNA topoisomerases in cancer chemotherapy: using enzymes to generate selective DNA damage. Curr Opin Investig Drugs 3(10):1512–1516
Zuckerman V, Wolyniec K, Sionov RV, Haupt S, Haupt Y (2009) Tumour suppression by p53: the importance of apoptosis and cellular senescence. J Pathol 219(1):3–15
Campisi J, d’Adda di Fagagna F (2007) Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 8(9):729–740
Attema JL, Pronk CJ, Norddahl GL, Nygren JM, Bryder D (2009) Hematopoietic stem cell ageing is uncoupled from p16 INK4A-mediated senescence. Oncogene 28(22):2238–2243
Mirzayans R, Andrais B, Scott A, Paterson MC, Murray D (2010) Single-cell analysis of p16(INK4a) and p21(WAF1) expression suggests distinct mechanisms of senescence in normal human and Li-Fraumeni syndrome fibroblasts. J Cell Physiol 223(1):57–67
Bernardo ME, Zaffaroni N, Novara F, Cometa AM, Avanzini MA, Moretta A, Montagna D, Maccario R, Villa R, Daidone MG, Zuffardi O, Locatelli F (2007) Human bone marrow derived mesenchymal stem cells do not undergo transformation after long-term in vitro culture and do not exhibit telomere maintenance mechanisms. Cancer Res 67(19):9142–9149
Izadpanah R, Trygg C, Patel B, Kriedt C, Dufour J, Gimble JM, Bunnell BA (2006) Biologic properties of mesenchymal stem cells derived from bone marrow and adipose tissue. J Cell Biochem 99(5):1285–1297
Rosland GV, Svendsen A, Torsvik A, Sobala E, McCormack E, Immervoll H, Mysliwietz J, Tonn JC, Goldbrunner R, Lonning PE, Bjerkvig R, Schichor C (2009) Long-term cultures of bone marrow-derived human mesenchymal stem cells frequently undergo spontaneous malignant transformation. Cancer Res 69(13):5331–5339
Giacinti C, Giordano A (2006) RB and cell cycle progression. Oncogene 25(38):5220–5227
Paggi MG, Giordano A (2001) Who is the boss in the retinoblastoma family? The point of view of Rb2/p130, the little brother. Cancer Res 61(12):4651–4654
Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW (2010) Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17(4):376–387
Laurie N, Mohan A, McEvoy J, Reed D, Zhang J, Schweers B, Ajioka I, Valentine V, Johnson D, Ellison D, Dyer MA (2009) Changes in retinoblastoma cell adhesion associated with optic nerve invasion. Mol Cell Biol 29(23):6268–6282
Haneline LS (2008) Redox regulation of stem and progenitor cells. Antioxid Redox Signal 10(11):1849–1852
Zanichelli F, Capasso S, Cipollaro M, Pagnotta E, Carteni M, Casale F, Iori R, Galderisi U (2012) Dose-dependent effects of R-sulforaphane isothiocyanate on the biology of human mesenchymal stem cells, at dietary amounts, it promotes cell proliferation and reduces senescence and apoptosis, while at anti-cancer drug doses, it has a cytotoxic effect. Age (Dordr) 34(2):281–293
Zanichelli F, Capasso S, Di Bernardo G, Cipollaro M, Pagnotta E, Carteni M, Casale F, Iori R, Giordano A, Galderisi U (2012) Low concentrations of isothiocyanates protect mesenchymal stem cells from oxidative injuries, while high concentrations exacerbate DNA damage. Apoptosis 17(9):964–974
Guo Y, Einhorn L, Kelley M, Hirota K, Yodoi J, Reinbold R, Scholer H, Ramsey H, Hromas R (2004) Redox regulation of the embryonic stem cell transcription factor oct-4 by thioredoxin. Stem Cells 22(3):259–264
Kuilman T, Peeper DS (2009) Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer 9(2):81–94
Nandy C, Mrazek J, Stoiber H, Grasser FA, Huttenhofer A, Polacek N (2009) Epstein–Barr virus-induced expression of a novel human vault RNA. J Mol Biol 388(4):776–784
Montanaro L, Trere D, Derenzini M (2008) Nucleolus, ribosomes, and cancer. Am J Pathol 173(2):301–310
White RJ (1997) Regulation of RNA polymerases I and III by the retinoblastoma protein: a mechanism for growth control? Trends Biochem Sci 22(3):77–80
Helmbold H, Galderisi U, Bohn W (2011) The switch from Rb1/p105 to Rb2/p130 in DNA damage and cellular senescence. J Cell Physiol 227(2):508–513
Takahashi P, Polson A, Reisman D (2011) Elevated transcription of the p53 gene in early S-phase leads to a rapid DNA damage response during S-phase of the cell cycle. Apoptosis 16(9):950–958
Grimmler M, Wang Y, Mund T, Cilensek Z, Keidel EM, Waddell MB, Jakel H, Kullmann M, Kriwacki RW, Hengst L (2007) Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell 128(2):269–280
Kapic A, Helmbold H, Reimer R, Klotzsche O, Deppert W, Bohn W (2006) Cooperation between p53 and p130(Rb2) in induction of cellular senescence. Cell Death Differ 13(2):324–334
Odell A, Askham J, Whibley C, Hollstein M (2010) How to become immortal: let MEFs count the ways. Aging (Albany NY) 2(3):160–165
Helmbold H, Deppert W, Bohn W (2006) Regulation of cellular senescence by Rb2/p130. Oncogene 25(38):5257–5262
Helmbold H, Komm N, Deppert W, Bohn W (2009) Rb2/p130 is the dominating pocket protein in the p53–p21 DNA damage response pathway leading to senescence. Oncogene 28(39):3456–3467
Hurford RK Jr, Cobrinik D, Lee MH, Dyson N (1997) pRB and p107/p130 are required for the regulated expression of different sets of E2F-responsive genes. Genes Dev 11(11):1447–1463
Jackson JG, Pereira-Smith OM (2006) Primary and compensatory roles for RB family members at cell cycle gene promoters that are deacetylated and downregulated in doxorubicin-induced senescence of breast cancer cells. Mol Cell Biol 26(7):2501–2510
Zhu L (2005) Tumour suppressor retinoblastoma protein RB: a transcriptional regulator. Eur J Cancer 41(16):2415–2427
Hansen JB, te Riele H, Kristiansen K (2004) Novel function of the retinoblastoma protein in fat: regulation of white versus brown adipocyte differentiation. Cell Cycle 3(6):774–778
Richon VM, Lyle RE, McGehee RE Jr (1997) Regulation and expression of retinoblastoma proteins p107 and p130 during 3T3-L1 adipocyte differentiation. J Biol Chem 272(15):10117–10124
Higgins C, Chatterjee S, Cherington V (1996) The block of adipocyte differentiation by a C-terminally truncated, but not by full-length, simian virus 40 large tumor antigen is dependent on an intact retinoblastoma susceptibility protein family binding domain. J Virol 70(2):745–752
Chen PL, Riley DJ, Chen Y, Lee WH (1996) Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev 10(21):2794–2804
Fajas L, Egler V, Reiter R, Hansen J, Kristiansen K, Debril MB, Miard S, Auwerx J (2002) The retinoblastoma-histone deacetylase 3 complex inhibits PPARgamma and adipocyte differentiation. Dev Cell 3(6):903–910
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2012_1224_MOESM3_ESM.tif
Supplemental File 3. Long-term culture of shR2 cells. A: Contrast phase microscopic field of shR2 cells at different time points. B: Polyacrylamide gel electrophoresis of TRAP assay products obtained from shR2-MSCs at different time points. The image shows the PCR-amplified products indicating telomerase activity. The assay measures the enzymatic activity of telomerase. In the first step of the reaction, active telomerase in cell extracts adds a varying number of telomeric repeats (TTAGGG) onto the 3′ end of a substrate oligonucleotide. Next, PCR is used to amplify the extended products. Products are then electrophoresed on a polyacrylamide gel (see details of the method in Supplementary File 1). C: Differentiation potential of shR2 cells that were cultivated for 12 months. Cells were induced to differentiate into adipocytes, osteocytes, and chondrocytes, as described in the Materials and methods section. The picture on the left shows adipocytes stained with Oil Red. In the middle, differentiated chondrocytes were revealed by staining with Fast Green. On the right, differentiated osteocytes were revealed by staining with Alizarin Red. (TIFF 4381 kb)
18_2012_1224_MOESM4_ESM.tif
Supplemental File 4. Annexin assay. The micrographs show a higher magnification of figure 3A, with representative fields of cells stained with Annexin V (green). Nuclei were counterstained with Hoechst 33342 (blue). Arrows indicate Annexin-positive cells. (TIFF 13091 kb)
18_2012_1224_MOESM5_ESM.tif
Supplemental File 5. Senescence assay. The micrographs show a higher magnification of figure 3B, with representative fields acid beta-galactosidase (blue) in cells with silenced retinoblastoma proteins. (TIFF 15667 kb)
18_2012_1224_MOESM6_ESM.tif
Supplemental File 6. H2AX staining. The micrographs show a higher magnification of figure 3C, with representative fields of cells stained with anti-H2AX (green) and Hoechst 33342 (blue). Arrows indicate double-stained cells. (TIFF 3460 kb)
18_2012_1224_MOESM7_ESM.tif
Supplemental File 7. Anti-8-oxo-dG staining. The micrograph shows a higher magnification of figure 3D, with representative fields of cells stained with anti-8-oxo-dG (green) and Hoechst 33342 (blue). Arrows indicate double-stained cells. (TIFF 25240 kb)
18_2012_1224_MOESM10_ESM.tif
Supplemental File 10. Real-time RT-PCR validation of microarray data. We selected some genes that showed significant changes in their expression according to microarray analysis to validate microarray data by real-time RT-PCR. In cells expressing either shR1, shR2, sh107, or shCTRL, the mRNA levels of genes under analysis were normalized with respect to HPRT, chosen as an internal control. The histogram shows the ratio of gene expression between treated and control cells (shCTRL). The mean expression values (±SD, n = 3; * p < 0.05) of each gene are presented. (TIFF 95 kb)
Rights and permissions
About this article
Cite this article
Alessio, N., Bohn, W., Rauchberger, V. et al. Silencing of RB1 but not of RB2/P130 induces cellular senescence and impairs the differentiation potential of human mesenchymal stem cells. Cell. Mol. Life Sci. 70, 1637–1651 (2013). https://doi.org/10.1007/s00018-012-1224-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-012-1224-x