Abstract
Cyclotides are disulfide-rich peptides from plants that are exceptionally stable as a result of their unique cyclic cystine knot structural motif. Their natural role is thought to be as plant defence agents, most notably against insect pests, but they also have potential applications in drug design and agriculture. This article identifies gaps in current knowledge on cyclotides and suggests future directions for research into this fascinating family of ultra-stable mini-proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 1999 the term cyclotides (cyclo peptides) was introduced to describe a newly discovered family of plant-based mini-proteins, approximately 30 amino acids in size, that are characterised by a topologically unusual structural motif called the cyclic cystine knot (CCK) [1]. As its name suggests, the CCK motif comprises a head-to-tail cyclised peptide backbone that is cross-braced by a knotted arrangement of disulfide bonds, formed from six absolutely conserved cysteine residues (Fig. 1). Cyclotides occur in a large number of plants from the Violaceae (violet) and Rubiaceae (coffee) families and one member of the Cucurbitaceae (squash) family, and recent studies suggest that they might also occur in other plant families [2]. Indeed, it seems probable that cyclotides will ultimately turn out to be a particularly large class of proteins. Cyclotides are found in a range of plant tissues, including leaves, flowers, stems and roots, with individual plants expressing a suite of many different cyclotides [3, 4].
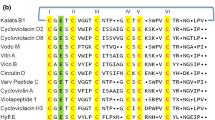
Schematic overview of the prototypic cyclotide kalata B1 and two precursor proteins, Oak1 and Oak4. (Oak refers to O ldenlandia a ffinis kalata). a Sequence of kalata B1 showing disulfide connectivities, loops between conserved Cys residues and the ligation point (arrow) involved in peptide cyclisation. b A ribbon model of kalata B1 highlighting the CCK motif of three disulfide bonds and cyclic backbone. c A surface-rendered model of kalata B1 showing the hydrophobic patch (green) which associates with membranes. d Precursor proteins for kalata B1 (Oak1) and kalata B2 (Oak4). Oak4 is shown to highlight the fact that some cyclotide genes encode multiple copies of cyclotides (as is the case for kalata B2) [16]. The cyclotide C-terminal residue (Asn or Asp), thought to be targeted by AEP, is marked with an arrow
Initial interest in macrocyclic peptides from plants arose because of their bioactivities, which include uterotonic [5] and anti-HIV activities [6], amongst others [7, 8], although in the early studies the few peptides discovered had not yet been recognised as a family. This article reflects on current cyclotide research and speculates about future directions for the study of this structurally fascinating family of proteins. A reflective review with some future visions is timely because 2009 represents the 10th anniversary of the naming of the cyclotide family. Readers are also referred to other recent reviews for more detailed information on cyclotides [9–15].
The first clue to the biosynthetic origin of cyclotides came in 2001 when it was reported that they are gene-encoded peptides [16]. This finding stood in contrast to most previously known naturally occurring cyclic peptides, which are typically smaller in size (<12 amino acids) and biosynthesised via non-ribosomal peptide synthetases [17]. The genes for cyclotides encode precursor proteins of 11–14 kDa, which contain one, two or three mature cyclotide domains, along with an endoplasmic reticulum (ER) signal sequence, a pro-domain and a C-terminal region of hydrophobic residues (Fig. 1). The ribosomal synthesis of cyclotides via precursor proteins raises the question of how the nascent cyclotide termini become joined after excision of the mature sequences from the precursor proteins. Some aspects of this process are now understood, as described later in this article [18, 19], but fully understanding this process remains an important topic for future research.
Cyclotides are plant-derived cyclic peptides, but other examples of gene-encoded cyclic peptides have also been discovered over the last decade in cyanobacteria, bacteria, fungi, plants and animals [20–25], suggesting that post-translational cyclisation is a strategy that has evolved in multiple kingdoms of life. What are the advantages of these cyclic proteins compared to conventional acyclic proteins? Initially it might seem likely that cyclisation would involve the need for extra biosynthetic infrastructure over that required for conventional proteins and so it is reasonable to suggest that there must be benefits to these organisms that offset this extra metabolic burden. Indeed one advantage is fairly clear as cyclic peptides, in general, are much more stable than their linear counterparts, and cyclotides most probably evolved because of this stability. Interestingly, and in contrast to initial expectations, recent studies suggest that such benefits may have come at little extra cost to the organism, with cyclisation able to be done in planta by enzymes that are present anyway for other purposes [18, 19].
Circling the enemy: pesticidal activities of cyclotides
Cyclotides have a range of biological activities, and it is these activities that were responsible for their discovery, initially from indigenous medicine applications [5, 26] and later during pharmaceutically oriented screening programs seeking anti-HIV [6] or neurotensin-inhibitory activities [8]. Other approaches that focussed on identifying proteins from plant biomass took advantage of the unique biophysical properties of cyclotides [1, 27, 28]. Most of the originally reported activities seem to have little physiological relevance to a plant. For example, why would a plant need to produce an anti-HIV agent? The answer of course is that this is a fortuitous activity and that plants produce cyclotides for another purpose. Indeed, like other ribosomally synthesised cyclic peptides, such as bacteriocins [22] and θ-defensins [20], from bacteria and mammals, respectively, cyclotides appear to be produced as part of the host defence armoury [29].
The most extensively studied defence-related role of cyclotides is their insecticidal activity [16, 30, 31]. Other miscellaneous activities have also been reported for individual cyclotides, including antimicrobial [32], antifouling [33], cytotoxic [34], molluscicidal [35] and nematocidal activities [36–38]. The apparent antimicrobial activity requires further investigation because synthetic kalata B1 was reported to be inactive against Escherichia coli by Tam et al. [32], but native kalata B1 was found to be active in a recent study by Gran et al. [39]. Similarly, kalata B1 was shown to be active under low salt conditions against Staphylococcus aureus in the former study but inactive in the latter study. The differences have been attributed to the use of different bacterial strains, but additional studies are underway to resolve this issue [39]. It appears that bacterial strains that might be involved in plant pathogenesis have not yet been tested for their susceptibility to cyclotides, and this represents an important area for future study.
The diverse range of cyclotide activities despite their conserved core structural motif prompts the question: Is there a common mechanism of action? A number of biophysical studies have now clearly established that cyclotides bind to membranes, and it appears that their diverse actions can be explained on the basis of this binding. Surface plasmon resonance studies [40] have shown that cyclotides bind to immobilised phospholipid bilayers, and NMR experiments have been used to define the binding orientation of the cyclotides kalata B1 and kalata B7 on dodecylphosphocholine (DPC) micelles, used as a model for biological membranes [41, 42]. These NMR studies show that cyclotides orient on the surface of the DPC micelle, with a hydrophobic patch on the surface of the cyclotides (Fig. 1) slightly embedded at the micelle surface. Another recent study has shown that different cyclotides have their hydrophobic patches in different orientations with respect to the cystine knot and confirmed that it is the hydrophobic patch that determines the binding orientation [43].
However, in our opinion it is likely that more than simple membrane binding of cyclotide monomers is required for cyclotides to exert biological effects at membranes. One study has shown that some cyclotides have a tendency to self-associate in solution [44]. We have suggested that this self-association is mediated by a patch of adjacent hydrophilic residues on one face of the prototypical cyclotide kalata B1 [45]. When any of these residues are mutated to an alanine, kalata B1 loses insecticidal activity. We hypothesize that cyclotides self-associate by this hydrophilic “bioactive face” then embed in the membrane via the hydrophobic face, ultimately forming pores that disrupt cell membranes of the pest or pathogen [45, 46]. This mode of action may be responsible for the observations made recently using electron microscopy that showed major disruption to the midgut membranes of Helicoverpa species when fed an artificial diet containing cyclotides [30]. An earlier study established a link between cytotoxic activity of cycloviolacin O2 against a human lymphoma cell line and membrane disruption [47].
In summary, although it remains to be determined whether cyclotides act via a stereospecific protein-based receptor, the likelihood is that they do not, and that their actions are mediated purely, or at least mainly, by membrane binding, by which they cause disruption and leakage of biological membranes. Interestingly, an intact cyclic backbone seems to be essential for the bioactivities tested for so far, and synthetic acyclic variants of cyclotides have been shown to be inactive in haemolytic and insecticidal assays [30, 48, 49]. It is not known if this lack of activity results from the acyclic peptides no longer interacting with membranes or whether changes in stability affect the activity, and this is another area that warrants future investigation.
Distribution: revolution or evolution?
Initially, it was surprising that only a limited number of cyclotides had been found in sporadically distributed members of the Rubiaceae, yet in every member of the Violaceae we examined, we found cyclotides [2, 3, 50]. The Violaceae and Rubiaceae are not closely related phylogenetically, and in early studies there was no evidence of cyclotides in other plant families. However, a recent study screened more than 250 Rubiaceae species and found a hit rate of 5–10% for cyclotides [2]. Evidence is also emerging that cyclotides are likely to occur in other families, including the Apocynaceae [2], thus it is our prediction that cyclotides will be found in a much wider group of plant families than is currently known. The lack of discovery so far perhaps reflects the fact that cyclic peptides are intrinsically more difficult to sequence than conventional linear peptides, and there have been relatively few screens of plant species for peptide components. Alternatively, cyclotides might be present in lower abundance in some plant families.
Two recent reports have suggested that cyclotide-like nucleic acid sequences are present in monocotyledonous plants such as rice, wheat and corn, which are part of the Poaceae family [51, 52]. A bioinformatics-based study [51] looking at nucleic acid sequences from these plants found, for example, ~20 cyclotide-like sequences that encode the signature cysteine spacing of cyclotides and a conserved glutamic acid that is a characteristic of cyclotides [53, 54]. Basse [52] found upregulation of a similar sequence in response to fungal infection in corn, supporting the suggestion that cyclotides (or cyclotide-like sequences) have a role in host defence. However, none of these nucleic acid sequences have been detected at the peptide level, and it remains to be discovered whether these mRNAs are translated to protein, and, if so, whether they are processed into linear or cyclic peptides. Their sequences lack a key Asn (or Asp) residue involved in processing, as well as residues conserved on either side of the mature cyclotide sequence, and our prediction is that these are linear ancestral cyclotide-like molecules. One of the important areas for future studies of cyclotides will be examining Poaceae species to see if expressed proteins corresponding to these sequences can be found, and, if so, their functions determined.
Finally, the title of this section, “revolution or evolution”, is meant to reflect on the question: Did circular proteins arise as a revolutionary new class of molecules, or do they reflect a gradual evolution from linear precursors? The latter seems more probable, given the discovery of apparently linear cyclotide-like sequences in the Poaceae, together with the fact that a key Asn mutation is implicated in cyclisation, as described in the following section.
Cyclisation: making ends meet
At first sight, the formation of a peptide bond between the N- and C-termini of a linear precursor protein would seem to be intrinsically unfavourable from an entropic perspective, as would formation of a cystine knot. Recent studies have identified two key enzymes (an asparaginyl endopeptidase and a protein disulfide isomerise) that appear to be involved in the processing of cyclotides, from cyclisation and folding perspectives, respectively. Specifically, Saska et al. [18] and Gillon et al. [19] have shown that an asparaginyl endopeptidase (AEP) enzyme is involved in the cyclisation of cyclotides. AEPs are ubiquitous enzymes in plants, and their normal role is proteolytic cleavage of substrate proteins after Asn residues. However, it appears that cyclotide precursors are able to hijack these enzymes and utilise them not only for proteolytic cleavage but also for the formation of peptide bonds, i.e., the reverse of the normal proteolytic process. The use of enzymes “in reverse” is a well-established thermodynamic principal, but it is something that has not yet been seen widely in practice. However, from an evolutionary perspective, it makes sense that the cyclisation process would not spontaneously arise as a “revolution”, rather it would arise incrementally by recruiting an existing enzyme infrastructure for fortuitously performing a cyclisation task that leads to advantages over the linear precursor protein. We wonder whether cyclisation as a stabilising post-translation modification may be under-discovered since cyclisation often frustrates most techniques typically employed to characterise proteins, as they rely on protein fragmentation. More generally, we recently suggested that protease-catalysed protein splicing, of which cyclisation is one example, might be a more widespread post-translational modification than originally thought [55].
Cyclotides can be readily synthesised in vitro [32, 56] using an adaptation of native chemical ligation technology [57]. In this process the backbone is first cyclised and then the folding reaction occurs subsequently. We suspect that in planta the opposite occurs, i.e., the oxidation and folding of the precursor protein occurs first, and the mature peptide is subsequently excised and cyclised.
We recently isolated a protein disulfide isomerase (PDI) from O. affinis leaves and have shown in vitro at least that it can increase the folding yield of cyclotide-related molecules, including a linear cyclotide and a reduced cyclic molecule [58]. The relevance of this mechanism in vivo remains to be established. Presumably the PDI interacts with the precursor protein, rather than with processed cyclotide domains. However, it is possible that the PDI has a secondary function of mopping up any misfolded cyclotides and can refold them appropriately. Some recent work has shown that one of the twelve PDIs in Arabidopsis, PDI5, directly interacts and traffics with a Cys-protease RD21 [59]. If these implications were extendable to cyclotide synthesis, it is possible a more intimate relationship between AEP (also a Cys protease) and PDI may be responsible for cyclotide biosynthesis.
Synthetic engineered cyclotides: reinventing the wheel
How can designer cyclotides be made? Because cyclotides are relatively small proteins of only ~30 amino acids, they are amenable to solid phase peptide synthesis (SPPS), and this has been our preferred method to produce them for a range of biochemical, mechanistic, and protein engineering studies [60]. The use of SPPS compared to bacterial expression is preferred because, until recently, cyclotides have been unable to be expressed conveniently in bacterial cells. Recent work has shown that cyclotides can be expressed in bacteria using intein-based expression systems [61, 62]; however, the yields are currently quite low. Thus, SPPS remains the most useful method for making cyclotides, although the intein approach is attractive, as scale up in SPPS is relatively expensive compared with scale up of bacterial fermentation systems. A recently developed chemo-enzymatic approach [63] in which SPPS is used to make a linear cyclotide precursor that subsequently is cyclised in an enzyme-catalysed reaction shows great promise as another way of making cyclotides.
The reason that synthetic methods for producing cyclotides are important is that they can be utilised to make modified cyclotides, including examples where small peptide epitopes with desired biological functions can be engineered into the cyclotide framework, essentially using it as a delivery vehicle for stabilising peptides for pharmaceutical applications [11]. Several examples of bioactive peptide grafting have been reported recently, using both cyclotides themselves, including kalata B1 [64] and MCoTI-II [65], and linear cystine knot molecules such as EETI [66–68]. Although promising results have been obtained, more work needs to be done in this area to confirm the viability of the CCK framework in drug design. For instance, pharmacokinetic properties and immunogenicity of grafted cyclotides are two key features that need to be explored.
If the CCK framework does prove to be useful in drug design, the issue of manufacture of cyclotide-like molecules will arise. As well as the SPPS, intein-based, and chemo-enzymatic methods described above, the use of cultured plant cells as production factories for cyclotides has been reported recently [69–71]. The latter method looks particularly exciting, producing yields of 0.4 g/kg of kalata B1, similar to that seen in whole plants (1–2 g cyclotides/kg plant material). In the longer term, since plants themselves can produce cyclotides in very high yields, it could be economically advantageous to produce pharmaceutically modified cyclotides, using whole plants as factories. Initial studies have shown that cyclotide genes can be transformed into Arabidopsis and tobacco and can produce cyclic peptides; however, the yields are still low and the technology is associated with the production of large amounts of misprocessed (linear) cyclotide derivatives [18, 19]. All of these studies so far have relied on the insertion of only a single gene into plants. Co-expression with auxiliary proteins such as the PDI or AEP from cyclotide-bearing plants has the potential to increase yields.
Future perspectives
There remains much to be done in the field of cyclotide research. In particular, although there have been promising starts to understand the biosynthesis of cyclotides, so far, nothing is known of the N-terminal processing events that precede cyclisation. It seems probable that there must be processing at the N-terminus to facilitate its nucleophilic attack on the C-terminal Asn residue and the subsequent transpeptidation reaction driven by AEP activity. Likewise, the sites within plant cells for the various processing reactions, and indeed the fate within the cell of cyclotides themselves, has not yet been reported. Are they stored in vacuoles or are they secreted?
The extent to which cyclotides are present in the plant kingdom is still not clear, with cyclotides only confirmed so far in three plant families. The current suite of known cyclotides is likely to be biased by the extraction procedures that have been commonly used and the focus on late eluting peaks on RP-HPLC. Searches for more hydrophilic peptides might yield novel cyclotides. Indeed, two hydrophilic cyclotides have already been found in the tropical vine Momordica cochinchinensis [72]. Studies of plant seeds are warranted and might be important for discovering novel cyclotides because the M. cochinchinensis cyclotides were found in seeds whereas the majority of other studies have focussed on other plant tissues, principally on leaves. Furthermore, a greater emphasis on screening for cyclotides at the nucleic acid rather than peptide level is likely to expand the range of cyclotides significantly. Several recent studies have emphasized the promise of this approach [3, 50, 73, 74]. However, improvements in methods for the detection of cyclotides at the peptide level will continue to be important [75].
The mechanism of action of cyclotides needs further investigation, although it seems fairly clear that membrane binding is likely to be the preferred common feature amongst the activities of many cyclotides [46, 47]. The exact details of how they disrupt biological membranes remains to be established. Studies that determine the geometry of self-association and the geometry of any pores formed in membranes will provide insight in this area.
Because cyclotides have so far not been conveniently amenable to bacterial expression, they have not been able to be isotopically labelled to assist in advanced NMR applications, for example. The recent demonstration [76] that uniformly labelled 15N cyclotides can be isolated from O. affinis paves the way for such studies.
In terms of applications of cyclotides, a promising start has been made in the production of transgenic plants that express cyclotides [18, 19]. Such plants could have applications in two areas: either as pharmaceutical production factories for producing high value molecules such as pharmaceutically grafted cyclotides, or agricultural applications, where the natural insecticidal or nematocidal activity of cyclotides is transferred to crop plants for example, as highlighted in Fig. 2.
Overview of potential pharmaceutical and agricultural applications of cyclotides. Oldenlandia affinis (centre) represents an example of a plant that naturally produces cyclotide genes (green arrow) and corresponding cyclotide peptides (red arrow). The upper (red) panel of the diagram represents pharmaceutical applications that involve grafting biologically active peptide epitopes (e.g., the indicated helical peptide) into the CCK framework of natural cyclotides. These modified cyclotides can be synthesised using solid phase peptide synthesis, chemo-enzymatic synthesis or inteins. Alternatively, pharmaceutically modified cyclotides might, in future, be produced (“pharmed”) in plants or in plant cell culture [71] as shown in the middle of the diagram, by transformation with genes encoding modified cyclotides. The lower (green) panel of the diagram represents agricultural applications, whereby cyclotide gene sequences can be expressed in crop plants (e.g., corn as indicated) to confer resistance to pests. The dashed arrows are shown to highlight the linkage between pharmaceutical and agricultural applications schematically. For example, lessons learned from synthetic chemical grafting studies to produce pharmaceutically active cyclotides can be combined with lessons learned from the recombinant expression of insecticidal cyclotide genes in plants, to undertake molecular pharming studies, i.e., the production of pharmaceutically active cyclotides in plants
Finally, there are many questions that can be raised about the distribution, mechanism of action and evolution of cyclotides: (1) Why do individual plants produce so many different cyclotides? Sometimes more than 100 cyclotides are found in a single plant. Perhaps this is a mechanism for overcoming the development of resistance. (2) Do all cyclotides act via the same general mechanism or are there some that interact with receptors? (3) How many other classes of circular proteins exist and are their mechanisms of processing similar to those of cyclotides? Already it is clear from the examination of the sequences of plant, bacterial and mammalian cyclic proteins that there are major differences in the amino acid residues at the processing sites. Even if the broad mechanisms of cyclisation are similar, the biochemical details are likely to be quite different in bacteria, plants and mammals.
References
Craik DJ, Daly NL, Bond T, Waine C (1999) Plant cyclotides: a unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J Mol Biol 294:1327–1336
Gruber CW, Elliott AG, Ireland DC, Delprete PG, Dessein S, Göransson U, Trabi M, Wang CK, Kinghorn AB, Robbrecht E, Craik DJ (2008) Distribution and evolution of circular miniproteins in flowering plants. Plant Cell 20:2471–2483
Trabi M, Mylne JS, Sando L, Craik DJ (2009) Circular proteins from Melicytus (Violaceae) refine the conserved protein and gene architecture of cyclotides. Org Biomol Chem 7:2378–2388
Trabi M, Svangard E, Herrmann A, Göransson U, Claeson P, Craik DJ, Bohlin L (2004) Variations in cyclotide expression in Viola species. J Nat Prod 67:806–810
Gran L (1970) An oxytocic principle found in Oldenlandia affinis DC. Medd Nor Farm Selsk 12:173–180
Gustafson KR, Sowder RCI, Henderson LE, Parsons IC, Kashman Y, Cardellina JHI, McMahon JB, Buckheit RWJ, Pannell LK, Boyd MR (1994) Circulins A and B: novel HIV-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J Am Chem Soc 116:9337–9338
Schöpke T, Hasan Agha MI, Kraft R, Otto A, Hiller K (1993) Hämolytisch aktive komponenten aus Viola tricolor L. und Viola arvensis Murray. Sci Pharm 61:145–153
Witherup KM, Bogusky MJ, Anderson PS, Ramjit H, Ransom RW, Wood T, Sardana M (1994) Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J Nat Prod 57:1619–1625
Ireland DC, Wang CK, Wilson JA, Gustafson KR, Craik DJ (2008) Cyclotides as natural anti-HIV agents. Biopolymers Pept Sci 90:51–60
Cemazar M, Gruber CW, Craik DJ (2008) Oxidative folding of cyclic cystine knot proteins. Antioxid Redox Signal 10:103–111
Craik DJ, Clark RJ, Daly NL (2007) Potential therapeutic applications of the cyclotides and related cystine knot mini-proteins. Expert Opin Investig Drugs 16:595–604
Craik DJ, Cemazar M, Daly NL (2007) The chemistry and biology of cyclotides. Curr Opin Drug Discovery Dev 10:176–184
Gruber CW, Cemazar M, Anderson MA, Craik DJ (2007) Insecticidal plant cyclotides and related cystine knot toxins. Toxicon 49:561–575
Pelegrini PB, Quirino BF, Franco OL (2007) Plant cyclotides: an unusual class of defense compounds. Peptides 28:1475–1481
Göransson U, Svangard E, Claeson P, Bohlin L (2004) Novel strategies for isolation and characterization of cyclotides: the discovery of bioactive macrocyclic plant polypeptides in the Violaceae. Curr Protein Pept Sci 5:317–329
Jennings C, West J, Waine C, Craik D, Anderson M (2001) Biosynthesis and insecticidal properties of plant cyclotides: the cyclic knotted proteins from Oldenlandia affinis. Proc Natl Acad Sci USA 98:10614–10619
Tan N-H, Zhou J (2006) Plant cyclopeptides. Chem Rev 106:840–895
Saska I, Gillon AD, Hatsugai N, Dietzgen RG, Hara-Nishimura I, Anderson MA, Craik DJ (2007) An asparaginyl endopeptidase mediates in vivo protein backbone cyclisation. J Biol Chem 282:29721–29728
Gillon AD, Saska I, Jennings CV, Guarino RF, Craik DJ, Anderson MA (2008) Biosynthesis of circular proteins in plants. Plant J 53:505–515
Tang Y-Q, Yuan J, Ösapay G, Ösapay K, Tran D, Miller CJ, Ouellette AJ, Selsted ME (1999) A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated α-defensins. Science 286:498–502
Trabi M, Craik DJ (2002) Circular proteins—no end in sight. Trends Biochem Sci 27:132–138
Maqueda M, Galvez A, Bueno MM, Sanchez-Barrena MJ, Gonzalez C, Albert A, Rico M, Valdivia E (2004) Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr Protein Pept Sci 5:399–416
Craik DJ (2006) Seamless proteins tie up their loose ends. Science 311:1563–1564
Hallen HE, Luo H, Scott-Craig JS, Walton JD (2007) Gene family encoding the major toxins of lethal Amanita mushrooms. Proc Natl Acad Sci USA 104:19097–19101
Donia MS, Ravel J, Schmidt EW (2008) A global assembly line for cyanobactins. Nat Chem Biol 4:341–343
Gran L, Sandberg F, Sletten K (2000) Oldenlandia affinis (R&S) DC: a plant containing uteroactive peptides used in African traditional medicine. J Ethnopharmacol 70:197–203
Claeson P, Göransson U, Johansson S, Luijendijk T, Bohlin L (1998) Fractionation protocol for the isolation of polypeptides from plant biomass. J Nat Prod 61:77–81
Göransson U, Luijendijk T, Johansson S, Bohlin L, Claeson P (1999) Seven novel macrocyclic polypeptides from Viola arvensis. J Nat Prod 62:283–286
Craik DJ (2009) Circling the enemy: cyclic proteins in plant defence. Trends Plant Sci 14:328–335
Barbeta BL, Marshall AT, Gillon AD, Craik DJ, Anderson MA (2008) Plant cyclotides disrupt epithelial cells in the midgut of lepidopteran larvae. Proc Natl Acad Sci USA 105:1221–1225
Jennings CV, Rosengren KJ, Daly NL, Plan M, Stevens J, Scanlon MJ, Waine C, Norman DG, Anderson MA, Craik DJ (2005) Isolation, solution structure, and insecticidal activity of kalata B2, a circular protein with a twist: do Möbius strips exist in nature? Biochemistry 44:851–860
Tam JP, Lu YA, Yang JL, Chiu KW (1999) An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc Natl Acad Sci USA 96:8913–8918
Göransson U, Sjogren M, Svangard E, Claeson P, Bohlin L (2004) Reversible antifouling effect of the cyclotide cycloviolacin O2 against barnacles. J Nat Prod 67:1287–1290
Lindholm P, Göransson U, Johansson S, Claeson P, Gulbo J, Larsson R, Bohlin L, Backlund A (2002) Cyclotides: a novel type of cytotoxic agents. Mol Cancer Ther 1:365–369
Plan MR, Saska I, Cagauan AG, Craik DJ (2008) Backbone cyclised peptides from plants show molluscicidal activity against the rice pest Pomacea canaliculata (golden apple snail). J Agric Food Chem 56:5237–5241
Colgrave ML, Kotze AC, Huang YH, O’Grady J, Simonsen SM, Craik DJ (2008) Cyclotides: natural, circular plant peptides that possess significant activity against gastrointestinal nematode parasites of sheep. Biochemistry 47:5581–5589
Colgrave ML, Kotze AC, Ireland DC, Wang CK, Craik DJ (2008) The anthelmintic activity of the cyclotides: natural variants with enhanced activity. ChemBioChem 9:1939–1945
Colgrave ML, Kotze AC, Kopp S, McCarthy JS, Coleman GT, Craik DJ (2009) Anthelmintic activity of cyclotides: in vitro studies with canine and human hookworms. Acta Trop 109:163–166
Gran L, Sletten K, Skjeldal L (2008) Cyclic peptides from Oldenlandia affinis DC: molecular and biological properties. Chem Biodivers 5:2014–2022
Kamimori H, Hall K, Craik DJ, Aguilar MI (2005) Studies on the membrane interactions of the cyclotides kalata B1 and kalata B6 on model membrane systems by surface plasmon resonance. Anal Biochem 337:149–153
Shenkarev ZO, Nadezhdin KD, Sobol VA, Sobol AG, Skjeldal L, Arseniev AS (2006) Conformation and mode of membrane interaction in cyclotides: spatial structure of kalata B1 bound to a dodecylphosphocholine micelle. FEBS J. 273:2658–2672
Shenkarev ZO, Nadezhdin KD, Lyukmanova EN, Sobol VA, Skjeldal L, Arseniev AS (2008) Divalent cation coordination and mode of membrane interaction in cyclotides: NMR spatial structure of ternary complex Kalata B7/Mn2+/DPC micelle. J Inorg Biochem 102:1246–1256
Wang CK, Colgrave ML, Ireland DC, Kaas Q, Craik DJ (2009) Despite a conserved cystine knot motif, different cyclotides have different membrane binding modes. Biochem J 97:1471–1481
Nourse A, Trabi M, Daly NL, Craik DJ (2004) A comparison of the self-association behavior of the plant cyclotides kalata B1 and kalata B2 via analytical ultracentrifugation. J Biol Chem 279:562–570
Simonsen SM, Sando L, Rosengren KJ, Wang CK, Colgrave ML, Daly NL, Craik DJ (2008) Alanine scanning mutagenesis of the prototypic cyclotide reveals a cluster of residues essential for bioactivity. J Biol Chem 283:9805–9813
Huang YH, Colgrave ML, Daly NL, Keleshian A, Martinac B, Craik DJ (2009) The biological activity of the prototypic cyclotide kalata B1 is modulated by the formation of multimeric pores. J Biol Chem 284:20699–20707
Svangard E, Burman R, Gunasekera S, Lovborg H, Gullbo J, Göransson U (2007) Mechanism of action of cytotoxic cyclotides: cycloviolacin O2 disrupts lipid membranes. J Nat Prod 70:643–647
Barry DG, Daly NL, Clark RJ, Sando L, Craik DJ (2003) Linearization of a naturally occurring circular protein maintains structure but eliminates hemolytic activity. Biochemistry 42:6688–6695
Daly NL, Craik DJ (2000) Acyclic permutants of naturally occurring cyclic proteins: characterization of cystine knot and beta-sheet formation in the macrocyclic polypeptide kalata B1. J Biol Chem 275:19068–19075
Herrmann A, Burman R, Mylne JS, Karlsson G, Gullbo J, Craik DJ, Clark RJ, Göransson U (2008) The alpine violet, Viola biflora, is a rich source of cyclotides with potent cytotoxicity. Phytochemistry 69:939–952
Mulvenna JP, Mylne JS, Bharathi R, Burton RA, Shirley NJ, Fincher GB, Anderson MA, Craik DJ (2006) Discovery of cyclotide-like protein sequences in graminaceous crop plants: ancestral precursors of circular proteins? Plant Cell 18:2134–2144
Basse CW (2005) Dissecting defense-related and developmental transcriptional responses of maize during Ustilago maydis infection and subsequent tumor formation. Plant Physiol 138:1774–1784
Rosengren KJ, Daly NL, Plan MR, Waine C, Craik DJ (2003) Twists, knots, and rings in proteins. Structural definition of the cyclotide framework. J Biol Chem 278:8606–8616
Herrmann A, Svangard E, Claeson P, Gullbo J, Bohlin L, Goransson U (2006) Key role of glutamic acid for the cytotoxic activity of the cyclotide cycloviolacin O2. Cell Mol Life Sci 63:235–245
Saska I, Craik DJ (2008) Protease-catalysed protein splicing: a new post-translational modification? Trends Biochem Sci 33:363–368
Daly NL, Love S, Alewood PF, Craik DJ (1999) Chemical synthesis and folding of large cyclic polypeptides: studies of the cystine knot polypeptide kalata B1. Biochemistry 38:10606–10614
Kent SB (2009) Total chemical synthesis of proteins. Chem Soc Rev 38:338–351
Gruber CW, Cemazar M, Clark RJ, Horibe T, Renda RF, Anderson MA, Craik DJ (2007) A novel plant protein-disulfide isomerase involved in the oxidative folding of cystine knot defense proteins. J Biol Chem 282:20435–20446
Andeme-Ondzighi C, Christopher DA, Cho EJ, Chang SC, Staehelin LA (2008) Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell 20:2205–2220
Gunasekera S, Daly NL, Anderson MA, Craik DJ (2006) Chemical synthesis and biosynthesis of the cyclotide family of circular proteins. IUBMB Life 58:515–524
Kimura RH, Tran A-T, Camarero JA (2006) Biosynthesis of the cyclotide kalata B1 by using protein splicing. Angew Chem Int Ed Engl 118:987–990
Camarero JA, Kimura RH, Woo Y-H, Shekhtman A, Cantor J (2007) Biosynthesis of a fully functional cyclotide inside living bacterial cells. ChemBioChem 8:1363–1366
Thongyoo P, Jaulent AM, Tate EW, Leatherbarrow RJ (2007) Immobilized protease-assisted synthesis of engineered cysteine-knot microproteins. ChemBioChem 8:1107–1109
Gunasekera S, Foley FM, Clark RJ, Sando L, Fabri LJ, Craik DJ, Daly NL (2008) Engineering stabilized vascular endothelial growth factor-A antagonists: synthesis, structural characterization, and bioactivity of grafted analogues of cyclotides. J Med Chem 51:7697–7704
Thongyoo P, Roque-Rosell N, Leatherbarrow RJ, Tate EW (2008) Chemical and biomimetic total syntheses of natural and engineered MCoTI cyclotides. Org Biomol Chem 6:1462–1470
Werle M, Kafedjiiski K, Kolmar H, Bernkop-Schnurch A (2007) Evaluation and improvement of the properties of the novel cystine-knot microprotein McoEeTI for oral administration. Int J Pharm 332:72–79
Reiss S, Sieber M, Oberle V, Wentzel A, Spangenberg P, Claus R, Kolmar H, Losche W (2006) Inhibition of platelet aggregation by grafting RGD and KGD sequences on the structural scaffold of small disulfide-rich proteins. Platelets 17:153–157
Werle M, Schmitz T, Huang HL, Wentzel A, Kolmar H, Bernkop-Schnurch A (2006) The potential of cystine-knot microproteins as novel pharmacophoric scaffolds in oral peptide drug delivery. J Drug Target 14:137–146
Seydel P, Dörnenburg H (2006) Establishment of in vitro plants, cell and tissue cultures from Oldenlandia affinis for the production of cyclic peptides. Plant Cell Tissue Organ Cult 85:247–255
Seydel P, Gruber CW, Craik DJ, Dörnenburg H (2007) Formation of cyclotides and variations in cyclotide expression in Oldenlandia affinis suspension cultures. Appl Microbiol Biotechnol 77:275–284
Dörnenburg H (2008) Plant cell culture technology-harnessing a biological approach for competitive cyclotides production. Biotechnol Lett 30:1311–1321
Hernandez JF, Gagnon J, Chiche L, Nguyen TM, Andrieu JP, Heitz A, Trinh Hong T, Pham TT, Le Nguyen D (2000) Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry 39:5722–5730
Dutton JL, Renda RF, Waine C, Clark RJ, Daly NL, Jennings CV, Anderson MA, Craik DJ (2004) Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J Biol Chem 279:46858–46867
Zhang J, Liao B, Craik DJ, Li J-T, Hu M, Shu W-S (2009) Identification of two suites of cyclotide precursor genes from metallophyte Viola baoshanensis: cDNA sequence variation, alternative RNA splicing and potential cyclotide diversity. Gene 431:23–32
Saska I, Colgrave ML, Jones A, Anderson MA, Craik DJ (2008) Quantitative analysis of backbone-cyclised peptides in plants. J Chromatogr B Analyt Technol Biomed Life Sci 872:107–114
Mylne JS, Craik DJ (2008) 15N cyclotides by whole plant labeling. Biopolym Pept Sci 90:575–580
Acknowledgments
Work in our laboratory on cyclotides is supported by grants from the Australian Research Council and the National Health and Medical Research Council (NHMRC). D.J.C. is a NHMRC Professorial Fellow, N.L.D. is a Queensland Smart State Fellow, J.S.M. is an ARC QEII Fellow. We thank colleagues from our laboratory and collaborators listed in the references for their valuable contributions to cyclotide research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Craik, D.J., Mylne, J.S. & Daly, N.L. Cyclotides: macrocyclic peptides with applications in drug design and agriculture. Cell. Mol. Life Sci. 67, 9–16 (2010). https://doi.org/10.1007/s00018-009-0159-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-009-0159-3