Abstract
The trafficking of macromolecules between the cytoplasm and the nucleus is controlled by the nuclear pore complexes (NPCs) and various transport factors that facilitate the movement of cargos through the NPCs and their accumulation in the target compartment. While their functions in transport are well established, an ever-growing number of observations have also linked components of the nuclear transport machinery to processes that control chromosome segregation during mitosis, including spindle assembly, kinetochore function, and the spindle assembly checkpoint. In this review, we will discuss this evolving area of study and emerging hypotheses that propose key roles for components of the nuclear transport apparatus in mitotic progression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In eukaryotes, the contents of the nucleus (nucleoplasm) are separated from the surrounding cytoplasm by a double membrane nuclear envelope (NE). All transport across this physical barrier is controlled by elaborate structures termed nuclear pore complexes (NPCs), with a typical mammalian nucleus containing several thousand NPCs. Each NPC is ~65 MDa in mass and consists of ~30 different proteins (termed nucleoporins or nups) present in multiple copies (Fig. 1). The NPCs act as gateways that control the movement of transport proteins, many termed karyopherins or kaps (also termed importins or exportins), and their attached cargos across the NE. These transport processes are regulated by Ran, a small Ras-related GTPase. Through this role in regulating transport into and out of the nucleus, the NPCs and other components of the transport machinery influence a variety of cellular processes, notably gene expression and the maintenance of genome integrity (reviewed in [1, 2]). However, in recent years, it has further become apparent that components of the nuclear transport apparatus also function in other capacities, most notably in events occurring during mitosis.
Schematic representation of the nuclear pore complex (NPC) and its constituents (the nucleoporins/Nups) in vertebrates and in the yeast Saccharomyces cerevisiae. Simplified model of the organization of nucleoporins within the NPC framework in vertebrates (left) and in budding yeast (right). Nucleoporins that belong to a stable sub-complex are enclosed in boxes. Proteins or complexes that are localized at kinetochores during mitosis are circled in red. Other nucleoporins relevant to this review are written in red. Enzymes of the SUMO pathways are highlighted in orange
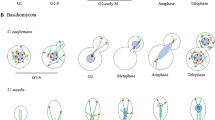
During mitosis in metazoan cells, the NE is either partially or completely disassembled, and components of the NPC are dispersed [2, 3]. At this time, chromosomes condense into visibly definable structures and are exposed to the cytoplasm and the forming mitotic spindle. Spindle assembly is initiated by the nucleation of microtubules from the centrosomes or microtubule organizing centres (MTOCs), and to a lesser extent in the vicinity of chromosomes. In many fungi, however, the MTOCs, called spindle pole bodies, are embedded in the nuclear envelope and the mitotic spindle assembles within the nucleus (Fig. 2). In both open and closed mitoses, the spindle microtubules proceed to engage kinetochores, proteinaceous scaffolds that assemble around a specialized region of each chromosome known as the centromere (reviewed in [4–6]). At the kinetochores, a safety device known as the spindle assembly checkpoint (SAC) monitors microtubule attachment and prevents the separation of the sister chromatids until all kinetochores are bound to microtubules (for review see [7]). Once this requirement has been met, the stage is set for segregation of the sister chromosomes into two forming daughter cells. In metazoans, this is followed by the reassembly of the NE and NPCs. The analysis of this complex series of events has revealed an important role for the nuclear transport machinery, including nucleoporins, karyopherins, and molecules that regulate the GTP-bound state of Ran. While the role of Ran and karyopherins in mitotic spindle assembly has been extensively reviewed [8, 9], we will here mainly focus on proteins that share a dual localization at the NPCs in interphase and at the kinetochores in mitosis. These include: (1) well-established SAC factors, Mad1 and Mad2, that interact with nucleoporins localized at the nuclear side of the NPCs, (2) RanBP2 and Ran-GAP1, two regulators of Ran that are localized on the cytoplasmic NPC filaments in interphase, and (3) the vertebrate Nup107-160 complex, a major structural module of nuclear pores. Current data and hypotheses in this emerging field will be presented.
Mlp-like proteins are binding sites for SAC proteins. a In metazoan cells the nuclear envelope (gray) disassembles during mitosis and Tpr (human)/Mtor (Drosophila), with attached Mad1 and Mad2, are released from their interphase localization at NPCs (not shown). Tpr or Mtor, potentially within a spindle matrix (pink), appears to play a role in the binding of Mad1, Mad2, and Msp1 to unattached kinetochores (red). Tpr may also bind to kinetochores. b In Aspergillus nidulans, NPCs partially disassembles during mitosis increasing the permeability of the nuclear envelope. However, during this period, AnMlp, AnMad1, and AnMad2 remain intranuclear and are associated with what is thought to be the spindle matrix (pink), and, potentially, kinetochores (not shown). From this location, AnMlp may facilitate the association of AnMad1 and AnMad2 with unattached kinetochores. c S. cerevisiae undergo a closed mitosis, with the nuclear envelope and NPCs remaining intact. Mlp1 and Mlp2 remain associated with the NPC during mitosis and activation of the SAC. SAC activation leads to the recruitment of Mad1 and Mad2 to kinetochores and the loss of detectable Mad2 at NPCs. During SAC arrest, Mad1 cycles between NPCs and kinetochores. Note, in all panels, spindles are depicted as dashed black lines attached to centrosomes/spindle pole bodies
Nuclear pore complexes and the spindle assembly checkpoint
The spindle assembly checkpoint (SAC) functions to ensure accurate segregation of mitotic chromosomes by delaying anaphase onset until each kinetochore has properly attached to the mitotic spindle (reviewed in [7]). Under conditions where kinetochore/microtubule attachment is compromised, various proteins functioning in the SAC, among them Mad1 and Mad2, contribute to the formation of a mitotic checkpoint complex containing BubR1/Mad3, Bub3, Mad2, and Cdc20. Key to the role of this complex is the interaction between Mad2 and Cdc20, which negatively regulates the interaction of Cdc20 with the anaphase promoting complex/cyclosome (APC/C). Once functional kinetochore/microtubule interactions are established, the SAC is satisfied allowing Cdc20 to bind the APC/C and initiate its role in the destruction of cyclin B and securin, a key regulator of sister chromatid cohesion, and ultimately progression into anaphase. A number of years ago, the surprising observation was made that two key components of the SAC machinery, Mad1 and Mad2, were positioned at NPCs during interphase. The significance of this association and the relationships between the SAC and the nuclear transport machinery have been the focus of a number of recent studies summarized here.
An evolutionarily conserved NPC localization
Since their initial identification as checkpoint proteins in budding yeast [10], Mad1 and Mad2 have been the focus of many studies in various organisms, most of which examined their function in the SAC, including their role in transmitting a metaphase delay signal from kinetochores lacking association with microtubules [7]. Once all kinetochores are engaged with microtubules, the SAC is satisfied, and Mad1 and Mad2 are no longer detected at kinetochore. While apparently functional only during active SAC, early studies in yeast revealed that Mad1 was present in cells throughout the cell cycle and visibly concentrated at the nuclear envelope [11], suggesting it might function in processes outside the SAC. The nature of its interaction with the nuclear envelope remained unclear until later studies established that yeast Mad1, in complex with Mad2, was bound to the NPC [12]. Both the localization of fluorescent chimeras and copurification of Mad1 and Mad2 with NPC proteins supported this conclusion.
Several studies have shed light on the molecular basis for the interactions of the Mad1/Mad2 complex with the yeast NPC. Through the analysis of deletion mutants in budding [12] and fission [13] yeast, it was shown that cells lacking Mad1 fail to accumulate Mad2 at the nuclear periphery, while loss of Mad2 does not inhibit Mad1 binding to the NPC [12]. Based on these data, it was concluded that Mad1 mediates the association of the Mad1/Mad2 complex to the NPC. Studies in Aspergillus nidulans resulted in similar conclusions [14]. Which NPC proteins interact with the Mad1/Mad2 complex appears to be more complex, and several nucleoporins have been identified as potential binding partners. In Saccharomyces cerevisiae, these include members of the Nup53-containing NPC subcomplex [12]. When affinity purified from cell extracts, several members of this NPC subcomplex, including Nup53, Nup170, and Nup157, are associated with the Mad1/Mad2 complex. Nup53 can also directly bind to Mad1 in vitro and, furthermore, the region of Mad1 that mediates its interactions with the NPC in vivo also binds Nup53 [15]. These interactions, however, do not completely explain how these SAC proteins bind to the NPC, as a deletion mutation in the NUP53 gene, while attenuating the binding of Mad1 to the NPC does not abolish it [12]. Further studies revealed that loss of Mad1/Mad2 binding to the nuclear periphery occurs only after the depletion of two homologous proteins, Mlp1 and Mlp2 [15]. These proteins are believed to form filaments that extend into the nucleus from the nucleoplasmic face of the NPC core. It was thus inferred from these observations that the predominant binding site for the Mad1/Mad2 complex during interphase is on the nucleoplasmic face of the NPC. Similar conclusions have also been drawn from studies in A. nidulans [14]. Consistent with this intranuclear localization, nuclear transport factors and the GTPase Ran were shown to play a role in the targeting of Mad1 and Mad2 to the nuclear face of the NPC in S. cerevisiae [15, 16].
The nuclear envelope association of Mad1 and Mad2 is not restricted to yeast and was also detected in higher eukaryotes during interphase, first in Xenopus and human cells [17, 18], and later in Drosophila cells [19]. As in yeast, the functional relevance of this localization pattern remained unclear until the association of Mad1 and Mad2 with the NPC was demonstrated [20]. In these studies, both immunofluorescence and immunoelectron microscopy placed human Mad1, and by inference Mad2, at the nucleocytoplasmic face of the NPC during interphase. Consistent with these observations, recent studies have demonstrated a physical link between the mammalian counterpart of the Mlp proteins, Tpr, and Mad1 and Mad2, and revealed that siRNA depletion of Tpr leads to a decrease in the levels of NE bound Mad1 and Mad2 [21]. Unlike in yeast, however, depletion of Mad1 did not change the NPC localization of Mad2, a result likely reflecting the existence of two distinct binding sites on Tpr for Mad2 and Mad1 [21]. Interactions between Mlp-like proteins are also likely to contribute to NPC binding of Mad1 and Mad2 in other metazoan cells, including Drosophila. In support of this hypothesis, in Drosophila embryos, Mad1 and Mad2 are recruited to the NE following mitosis at times coincident with the appearance of the Mlp-like protein, Mtor, at the NPCs [22]. Furthermore, recent studies have shown that Mtor coimmunoprecipitates with Mad2 in early Drosophila embryos [23]. Whether the Mlp-like proteins are the sole binding partners for these SAC proteins at the NPCs and whether other interacting nucleoporins or transport factors contribute, as in yeast, to this final targeting remains to be directly established. However, the latter may be the case, as depletion of Nup53 in HeLa cells causes a corresponding decrease in NE-associated Mad1 [24]. Moreover, the loss of nuclear Ran-GTP leads to the release of Mad2 from vertebrate NPCs [16], raising further questions about the nature of its interactions with the NPC.
Functional implications for the association of Mad1/Mad2 with NPC proteins
Several recent publications have shed some light on the potential function of the association of Mad1 and Mad2 with the Mlp-like proteins in various species. Studies focusing on the role of these interactions in SAC function support the conclusion that Mlp-like proteins function as a scaffold for Mad1 during mitosis. However, they point to potential differences in where and how Mlp proteins may function during SAC arrest.
During mitosis in Drosophila, Mtor is recruited to a matrix structure that surrounds the spindle and is distinct from the microtubules ([23, 25]; see Fig. 1). Several observations suggest that, from its position within the spindle matrix, Mtor regulates the association of Mad2 and the SAC kinase Mps1 with kinetochores. Depletion of Mtor, while not affecting spindle structure, results in abnormal chromosome congression and accelerated entry into anaphase, a defect consistent with a role for Mtor in regulating the SAC response [23]. This effect likely arises from a loss of Mad2 accumulation at kinetochores. While Mtor does not appear to interact with kinetochores, it binds to Mad2, and depletion of Mtor causes a decrease in the amount of Mad2 detected at kinetochores. Interestingly, the decrease in kinetochore-bound Mad2 is not caused by a lack of kinetochore-associated Mad1, but rather by a decrease in the Mps1 kinase.
In vertebrates, NE and NPC disassembly during mitosis is accompanied by the release of Tpr into the cytoplasm [26]. Studies by Lee et al. [21] and Lince-Faria et al. [23] revealed that Tpr, Mad1, Mad2, and Mps1 co-precipitate in mitotic-enriched HeLa cell extracts which were devoid of microtubules and intact nuclear pores. Tpr-depleted cells exhibited chromosome segregation defects similar to those seen in cells depleted of Mad1 and Mad2. As in Drosophila, this was accompanied by a mild acceleration of mitotic progression, though less exaggerated as compared to the increase observed upon Mad2 depletion [21].
What changes may occur to the Tpr platform during mitotic progression and how they may influence the activity of Mad1 and Mad2 in the SAC still remains unclear (Fig. 1). Lee et al. [21] have suggested that, during SAC activation in HeLa cells, a portion of the Tpr protein is recruited onto kinetochores. Moreover, they showed that Tpr depletion reduces the levels of Mad1 visible at kinetochores. Under these same conditions, the levels of the Mad2/Cdc20 complex were also diminished, consistent with the loss of Mad1 at unattached kinetochores and its function at this location in the formation of the Mad2/Cdc20 complex. These results would suggest that the same framework that supports the Mad1/Mad2 complex at the NPCs during interphase is transmitted to kinetochores following SAC activation. However, this model was challenged by Lince-Faria et al. [23] who reported a microtubule-dependent association of a fraction of Tpr with the mitotic spindle but no detectable Tpr at kinetochores in human cells. Consistent with these observations, they found that, as in Drosophila, Tpr depletion leads to the decrease of kinetochore-associated Mad2, but not Mad1 [23]. In view of previous studies demonstrating the requirement of Mps1 in the targeting of Mad2, but not Mad1, to unattached kinetochores in human cells [27], they suggested the regulation of Mad2 association with kinetochores by Tpr may, as in Drosophila, be indirectly catalyzed by Mps1.
Evidence for a role of Mlp-like proteins in a spindle matrix has also been provided by studies by De Souza et al. in A. nidulans [14]. Like S. cerevisiae, A nidulans undergoes a closed mitosis (i.e., its NE remains intact during mitosis). However, in these cells, NPCs partially disassemble during mitosis, and the Mlp-like protein, An-Mlp1, dissociates from the NPC [14, 28]. In spite of the increased NE permeability caused by these events, An-Mlp1 remains in the nucleoplasm and, much like its Drosophila counterpart Mtor, it appears to contribute to a spindle matrix. In prophase, An-Mlp1 is concentrated in the vicinity of the kinetochores and forming spindle, and it remains around the spindle as it elongates during anaphase. Interestingly, the localization patterns of An-Mad1 and An-Mad2 in these mitotic cells are nearly identical to An-Mlp1. This is also the case in cells where the SAC was activated. Under these conditions, An-Mad1, An-Mad2, and An-Mlp1 all concentrate around, and, to a degree, overlap with, kinetochores. At these locations, An-Mad1 is likely present in two separate pools, one bound to An-Mlp1, both within the spindle matrix and, potentially, at kinetochores, and another bound to kinetochores at sites independent of An-Mlp1. This latter observation is similar to that made in Drosophila [23]. These results led De Souza et al. to conclude that An-Mlp1, within the spindle matrix, binds An-Mad1 and maintains it at a high concentration in the vicinity of the spindle and kinetochores where it would be poised to rapidly activate the SAC in respond to spindle damage. The spindle matrix also concentrates An-Mad2; however, it was not directly determined whether An-Mlp1 is required for the spindle matrix association of An-Mad2. In fact, other binding sites for An-Mad2 may exist in the spindle matrix. This indeed appears to be the case in Drosophila S2 cells where depletion of Mtor does not affect Mad2 localization in the spindle matrix [23]. An-Mlp1 may also perform a similar function later in mitosis as it is required to concentrate An-Mad1 near the spindle during telophase, a localization also shared by cyclin B [14]. It was also proposed that, in A. nidulans, the localization of these mitotic regulators between the reforming daughter nuclei may help to coordinate NPC reformation or post-mitotic nucleolar dynamics with other aspects of mitosis. Of note, a similar localization has also been reported for Mad2 and Mtor in Drosophila syncytial embryos [22, 29], suggesting that this behavior might be a common, but not exclusive [23], feature of syncytial mitoses.
A ‘fencing’ function performed by Mlp-like proteins to contain or control the activity of SAC proteins within the region of the spindle matrix would be most useful in cells that undergo an open mitosis or, as in the case of A. nidulans, that lack an effective NE barrier during mitosis. By contrast, this mechanism may provide little selective advantage in budding yeast where an intact nuclear envelope and NPCs could perform a functionally similar task during mitosis. This may explain why S. cerevisiae cells have failed to evolve a mechanism for releasing the Mlp proteins from the NPC, instead maintaining their association with the NPC during mitosis. In these cells, activation of the checkpoint induces the release of Mad1 and Mad2 from the NPC-associated Mlp proteins, and the accumulation of Mad1 and Mad2 at kinetochores [12, 15, 30]. While all the detectable Mad2 leaves the NPCs, much of Mad1 remains at the NPCs, thus creating two pools of Mad1, one bound to Mlps and the other at kinetochores [15]. This situation is, perhaps, functionally analogous to the dual binding of Mad1 and/or Mad2 to the spindle matrix associated Mlp-proteins and kinetochores in Drosophila and A. nidulans. Strikingly, in S. cerevisiae, most of the Mad1 (>60%) within these two pools is dynamic and exchanges rapidly between the two locations. This cycling process is controlled by the nuclear export factor, Xpo1 (the S. cerevisiae homologue of vertebrate Crm1), and RanGTP [31]. Xpo1 binds to Mad1 and both facilitates its targeting to kinetochores and its exchange between these structures and the Mlp sites at the NPC. This functional relationship is further supported by the observation that the NES of Mad1 is required for the association of Xpo1 with kinetochores. While Xpo1-mediated targeting to kinetochores is not essential for the execution of the SAC, it may improve the efficiency and fidelity of the response to spindle and kinetochore defects. It will be interesting to see whether in other species the functional counterparts of Xpo1 or other transport factors mediate the association of Mad1 with the spindle matrix-associated Mlp proteins and kinetochores.
Even with the advances in our understanding of the molecular basis for the association of Mad1 and Mad2 with the NPC and the potential roles of the Mlp proteins during mitosis, we are largely unaware of what functions Mad1 and Mad2, as well as Mps1, may perform while associated with the interphase NPC. One report suggests that deletion of the MAD1 gene in S. cerevisiae attenuated the rate of nuclear import mediated by the transport factor Kap121 [12]. This observation is intriguing, as conditions that activate the SAC (specifically nocodazole induced cell cycle arrest) induce structural changes in the NPC that inhibit Kap121-mediated import [32]. Among these changes are alterations in the binding partners of Nup53. Coincidental changes in the dynamics of the interactions of Mad1 with NPCs, as well as previously observed binding of Mad1 to Nup53, raise the question of whether the NPC-bound checkpoint proteins play a role in regulating the Kap121-mediated transport pathway. It is also reasonable to extrapolate from this concept that changes in the association of Mad1 or Mad2 with the NPC that occur in other species prior to or following mitosis may also serve to activate transport regulatory mechanisms. For example, the dissociation of Mad1 and Mad2 from the NE in prophase prior to NPC disassembly, their accumulation in the nucleoplasm in G1, and their recruitment back to nuclear periphery only by the later stages of G1 as observed in Drosophila, A. nidulans, and human cells may reflect changes in NPC structure that alter its transport properties [14, 18, 22].
The RanBP2–Ran-GAP1:SUMO1–Ubc9 complex and CRM1: from cytoplasmic NPC filaments to kinetochores
Ran cycle and the RanBP2/Nup358–Ran-GAP1:SUMO1–Ubc9 (RRSU) complex in interphase
One of the key regulators of nucleocytoplasmic transport is Ran, a small Ras-related GTPase. The primary function of Ran is to impose directionality on the transport process by coordinating the loading and unloading of Karyopherins [33]. Binding of cargo to nuclear import receptors is Ran-independent. However, unloading of the import cargo is mediated by binding of Ran-GTP to the karyopherin/importin. In the case of export complexes, cargo loading is stimulated by Ran-GTP binding to the export receptor. Thus, karyopherin/exportin-mediated nuclear export involves the assembly of a ternary complex, consisting of the karyopherin, Ran-GTP, and the cargo molecule, that is translocated across the NE via the NPC central channel. Unloading of the export receptor in the cytoplasm is stimulated by activation of the Ran-GTPase. It follows from this that Ran-GTP must be maintained at a high level within the nucleus while Ran-GDP should be the predominant form in the cytoplasm [34]. This Ran gradient across the NE is maintained by segregation of the Ran guanine nucleotide exchange factor (Ran-GEF) within the nucleus as a chromatin-associated protein. In fact, the RanGEF, known as RCC1 in vertebrates, is chromatin-associated at all stages of the cell cycle. Conversely, Ran-GTPase-activating proteins (Ran-GAPs), are restricted primarily to the cytoplasm [34]. In this way, Ran-GTP exiting the nucleus as part of an export complex will be converted to the GDP form leading to complex dissociation.
Activation of the Ran-GTPase occurs as soon as an export complex has traversed the central channel of the NPC. The cytoplasmic face of vertebrate nuclear pore complexes features eight seemingly flexible filaments that extend 50–100 nm into the cytoplasm [35]. In vertebrates, a major component of these filaments is Ran binding protein 2 (RanBP2) or Nup358 a very large multifunctional protein [36, 37]. The RanBP2/Nup358-containing filaments are anchored to the NPC via interaction with a complex of nucleoporins containing Nup214 and Nup88, the location of which in turn is restricted to the cytoplasmic side of the NPC (Fig. 1) [38]. RanBP2/Nup358 is a member of a family of nucleoporins, that includes Nup214, featuring multiple FG and FXFG repeats that represent binding sites for transport receptors as part of transport complexes [39]. Most significantly, vertebrate RanBP2/Nup358 forms a complex with Ran-GAP1, which is therefore concentrated at the nuclear periphery [40, 41]. In this way, RanBP2/Nup358 provides a platform on which Ran-GTPase activation may occur before an export complex has even left the vicinity of the NPC. RanBP2/Nup358 also contains four Ran-GTP binding domains, the presence of which facilitates the dissociation of Ran-GTP from transport complexes [37]. This in turn will further enhance Ran-GAP1-mediated GTP hydrolysis by Ran. In yeast, however, that lack any obvious RanBP2/Nup358 homologue, Ran-GAP1 has a diffuse localization in the cytoplasm, with no significant enrichment at the NE.
The targeting of Ran-GAP1 to RanBP2/Nup358 highlights a fundamental aspect of the relationship between these two proteins. In vertebrates, Ran-GAP1 is a substrate for conjugation with SUMO (small ubiquitin-like modifier) 1 [42], and it is only the SUMOylated form of Ran-GAP1 that will associate with RanBP2/Nup358 [40, 41, 43]. Four classes of enzymes are involved in the SUMOylation process (reviewed in [42, 44]). Newly synthesized SUMO undergoes C-terminal proteolytic maturation catalyzed by members of the SENP family, of which some are also capable of de-conjugating SUMO-modified proteins. Conjugation of mature SUMO to target proteins involves the sequential action of a heterodimeric E1-activating enzyme (Uba2/Aos1), an E2-conjugating enzyme (Ubc9) and a SUMO E3 ligase. In budding yeast, there are four E3 enzymes whereas in mammals there are at least seven and potentially nine. The binding region on RanBP2/Nup358 for Ran-GAP1 features a pair of internal repeat (IR) motifs near the C-terminus. Fragments of RanBP2/Nup358 containing this repeat region display SUMO1 E3 ligase activity [45]. Furthermore, this same region represents a site of interaction with Ubc9, a vertebrate SUMO E2-conjugating enzyme that is required for SUMOylation of Ran-GAP1. This ternary complex containing RanBP2/Nup358, Ran-GAP1:SUMO1 and Ubc9 (RRSU complex) is stable throughout the cell cycle [42, 46].
Interplay between the RRSU and Crm1 in mitosis
During the open mitosis of metazoans, as the nuclear envelope undergoes breakdown, the RRSU complex is released en bloc from NPCs [42]. At the same time, Ran-GAP1:SUMO is phosphorylated by cyclin B/Cdk-1 [47]. The significance of this modification remains uncertain. What is clear, however, is that the RRSU complex does not behave as a bystander following nuclear envelope breakdown. Instead, it plays an essential function in subsequent mitotic progression.
An active role for RRSU complex during mitosis was first suggested by the finding that both Ran-GAP1 and RanBP2/Nup358 become concentrated at kinetochores upon microtubule attachment, as well as at spindle poles, during vertebrate prometaphase [48]. Subsequent studies in mammalian cells employing RNA interference revealed that down-regulation of RanBP2/Nup358 caused abnormal spindle morphology, kinetochore dysfunction, and defects in chromosome congression and segregation [49]. This was consistent with findings in Caenorhabditis elegans embryos where depletion of Ran cycle components, including Ran itself, RanBP2, Ran-GAP1, and RCC1, all caused spindle assembly and chromosome segregation abnormalities [50]. Evidence of an essential role for RanBP2/Nup358 and Ran-GAP1 at kinetochores was ultimately provided by Joseph and colleagues [51]. Depletion of proteins required for microtubule interactions with kinetochores, such as Hec1/Ndc80 and Nuf2, resulted in a loss of kinetochore-localized RanBP2/Nup358 and Ran-GAP1. Conversely, depletion of RanBP2/Nup358 caused the mis-localization of Ran-GAP1 as well as that of other kinetochore-associated proteins including Mad1, Mad2, Zw10, CENP-E, CENP-F, and dynein. Of particular significance was the observation that RanBP2/Nup358 depletion was associated with a loss of cold-stable kinetochore microtubule interactions. These findings suggest that the concentration of RanBP2/Nup358 and Ran-GAP1 at kinetochores is at once both facilitated by and required for functional connections between kinetochore and microtubules.
The role of RanBP2/Nup358 and Ran-GAP1 in kinetochore-MT attachment is intimately linked to the mechanisms by which the RRSU complex is targeted to kinetochores. A key observation is that Crm1, a karyopherin family member that is the export receptor for proteins bearing leucine-rich nuclear export sequences (NESs), is itself localized to kinetochores [52]. In interphase, Crm1 would form a ternary export complex with an export substrate and Ran-GTP, whose assembly is efficiently inhibited by the drug leptomycin B. In mitotic cells, leptomycin B treatment or depletion of Ran-GTP through inactivation of a temperature-sensitive form of RCC1 have only a marginal effect on the kinetochore localization of Crm1 [52]. Thus, the localization of Crm1 at kinetochores appears independent of Ran-GTP or NES-containing proteins. This situation in metazoans is in contrast to that observed during closed mitosis in S. cerevisiae where, as mentioned in the previous section, SAC-checkpoint-induced association of Xpo1 with kinetochores is dependent on the NES-containing Mad1 [31]. Recent studies in metazoan cells instead suggest that Crm1 is anchored to the kinetochore, either directly or indirectly through an interaction with another group of NPC components, the Nup107-160 complex ([53]; Fig. 3; see also next section). Extension of these observations to RanBP2/Nup358 and Ran-GAP1 reveals that both proteins are lost from the kinetochore following either leptomycin B treatment or inactivation of tsRCC1. The implication here is that Crm1 forms a ternary export-type complex, potentially with either RanBP2/Nup358 or Ran-GAP1, both of which feature leucine-rich NES-like sequences [52, 54].
Schematic representation of the network of interactions between vertebrate NPC-associated and centromere/kinetochore constituents (see text for details). Proteins or complexes that are localized at NPCs during interphase are circled in red (see also Fig. 1). Black outline arrows indicate direct or indirect interactions between the proteins or complexes contributing to their recruitment at kinetochores or centromeres. Red outline arrows indicate NES-mediated recruitment by CRM1 (note that an interaction with CRM1 appears to be required for the initial targeting, but not the maintenance of the CPC at centromeres). Spindle assembly factors whose activity is regulated by RanGTP and importin a/β or importin β are listed (asterisks indicates proteins interacting with importin β only). SUMOylated proteins are also listed. Substrates of Aurora B phosphorylation are also shown (green P). Note that Mad1, Mad2, and Mps1 have not been included in this scheme (see Fig. 2)
The localization of RanBP2/Nup358 and Ran-GAP1 to kinetochores in a Crm1-dependent manner seems at first sight paradoxical. Binding of the RRSU complex to Crm1 requires Ran-GTP, yet Ran-GAP1 in association with RanBP2/Nup358 will promote the conversion of Ran-GTP to its GDP bound form leading to unloading of Crm1 (Fig. 3). In essence, these proteins will function in a futile cycle or autoregulatory loop at the kinetochore [55]. The situation makes more sense once microtubules and Ran-mediated regulation of spindle assembly factors (SAFs) enter the equation.
Microtubule-dependent mitotic functions of the RRSU
Ran-mediated regulation of spindle assembly factors (SAFs)
The function of Ran cycle components at kinetochores can best be appreciated by first considering the role of Ran itself in mitotic progression. Ran, importins α and β, the prototypic import receptors subunits, and, more recently, another transport receptor of the karyopherin family, transportin, were implicated in spindle assembly (reviewed in [8, 9]; see also [56]). During interphase, importin β, most frequently in conjunction with importin α, binds transport substrates in the cytoplasm, which are then unloaded in the nucleus in a Ran-GTP-dependent manner. Ran and importin α/β conform to the same paradigm during mitosis. Importin α/β or importin β alone binds to nuclear localization sequences on spindle assembly factors (SAFs) within the mitotic cytoplasm thereby inhibiting their activities [8, 9]. In the presence of Ran-GTP, SAFs will be released from importin α/β becoming competent to promote spindle assembly. Since RCC1, the exchange factor for Ran, remains chromatin-bound during mitosis, Ran-GTP concentrations and hence SAF activity will be highest in the vicinity of mitotic chromosomes. Studies in Xenopus egg extracts have revealed several SAFs that are regulated by importins and Ran-GTP (reviewed in [9, 57]). These include TPX2 and NuMA (nuclear mitotic apparatus protein) as well as the kinesin family member HSET/XCTK2, that are primarily involved in spindle pole formation [58, 59]. Other SAFs sequestered in this way include NuSAP and HURP, which promote microtubule bundling and stabilization in chromatin proximal regions of the spindle [59–61]. The beauty of this system is that, in interphase, all these SAFs are sequestered in the nucleus where there is no chance of them promoting premature spindle assembly.
Rae1 is a noteworthy member of the growing list of Ran-regulated importin β cargo and MT-binding proteins that contribute to spindle assembly. Indeed, during interphase, a fraction of Rae1 is localized to the NPC in a Nup98-dependent fashion where it has been suggested to play a role in nuclear mRNA export [62, 63]. However, following early studies revealing that Rae1 binds MTs [64], Blower et al. [65] demonstrated that during mitosis Rae1 actually exists in a large ribonucleoprotein complex that controls microtubule dynamics in a RanGTP/importin β-regulated manner. These authors further showed that correct spindle formation in Xenopus egg extracts is RNA-dependent, suggesting a general, translation-independent role of RNA in mitotic spindle assembly. Finally, a mitotic-specific interaction between Rae1 and another SAF, NuMA, was shown to be critical for normal spindle formation in mitosis [66]. In addition to this role in spindle assembly, studies in mice have revealed that reduced levels of Rae1 and Nup98 lead to premature degradation of securin, an important regulator of sister chromatid cohesion. This in turn allowed premature sister chromatid separation with resultant aneuploidy [67]. It was proposed that Rae1 and Nup98 would exert this function through their association with Cdh1-activated APC [67, 68]. However, this hypothesis appears difficult to reconcile with the established view that the destruction of cyclin B and securin at the metaphase/anaphase transition is mediated by Cdc20- and not Cdh1-activated APC [69, 70]. This issue clearly deserves further analysis.
The RRSU complex, a regulator of kinetochore microtubule outgrowth?
As a modulator of Ran-GTP levels, the RRSU complex will locally contribute to the regulation of spindle assembly factors (SAFs), and consequently, to the establishment of functional kinetochore-microtubule attachments. There are two pathways by which stable kinetochore microtubules can be formed. The first involves growth from centrosomes and capture by kinetochores (centrosomal pathway). The second involves seeding from kinetochores (chromatin pathway). In mammalian systems, transient microtubule nucleation at kinetochores is observed during microtubule regrowth following either nocodozole treatment or cold treatment combined with centrosome impairment by RNAi-mediated depletion of polo-like kinase (Plk) 1 ([71] and references therein).
Nocodazole-induced depolymerization of spindle microtubules results in the loss of Ran-GAP1 (and presumably other RRSU complex components) from kinetochores [48]. Following nocodazole washout, there is a brief period of microtubule nucleation at kinetochores. These microtubules are stabilized by HURP [71]. In the absence of the RRSU complex, chromatin-associated RCC1 will promote the accumulation of Ran-GTP in the vicinity of kinetochores. This in turn will induce the unloading of HURP from importin a/β. HURP will then be free to bind to and stabilize the nascent kinetochore-associated microtubules. The appearance of these microtubules, however, will promote the loading of the RRSU complex at the kinetochore through interactions with Crm1. The presence of Ran-GAP1 in the RRSU complex will obviously lead to a local reduction in the concentration of Ran-GTP with a concomitant depression of HURP availability. In this way, the RRSU complex serves to put the brake on kinetochore microtubule stabilization and acts to regulate the growth of microtubules from the kinetochore versus those from the centrosome. It is possible that this is a prerequisite for maturation of the microtubule-kinetochore attachment. A prediction here is that leptomycin B, by releasing the RRSU complex, should promote the growth of microtubules from kinetochores following cold-induced depolymerization, even in the presence of competing centrosomal nucleation. This in fact turns out to be the case [71].
RanBP2/Nup358 clearly has an essential function in the regulation of kinetochore microtubule attachment by virtue of its role as a binding partner for Ran-GAP1 and, consequently, as a modulator of Ran-GTP levels. What has become increasingly evident in recent years is that RanBP2/Nup358 also regulates aspects of mitotic progression via its activity as a SUMO E3 ligase.
The RSSU as a regulator of SUMO pathway in mitosis
Multiple chromosome-associated proteins have been identified as targets for SUMOylation. These include CENP-E, a plus end-directed microtubule motor protein that is required for kinetochore function and mitotic progression [72], as well as both the condensin and cohesin complexes, which are essential organizers of mitotic chromosome architecture [42]. The condensin complex serves to maintain the compact structure of mitotic chromosomes, while the related cohesin complex mediates the close association of sister chromatids until anaphase onset. The regulation of condensin and cohesin complex activity by SUMOylation has been the subject of a recent in-depth review [42], and will not be described further here. Instead, our focus will be on the group of chromosome-associated proteins whose SUMOylation is linked to the RanBP2 E3 ligase activity.
Topoisomerase II
RanBP2/Nup358 is an essential protein. However, a single RanBP2/Nup358 hypomorphic allele (H) is sufficient for viability. Embryo fibroblasts derived from such RanBP2/Nup358-/H mice express approximately 26% of wild-type RanBP2/Nup358 levels. While these cells display no overt nuclear transport abnormalities, they do develop severe aneuploidy associated with chromosome segregation defects, including anaphase bridges. This phenomenon is also evident in splenocytes derived from 5 month-old RanBP2/Nup358-/H mice. It is interesting to note that RanBP2/Nup358-/H mice are significantly more prone to both carcinogen-induced and spontaneous tumorigenesis than are wild-type animals. This presumably is a reflection of increased chromosome instability [73]. The formation of anaphase chromatin bridges can be a consequence of failure in the decatenation of DNA strands at sister centromeres, an essential process in chromatid separation following cohesin complex inactivation. Decatenation at centromeres is facilitated by topoisomerase IIα (Topo IIα) [74]. In order to accomplish this crucial mitotic function Topo IIα must be targeted to the inner centromere region. This targeting is dependent upon Topo IIα SUMOylation by the RanBP2/Nup358 SUMO E3 ligase activity ([73], Fig. 3). It was therefore proposed that impaired SUMOylation of TopoIIα may contribute to mitotic defects occurring in RanBP2/Nup358-/H mice [73].
Noticeably absent in RanBP2/Nup358-/H cells were chromosome congression abnormalities and any evidence for kinetochore defects. Evidently, centromere decatenation is far more sensitive to RanBP2/Nup358 levels than is kinetochore function or interphase nucleocytoplasmic transport.
The chromosome passenger complex (CPC)
All the mitotic events described so far are influenced either directly or indirectly by another key mitotic regulator, the chromosome passenger complex (CPC) [75]. The CPC consists of four subunits, the Aurora B protein kinase, inner centromere protein (INCENP), survivin, and borealin (also known as Dasra B). INCENP functions as a regulatory scaffold for the other three proteins. Its binding to Aurora B activates the Aurora B kinase which phosphorylates INCENP, which in turn leads to increased Aurora B activation. Aurora B can also phosphorylate both survin and borealin (Fig. 3). However, the significance of this remains uncertain. The CPC displays a highly dynamic localization during mitosis. In prophase, the CPC can be found along the chromosome arms. However, its distribution progressively shifts towards the centromeres until by metaphase it is restricted exclusively to the inner centromeres. Targeting to the centromeres may be under the purview of the survivin and borealin subunits. At anaphase onset, the chromatids begin their migration to the spindle poles. However, the CPC remains behind at the spindle equator. Later on in anaphase, the CPC migrates to the equatorial cortex and subsequently concentrates in the midbody during telophase and cytokinesis.
The role of the CPC has been extensively reviewed by Ruchaud et al. [75]. In early mitosis, the CPC has been implicated in both the centrosome- and chromatin-driven pathways of spindle assembly. It is also required for kinetochore formation and is involved in the detection of aberrant kinetochore microtubule interactions. One of the targets for Aurora B at the kinetochore is Hec1, a protein required for microtubule attachment (Fig. 3). As an additional regulatory layer, the CPC and its Aurora B kinase activity is indispensable for operation of the spindle assembly checkpoint in the absence of tension at kinetochores. Finally, the CPC has a role in regulating the cohesion of sister chromatids. In particular, it cooperates with Plk1 in releasing cohesin from the chromosome arms during prophase and prometaphase.
A recent study has revealed that the borealin subunit of the CPC is conjugated with SUMO2 or SUMO3 during the early stages of mitosis ([76]; Fig. 3). As is the case for Topo IIα, RanBP2/Nup358 associates with the CPC during mitosis and stimulates the SUMOylation of borealin. This modification is reversed by the SUMO protease SENP3 coincident with the transfer of the CPC from the centromere to the spindle equator at anaphase onset. The significance of this modification is still uncertain. Overexpression of SENP2 leads to an extensive decline in SUMOylation. However, CPC subunits are still retained at centromeres under these circumstances [72, 76]. Klein et al. suggest that the RanBP2/Nup358-dependent SUMOylation of borealin might be required for the interaction of as-yet-unknown protein(s) with the CPC at the centromere [76]. Whether this might be linked to the kinetochore/congression defects that are linked to loss of RanBP2/Nup358 is unknown. However, this clearly represents an important question in our understanding of the function of RanBP2/Nup358 during mitosis.
The targeting and function of the CPC complex is also intimately linked to the Ran pathway. Indeed, prolonged treatment of cells with leptomycin B (LMB) during late G2 results in failure of the CPC to associate with centromeres [77]. This effect is mediated by the survivin subunit of the CPC. Mammalian survivin contains a highly conserved leucine rich NES that can recruit Crm1/RanGTP to the CPC. While this sequence is dispensable for CPC assembly, it is absolutely essential for appropriate CPC localization and function. Indeed, NES-deficient survivin cannot replace the wild-type molecule [77]. However, in contrast to the situation observed for the RRSU complex (see above), short (20–40 min) treatments of late G2 or of metaphase cells with LMB does not affect the localization of already assembled CPCs at the centromeres [77]. A more recent study reveals that survivin also interacts with RanGTP in a Crm1- and NES-independent fashion [78]. This association seems to have little if any role in either CPC assembly or localization. Rather, it is required for the delivery of the Ran-effector molecule and spindle assembly factor TPX2 to microtubules. Disruption of the survivin-Ran interaction is associated with aberrant spindle assembly and chromosome missegregation in tumor cells.
The CPC thus behaves as a shared target of SUMO and Ran-dependent regulations, and also a novel effector of Ran signaling. These data further highlight the complex network of interactions that takes place during mitosis between the RRSU, nuclear transport factors, and key mitotic regulators.
Functional role of core NPC constituents, the Nup107-160 complex, and ELYS/MEL-28 at various stages of mitosis
The Nup107-160 NPC sub-complex, composed in vertebrates of nine different polypeptides (Nup160, Nup133, Nup107, Nup96, Nup85, Nup43, Nup37, Sec13, and Seh1), is the major structural subunit of nuclear pores, both in size and complexity [79–81]. This complex (designated the Nup84 complex in budding yeast) is conserved in all eukaryotes, but with some variations: no orthologue of Nup43 has so far been identified in fungi. In addition, while orthologues of Nup37 exist in filamentous fungi and possibly in Schizosaccharomyces pombe, they have not been observed in S. cerevisiae [82]. In budding yeast, structural studies have now provided a refined view of the assembly of its seven members within a 0.6-MDa Y-shaped complex, an organization likely conserved in other organisms (reviewed in [83]). Recently, a novel protein of greater than 200 kD, ELYS, was shown to co-immunoprecipitate with the Nup107-160 complex in both human and Xenopus cell extracts [84–86]. ELYS is the vertebrate orthologue of C. elegans MEL-28/C38D4.3 and is encoded by the flo gene in zebrafish [87–89]. Affinity-purification of the Nup84-120 complex from A. nidulans led to the identification of a small (~36 kD) ELYS relative whose similarity with vertebrate ELYS is, however, restricted to a region that is absent from C. elegans MEL-28 [82]. An-ELYS has related sequences in multiple fungi including S. pombe but not S. cerevisiae; its function in fungi is still unknown.
During interphase, the Nup107-160 complex is found stably bound to both the cytoplasmic and nuclear faces of vertebrates NPCs ([79, 90]; see Fig. 1). In contrast, ELYS appears to be present only on the nuclear side of the pores (V. Doye, unpublished observations). As initially demonstrated in yeasts, the vertebrate Nup107-160 complex is required for mRNA export and de novo nuclear pore assembly in interphase, [80, 91–94]. However, the Nup107-160 complex and ELYS also play crucial roles at various stages of mitosis that will be detailed in the following paragraphs.
ELYS/MEL-28 and the Nup107-160 complex have a crucial role, along with the Ran pathway, in post-mitotic NPC assembly
At mitosis, as NPCs are dismantled, most of the constituents of the vertebrate Nup107-160 subcomplex remain stably associated but with two exceptions: proteasome-mediated downregulation of Nup96 in mitosis has been observed, with implications for G1/S progression, and the dissociation of a pool of Nup43 from the complex has also been reported [81, 95, 96]. From late anaphase on, nuclear re-assembly requires the coordinated step-wise recruitment of both nuclear membranes and NPC constituents around chromatin (for review, see [3, 97]). In vivo RNAi depletion experiments, mutant analysis in zebrafish, and in vitro approaches have revealed that the Nup107-160 complex and Mel28/ELYS are critically required in an early step of post-mitotic NPC reassembly in vertebrates and C. elegans [88, 89, 98, 99]. NPC formation was shown to be initiated on chromatin by the recruitment of ELYS/MEL-28, which subsequently recruits the Nup107-160 complex [84–86, 100]. Significantly, in vitro studies have also revealed that the key function of Ran-GTP in post-mitotic NPC reassembly lies in the dissociation of a subset of nucleoporins, notably ELYS and the Nup107-160 complex, from importin-β and/or transportin. This step is crucial for their recruitment as seeding points—or pre-pores—on chromatin [56, 101, 102]. Loading of ELYS and the Nup107-160 onto chromatin is a prerequisite for the recruitment of membrane vesicles containing the integral membrane nucleoporins Pom121 and Ndc1 [100]. In another noteworthy study, Gillespie et al. [86] detected a physical association between ELYS and the Mcm2-7 replication-licensing factor. These authors further showed that preventing Mcm2-7 from loading onto chromatin partially impaired recruitment of ELYS to chromatin. They suggested that ELYS may contribute to coordinate post-mitotic NPC and nuclear envelope reassembly with the requirement to shut down replication origin licensing prior to S-phase entry [86]. A potential role for ELYS in DNA replication was also uncovered in zebrafish. In this organism, a mutation in ELYS was shown to affect the maintenance of chromatin-bound Mcm2, but not Mcm3 or Mcm4 [103]. Davuluri et al. [103] thus proposed that Elys might be needed, independent of its role in NPC assembly, to resolve replication errors that normally occur in highly proliferating cells.
Specific localization of the Nup107-160 complex and ELYS at kinetochores and mitotic spindle
In addition to its recruitment to chromatin in late anaphase, when NPCs begin to reform on the chromatin surface, a fraction (5–10%) of the human Nup107-160 complex moves to the outer plate of kinetochores in prophase, where it persists until late anaphase [79, 81, 99]. Members of the Nup107-160 complex were also found at both spindle poles and proximal spindle fibers in prometaphase mammalian cells, as well as throughout reconstituted spindles in Xenopus egg extracts [95]. Analysis of the ELYS homologue, C. elegans MEL-28, similarly revealed its dual localization and function at the NPC during interphase and on chromosomes during mitosis, in a pattern suggestive of a kinetochore localization [87, 88]. This localization was supported by the observation that chromatin localization of MEL-28 is abolished upon depletion of hcp3/cenp-A [89], and by the kinetochore localization of its vertebrate ortholog, ELYS [84]. Studies of the dynamics of one of the Nup107-160 complex subunits, GFP-tagged Nup133, revealed the existence of two distinct fractions of the Nup107-160 complex at kinetochores [79]. Consistent with this idea, siRNA experiments in HeLa cells have shown that the recruitment of the Nup107-160 complex to kinetochores was primarily contingent upon the kinetochore Ndc80 complex, whereas CENP-F, which directly interacts with Nup133, likely provides a second microtubule-dependent anchoring site for Nup107-160 recruitment to kinetochores ([53]; see Fig. 3). This result was confirmed by the enhanced accumulation of the Nup107-160 complex as extended crescents around kinetochores upon nocodazole-induced microtubule depolymerization [53, 95].
Studies in other organisms have also revealed that a fraction of Nup107 is localized at kinetochores during mitosis in C. elegans but not in Drosophila [22, 88]. In the latter organism, however, a fraction of Nup107 persists around the mitotic spindle. In A. nidulans, in which NPCs partially disassemble, the An-Nup84-120 complex and An-ELYS remain part of the incomplete NPCs and do not localize to kinetochores, although a fraction could be found at spindle pole bodies [82, 104]. Similarly, the Nup107-160 complex could not be detected at kinetochores during the closed mitosis of fission yeast [92]. Neither could it be seen at kinetochores in the corn smut fungus Ustilago maydis that, in contrast to many fungi, undergoes an open mitosis [105]. This varied spatial and temporal localization of the Nup107-160 complex detected in these distinct organisms may indeed underlie species-specific differences in mitotic spindle assembly and chromosome segregation through evolution.
Functional implications for the association of the Nup107-160 complex and ELYS with the mitotic spindle and kinetochores
The first insights into the mitotic function of the Nup107-160 complex at mitosis came from in vitro studies that revealed its requirement for correct bipolar spindle assembly in Xenopus egg extracts [95]. This study further indicated that the Nup107-160 complex is not required for microtubule nucleation around sperm centrosomes, or for Ran-GTP-mediated microtubule assembly, but rather plays an important role at later times, in the assembly or maintenance of microtubule fibers between spindle poles and chromosomes. However, this study did not establish whether the defects were caused by the absence of the Nup107-160 complex from kinetochores, or by its more general function in stabilizing spindle microtubules.
A potential role for Mel-28/ELYS and the Nup107-160 complex at kinetochores emerged from observations of chromatin segregation defects during the first zygotic division of Mel28 RNAi-depleted C. elegans embryos [88, 89]. RNAi experiments in human cells indicated that the kinetochore localization of ELYS and the Nup107-160 complex are interdependent [95]. However, siRNAs-induced depletion of individual constituents of this complex, while efficiently depleting the soluble pool of the corresponding nucleoporin, only moderately affected their kinetochore localization and did not lead to any major defect in chromosome segregation [53, 84]. Moreover, while efficient depletion of the Nup107-160 complex from kinetochores could be achieved by combining siRNAs targeting several of its subunits, this treatment also severely affected post-mitotic NPC reassembly, thus complicating the phenotypic analysis. In contrast, depletion of one specific subunit of the Nup107-160 complex, Seh1, that had only minor consequences on NPC assembly [81], significantly reduced the kinetochore levels of the Nup107-160 complex in HeLa cells [53]. Interestingly, efficient depletion of the Nup107-160 complex from kinetochores, achieved either by combining siRNAs targeting several of its subunits, or by depleting Seh1 alone, induced a similar mitotic delay. Analysis of Seh1-depleted cells revealed impaired chromosome congression and the presence of fewer and less well-organized cold-stable microtubules, suggesting improper or unstable kinetochore-microtubule attachment. Depletion of the Nup107-160 complex from kinetochores impaired the recruitment of the Ran effector, Crm1, as well as RanGAP1-RanBP2 to these structures [53]. This study also revealed that the presence of the Nup107-160 complex at kinetochores, while dispensable for the recruitment of CENP-F (a protein contributing to chromosome congression and tension; reviewed in [106]) to these structures, is required for CENP-F maintenance at attached kinetochores. Accordingly, the RanBP2–Ran-GAP1:SUMO1–Ubc9 (RRSU) complex and CENP-F behave as downstream effectors of the Nup107-160 complex at kinetochores ([53]; see Fig. 3). However, a recent study revealed that Seh1 or ELYS/Mel-28 depletion also severely impairs the centromeric localization of the CPC at metaphase-aligned chromosomes [107]. In contrast, the CPC was still recruited to unattached centromeres present in Seh1-depleted cells, where it displayed a decreased turnover. Similarly, both mitotic centromere-associated kinesin (MCAK, a known Aurora B substrate) and Aurora B-phosphorylated MCAK (P-MCAK) signals were lost from metaphase-aligned chromosomes and were retained only on the centromeres of the misaligned chromosomes in Seh1-depleted cells. Since phosphorylation of MCAK by Aurora B inhibits MCAK’s ability to depolymerise microtubules at kinetochores, it was proposed that the increased stability of the CPC complex at unattached kinetochores may compromise the error–correction system that eliminates kMT mis-attachments, leading to a misaligned chromosome phenotype [107].
As previously highlighted by Orjalo et al., depletion of either the Nup107-160 complex or the CPC from Xenopus egg extracts leads to similar defects in spindle formation [95, 108]. The reduced density of cold-stable k-MTs observed upon Seh1 depletion [53] may thus be linked to altered CPC function, or reflect another function of the Nup107-160 complex in spindle assembly. Considering the size (0.6 MDa) and complexity (9-subunit) of the Nup107-160 complex, additional interaction partners likely contribute to the targeting and/or function of this nucleoporin complex at kinetochores. In mitosis, the Nup107-160 complex and ELYS may thus contribute to the integration of distinct kinetochore functions required for proper chromosome congression and segregation (Fig. 3).
Nucleoporins and cytokinesis
Partial depletion of either ELYS or Nup133 in human cells, while not affecting spindle assembly or chromosome segregation, induced cytokinesis defects [84]. More pronounced defects, notably an increased occurrence of binucleated cells resulting from cytokinetic furrow regression, were recorded upon depletion of Seh1 [107]. Seh1-depleted cells displayed decreased levels of the CPC complex and of phosphorylated MKLP1, a well-characterized Aurora B substrate, at both the central spindle in anaphase and the midbody in telophase. Although other hypotheses cannot be excluded, these data suggest that impaired localization of the CPC complex may underlie the various mitotic functions of the Nup107-160 complex [107]. In future, it will be important to understand the molecular mechanism that leads to the impaired localization and dynamics of the CPC complex in cells depleted for Nup107-160 complex constituents.
It is worth mentioning here that a delay during late stages of mitosis, accompanied by an increase in unresolved midbodies, was also reported upon partial depletion of Nup153, a nucleoporin dynamically associated with the NPC basket structure (see Fig. 1). More extensive depletion of this nucleoporin led to a pronounced defect early in mitosis and an accumulation of cells with multilobed nuclei [109]. In contrast to ELYS/MEL-28 and the Nup107-160 complex, Nup153 is not detected at kinetochores, nor is it seen at the midbody. Moreover, although Nup153 is required to anchor Tpr at the NE, expression of a Nup153 construct lacking its C terminus, while sufficient to restore Tpr localization to the NPC in Drosophila [110, 111], did not rescue the increase in midbodies seen after Nup153 depletion. This suggests that the functions in cytokinesis of Nup153 do not involve Tpr localization [109]. However, it may be significant that SENP2, the mammalian homologue of the yeast SUMO protease Ulp1p, also localizes to the nuclear side of NPCs and has been found to associate with the C-terminal domain of Nup153 [46, 112]. It is therefore conceivable that alteration of the SUMO pathway, known to affect mitosis (reviewed in [42]; see also previous section), may underlie this mitotic role for Nup153. Finally, since Nup153 was shown to interact with the Nup107-160 complex in pull-down experiments [80], a crosstalk between these nucleoporins may also explain some of their common mitotic defects.
Together, data gathered over these last years have thus revealed a complex network of interactions between the Nup107-160 complex, ELYS (and to some extent Nup153), and factors known to regulate various stages of mitotic progression: replication-licensing, chromosome segregation, cytokinesis, and NPC/nuclear envelope reassembly. Although this aspect has not yet been investigated, it is likely that phosphorylation of the Nup107-160 complex [113], and the corresponding interplay with mitotic kinases and phosphatases, may provide a further level of coordination of these mitotic events.
Summary and perspectives
Since first reported more than 10 years ago that a subset of proteins exhibits dual localization to both nuclear pores and the mitotic apparatus, a clearer picture has emerged of the molecular network that support the interactions of these proteins with these varied locations. This includes a crucial role of Mlp1-like proteins in the tethering of spindle assembly checkpoint factors, and an as-yet-incomplete understanding of the hierarchical targeting of the Nup107-160 complex, CRM1, and the RRSU complex to kinetochores. In addition, it is now clear that Ran and its effectors, as well as post-translational modifications, notably phosphorylation or SUMOylation events, contribute to regulate the relocalization of proteins between NPCs and the mitotic apparatus.
Clues are now being accumulated that shed light on the potential functions of these proteins at their varied locations. For instance, increasing evidence indicates that Mlp-like proteins behave, in all eukaryotes, as a scaffold for Mad1, Mad2, and Mps1 during mitosis. Conversely, changes in the association of these checkpoint factors with the NPC may also serve to regulate specific transport pathways. In vertebrates, and possibly other metazoans, the huge Nup107-160 complex likely behaves as a molecular scaffold within kinetochores. Finally, the RRSU complex contributes to the regulation of kinetochore-microtubule attachment by virtue of its role as a modulator of Ran-GTP levels, and to other aspects of mitotic progression via its SUMO E3 ligase activity.
While our understanding of each of these individual events is improving, we are still challenged to develop a vision of how individual proteins, in some cases with well-defined functions, are incorporated into processes as diverse as nuclear transport and chromosome segregation. An attractive, yet still speculative, hypothesis is that these proteins, through their relocalization, contribute, along with mitotic kinases and phosphatases and controlled protein degradation, to the spatio-temporal coordination of mitosis.
References
Tran EJ, Wente SR (2006) Dynamic nuclear pore complexes: life on the edge. Cell 125:1041–1053
D’Angelo MA, Hetzer MW (2008) Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol 18:456–466
Antonin W, Ellenberg J, Dultz E (2008) Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett 582:2004–2016
Chan GK, Liu ST, Yen TJ (2005) Kinetochore structure and function. Trends Cell Biol 15:589–598
Maiato H, DeLuca J, Salmon ED, Earnshaw WC (2004) The dynamic kinetochore-microtubule interface. J Cell Sci 117:5461–5477
Santaguida S, Musacchio A (2009) The life and miracles of kinetochores. EMBO J 28:2511–2531
Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8:379–393
Ciciarello M, Mangiacasale R, Lavia P (2007) Spatial control of mitosis by the GTPase Ran. Cell Mol Life Sci 64:1891–1914
Kalab P, Heald R (2008) The RanGTP gradient—a GPS for the mitotic spindle. J Cell Sci 121:1577–1586
Li R, Murray AW (1991) Feedback control of mitosis in budding yeast. Cell 66:519–531
Hardwick KG, Murray AW (1995) Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol 131:709–720
Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW (2002) The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol 159:807–819
Ikui AE, Furuya K, Yanagida M, Matsumoto T (2002) Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J Cell Sci 115:1603–1610
De Souza CP, Hashmi SB, Nayak T, Oakley B, Osmani SA (2009) Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol Biol Cell 20:2146–2159
Scott RJ, Lusk CP, Dilworth DJ, Aitchison JD, Wozniak RW (2005) Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol Biol Cell 16:4362–4374
Quimby BB, Arnaoutov A, Dasso M (2005) Ran GTPase regulates Mad2 localization to the nuclear pore complex. Eukaryot Cell 4:274–280
Chen RH, Shevchenko A, Mann M, Murray AW (1998) Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol 143:283–295
Chen RH, Waters JC, Salmon ED, Murray AW (1996) Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science 274:242–246
Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE (2005) Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol 15:856–861
Campbell M, Chan G, Yen T (2001) Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci 114:953–963
Lee SH, Sterling H, Burlingame A, McCormick F (2008) Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev 22:2926–2931
Katsani KR, Karess RE, Dostatni N, Doye V (2008) In vivo dynamics of Drosophila nuclear envelope components. Mol Biol Cell 19:3652–3666
Lince-Faria M, Maffini S, Orr B, Ding Y, Claudia F, Sunkel CE, Tavares A, Johansen J, Johansen KM, Maiato H (2009) Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J Cell Biol 184:647–657
Hawryluk-Gara LA, Shibuya EK, Wozniak RW (2005) Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell 16:2382–2394
Johansen KM, Johansen J (2007) Cell and molecular biology of the spindle matrix. Int Rev Cytol 263:155–206
Byrd DA, Sweet DJ, Pante N, Konstantinov KN, Guan T, Saphire AC, Mitchell PJ, Cooper CS, Aebi U, Gerace L (1994) Tpr, a large coiled coil protein whose amino terminus is involved in activation of oncogenic kinases, is localized to the cytoplasmic surface of the nuclear pore complex. J Cell Biol 127:1515–1526
Tighe A, Staples O, Taylor S (2008) Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J Cell Biol 181:893–901
De Souza CP, Osmani AH, Hashmi SB, Osmani SA (2004) Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr Biol 14:1973–1984
Qi H, Rath U, Wang D, Xu YZ, Ding Y, Zhang W, Blacketer MJ, Paddy MR, Girton J, Johansen J, Johansen KM (2004) Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol Biol Cell 15:4854–4865
Gillett ES, Espelin CW, Sorger PK (2004) Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J Cell Biol 164:535–546
Scott RJ, Cairo LV, Van de Vosse DW, Wozniak RW (2009) The nuclear export factor Xpo1p targets Mad1p to kinetochores in yeast. J Cell Biol 184:21–29
Makhnevych T, Lusk CP, Anderson AM, Aitchison JD, Wozniak RW (2003) Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell 115:813–823
Pemberton LF, Paschal BM (2005) Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6:187–198
Cole CN, Hammell CM (1998) Nucleocytoplasmic transport: driving and directing transport. Curr Biol 8:R368–R372
Fahrenkrog B, Aebi U (2003) The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol 4:757–766
Delphin C, Guan T, Melchior F, Gerace L (1997) RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol Biol Cell 8:2379–2390
Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E (1995) Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem 270:14209–14213
Bernad R, van der Velde H, Fornerod M, Pickersgill H (2004) Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol Cell Biol 24:2373–2384
Rexach M, Blobel G (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83:683–692
Matunis MJ, Coutavas E, Blobel G (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 135:1457–1470
Mahajan R, Delphin C, Guan TL (1997) A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97–107
Dasso M (2008) Emerging roles of the SUMO pathway in mitosis. Cell Div 3:5
Lee GW, Melchior F, Matunis MJ, Mahajan R, Tian Q, Anderson P (1998) Modification of Ran GTPase-activating protein by the small ubiquitin-related modifier SUMO-1 requires Ubc9, an E2-type ubiquitin-conjugating enzyme homologue. J Biol Chem 273:6503–6507
Palancade B, Doye V (2008) Sumoylating and desumoylating enzymes at nuclear pores: underpinning their unexpected duties? Trends Cell Biol 18:174–183
Pichler A, Gast A, Seeler JS, Dejean A, Melchior F (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109–120
Zhang H, Saitoh H, Matunis MJ (2002) Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol 22:6498–6508
Swaminathan S, Kiendl F, Korner R, Lupetti R, Hengst L, Melchior F (2004) RanGAP1*SUMO1 is phosphorylated at the onset of mitosis and remains associated with RanBP2 upon NPC disassembly. J Cell Biol 164:965–971
Joseph J, Tan SH, Karpova TS, McNally JG, Dasso M (2002) SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol 156:595–602
Salina D, Enarson P, Rattner JB, Burke B (2003) Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol 162:991–1001
Askjaer P, Galy V, Hannak E, Mattaj IW (2002) Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol Biol Cell 13:4355–4370
Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M (2004) The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol 14:611–617
Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, McNally J, Dasso M (2005) Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat Cell Biol 7:626–632
Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita JB, Fukagawa T, Shiekhattar R, Yen T, Doye V (2007) The human Nup107–160 nuclear pore sub-complex contributes to proper kinetochore functions. EMBO J 26:1853–1864
Arnaoutov A, Dasso M (2005) Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle 4:1161–1165
Dasso M (2006) Ran at kinetochores. Biochem Soc Trans 34:711–715
Lau CK, Delmar VA, Chan RC, Phung Q, Bernis C, Fichtman B, Rasala BA, Forbes DJ (2009) Transportin regulates major mitotic assembly events: from spindle to nuclear pore assembly. Mol Biol Cell 20:4043–4058
Walczak CE, Heald R (2008) Mechanisms of mitotic spindle assembly and function. Int Rev Cytol 265:111–158
Cai S, Weaver LN, Ems-McClung SC, Walczak CE (2009) Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol Biol Cell 20:1348–1359
Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW (2006) HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol 16:743–754
Ribbeck K, Groen AC, Santarella R, Bohnsack MT, Raemaekers T, Kocher T, Gentzel M, Gorlich D, Wilm M, Carmeliet G, Mitchison TJ, Ellenberg J, Hoenger A, Mattaj IW (2006) NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol Biol Cell 17:2646–2660
Wong J, Fang G (2006) HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol 173:879–891
Kraemer D, Blobel G (1997) mRNA binding protein mrnp 41 localizes to both nucleus and cytoplasm. Proc Natl Acad Sci USA 94:9119–9124
Pritchard CE, Fornerod M, Kasper LH, van Deursen JM (1999) RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol 145:237–254
Kraemer D, Dresbach T, Drenckhahn D (2001) Mrnp41 (Rae 1p) associates with microtubules in HeLa cells and in neurons. Eur J Cell Biol 80:733–740
Blower MD, Nachury M, Heald R, Weis K (2005) A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 121:223–234
Wong RW, Blobel G, Coutavas E (2006) Rae1 interaction with NuMA is required for bipolar spindle formation. Proc Natl Acad Sci USA 103:19783–19787
Jeganathan KB, Malureanu L, van Deursen JM (2005) The Rae1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature 438:1036–1039
Jeganathan KB, Baker DJ, van Deursen JM (2006) Securin associates with APCCdh1 in prometaphase but its destruction is delayed by Rae1 and Nup98 until the metaphase/anaphase transition. Cell Cycle 5:366–370
Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM (2000) Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell 11:1555–1569
Hagting A, Den Elzen N, Vodermaier HC, Waizenegger IC, Peters JM, Pines J (2002) Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J Cell Biol 157:1125–1137
Torosantucci L, De Luca M, Guarguaglini G, Lavia P, Degrassi F (2008) Localized RanGTP accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol Biol Cell 19:1873–1882
Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ (2008) SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell 29:729–741
Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, Shuai K, Grosschedl R, van Deursen JM (2008) Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell 133:103–115
Shamu CE, Murray AW (1992) Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J Cell Biol 117:921–934
Ruchaud S, Carmena M, Earnshaw WC (2007) Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol 8:798–812
Klein UR, Haindl M, Nigg EA, Muller S (2009) RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on borealin. Mol Biol Cell 20:410–418
Knauer SK, Bier C, Habtemichael N, Stauber RH (2006) The Survivin-Crm1 interaction is essential for chromosomal passenger complex localization and function. EMBO Rep 7:1259–1265
Xia F, Canovas PM, Guadagno TM, Altieri DC (2008) A survivin-ran complex regulates spindle formation in tumor cells. Mol Cell Biol 28:5299–5311
Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, Ellenberg J, Doye V (2001) An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol 154:1147–1160
Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ (2001) Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol 155:339–354
Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V (2004) The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell 15:3333–3344
Liu HL, De Souza CP, Osmani AH, Osmani SA (2009) The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol Biol Cell 20:616–630
Brohawn SG, Partridge JR, Whittle JR, Schwartz TU (2009) The nuclear pore complex has entered the atomic age. Structure 17:1156–1168
Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ (2006) ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci USA 103:17801–17806
Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W (2007) MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep 8:165–172
Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ (2007) ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol 17:1657–1662
Gunsalus KC, Ge H, Schetter AJ, Goldberg DS, Han JD, Hao T, Berriz GF, Bertin N, Huang J, Chuang LS, Li N, Mani R, Hyman AA, Sonnichsen B, Echeverri CJ, Roth FP, Vidal M, Piano F (2005) Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature 436:861–865
Galy V, Askjaer P, Franz C, Lopez-Iglesias C, Mattaj IW (2006) MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr Biol 16:1748–1756
Fernandez AG, Piano F (2006) MEL-28 is downstream of the Ran cycle and is required for nuclear-envelope function and chromatin maintenance. Curr Biol 16:1757–1763
Rabut G, Doye V, Ellenberg J (2004) Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 6:1114–1121
Doye V, Hurt E (1997) From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol 9:401–411
Bai SW, Rouquette J, Umeda M, Faigle W, Loew D, Sazer S, Doye V (2004) The fission yeast Nup107–120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol Cell Biol 24:6379–6392
Resendes KK, Rasala BA, Forbes DJ (2008) Centrin 2 localizes to the vertebrate nuclear pore and plays a role in mRNA and protein export. Mol Cell Biol 28:1755–1769
D’Angelo MA, Anderson DJ, Richard E, Hetzer MW (2006) Nuclear pores form de novo from both sides of the nuclear envelope. Science 312:440–443
Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, Briggs S, Dasso M, Forbes DJ (2006) The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell 17:3806–3818
Chakraborty P, Wang Y, Wei JH, van Deursen J, Yu H, Malureanu L, Dasso M, Forbes DJ, Levy DE, Seemann J, Fontoura BM (2008) Nucleoporin levels regulate cell cycle progression and phase-specific gene expression. Dev Cell 15:657–667
Kutay U, Hetzer MW (2008) Reorganization of the nuclear envelope during open mitosis. Curr Opin Cell Biol 20:669–677
Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, Mattaj IW, Doye V (2003) The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell 113:195–206
Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ (2003) Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell 11:853–864
Rasala BA, Ramos C, Harel A, Forbes DJ (2008) Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell 19:3982–3996
Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M (2003) RanGTP mediates nuclear pore complex assembly. Nature 424:689–694
Rotem A, Gruber R, Shorer H, Shaulov L, Klein E, Harel A (2009) Importin beta regulates the seeding of chromatin with initiation sites for nuclear pore assembly. Mol Biol Cell 20:4031–4042
Davuluri G, Gong W, Yusuff S, Lorent K, Muthumani M, Dolan AC, Pack M (2008) Mutation of the zebrafish nucleoporin elys sensitizes tissue progenitors to replication stress. PLoS Genet 4:e1000240
Osmani AH, Davies J, Liu HL, Nile A, Osmani SA (2006) Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol Biol Cell 17:4946–4961
Theisen U, Straube A, Steinberg G (2008) Dynamic rearrangement of nucleoporins during fungal “open” mitosis. Mol Biol Cell 19:1230–1240
Feng J, Huang H, Yen TJ (2006) CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma 115:320–329
Platani M, Santarella-Mellwig R, Posch M, Walczak R, Swedlow JR, Mattaj IW (2009) The Nup107-160 nucleoporin complex promotes mitotic events via control of the localization state of the chromosome passenger complex. Mol Biol Cell 20:5260–5275
Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H (2004) The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 118:187–202
Mackay DR, Elgort SW, Ullman KS (2009) The nucleoporin Nup153 has separable roles in both early mitotic progression and the resolution of mitosis. Mol Biol Cell 20:1652–1660
Sabri N, Roth P, Xylourgidis N, Sadeghifar F, Adler J, Samakovlis C (2007) Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport. J Cell Biol 178:557–565
Hase ME, Cordes VC (2003) Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell 14:1923–1940
Hang J, Dasso M (2002) Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem 277:19961–19966
Glavy JS, Krutchinsky AN, Cristea IM, Berke IC, Boehmer T, Blobel G, Chait BT (2007) Cell-cycle-dependent phosphorylation of the nuclear pore Nup107–160 subcomplex. Proc Natl Acad Sci USA 104:3811–3816
Mishra RK, Chakraborty P, Arnauoutov A, Fontoura BMA, Dasso M (2010) The Nup107-160 complex and γ-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol 12:164–169
Acknowledgments
The authors would like to thank “la Fondation des Treilles” (http://www.les-treilles.com) for organizing the first transport and cell cycle meeting in 2007 at the very time when most of the data and concepts now included in this review were emerging (http://www.les-treilles.com/ssimages/c_2007_09_Doye_Wozniak.html). The authors also wish to acknowledge the following agencies for funding: la Ligue Nationale contre le Cancer (V.D., Equipe Labellisée 2006–2009), the Agence Nationale de la Recherche (to V.D.), Canadian Institutes of Health Research (to R.W.W.), the Alberta Heritage Foundation for Medical Research (to R.W.W.), the Howard Hughes Medical Institute (to R.W.W.), and the National Institutes of Health (to B.B.).
Author information
Authors and Affiliations
Corresponding author
Additional information
R. Wozniak, B. Burke, and V. Doye contributed equally to this review.
Rights and permissions
About this article
Cite this article
Wozniak, R., Burke, B. & Doye, V. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell. Mol. Life Sci. 67, 2215–2230 (2010). https://doi.org/10.1007/s00018-010-0325-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-010-0325-7