Abstract
Objectives
Allergic rhinitis (AR) is a chronic inflammatory disease of nasal mucosa. Loss of function of Th17 cells and regulatory T (Treg) cells plays a role in the pathogenesis of AR. IL18, FOXP3, and IL13 are key genes in the development of AR. However, the genetic associations between IL18, FOXP3 and IL13 genes polymorphisms and AR risk were inconclusive yet.
Methods
A meta-analysis was performed by searching through Pubmed, EMBASE, web of science and CNKI databases. The ORs and 95%CIs were used to assess the genetic association between the allelic, dominant and recessive models of IL18, FOXP3 and IL13 genes polymorphisms and AR risk.
Results
A total of 15 articles (6 for FOXP3, 5 for IL18, and 5 for IL13) were enrolled in the present study. No association was detected between the IL18 rs187238, rs1946518, rs360721, FOXP3 rs2232365, rs3761548 and IL13 rs1800925 polymorphisms and AR risk (p > 0.05). Significant associations were observed between the allelic (p = 0.001, OR 1.32, 95% CI 1.12–1.56), dominant (p = 0.005, OR 1.43, 95% CI 1.11–1.83) and recessive models (p = 0.01, OR 1.64, 95% CI 1.13, 2.40) of IL13 rs20541 and AR risk. Subgroup analysis based on ethnicity revealed that the IL13 rs20541 was significantly associated with AR risk in Asian population (allelic model: p = 0.009, OR 1.36, 95% CI 1.13–1.63, dominant model: p = 0.005, OR 1.43, 95% CI 1.11–1.83; recessive model: p = 0.01, OR 1.64, 95% CI 1.13–2.40).
Conclusions
IL13 rs20541 may contribute to the risk of AR in Asian population. To confirm these results, larger number of case–control study with more subjects is necessary in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Allergic rhinitis (AR) is a chronic inflammatory disease of nasal mucosa. Allergic individuals who are exposed to allergens may release IgE mediated transmitters and immunologically active cells and cytokines [1,2,3]. The incidence of AR worldwide is between 10 and 25% [4]. AR is influenced by both environmental factors and genetic factors [5, 6]. Previously, most studies have reported that the imbalance of Th1 and Th2 cell subsets plays a role in AR immune disorder [7, 8]. Researchers have been trying to reverse the response of naive CD4+ T cells to Th1 instead of Th2 [9, 10], but this does not fully elucidate the immune mechanism of AR. Recently, Th17 cells and regulatory T (Treg) cells were also shown to be involved in the pathogenesis of AR [11, 12]. And, the imbalance of CD4+ Th cell subsets may lead to the excessive Th2 differentiation in AR [12]. As far as current studies are concerned, polymorphic loci of relevant genes in the pathogenesis of AR are one of the important ways to determine the pathogenesis and treatment of AR.
Forkhead transcription factor P3 (forkhead-box P3, FOXP3) is a characteristic transcription factor of regulatory T cells, which plays an important role in the pathogenesis of AR through the control of regulatory T cells [13]. Zhang et al. found that the FOXP3 rs3761548 allele was associated with AR in a case–control study with 193 AR patients and 191 healthy controls in China [14]. Similar results were observed by Fodor et al. in a Hungarian population in 2011 [15]. However, negative results were also detected in other Chinese population by Zhang et al. [16].
In addition, as the main factor of Th2 cells, IL-13 could stimulate the growth and differentiation of B cells, promote the generation of IgE, and inhibit the activity of Thl cells [17]. Huebner et al. found that interleukin 13 (IL-13) rsl800925 was associated with rhinitis in a cohort study of 923 children in the United States [18]. Llanes et al. found that the distribution of the TT genotype of IL-13 rsl800925 significantly decreased, which suggested that the IL-13 rsl800925 was protective for rhinitis and asthma with olive pollen allergy in Spain [19]. However, no association was detected between the IL13 rsl800925 and AR in Chinese Han [20] and Dutch [21] populations.
Furthermore, IL-18 is a major member of the IL-1 family. It can promote mast cells, T cells and basophils to secrete Th2 cytokines such as IL-13 and IL-4, which can enhance Th2 cell-mediated immune response as well as Th1 cell-mediated immune response [22]. Sebelova et al. have investigated three polymorphisms (− 607 C/A, − 137 G/C and − 133 C/G) of the IL-18 gene in 539 AR patients and 312 healthy controls, and found no difference in frequencies of allele and genotype of the three polymorphisms [23]. While the results of the genetic associations between IL-18 − 607 C/A, − 137 G/C and − 133 C/G and AR susceptibility was not inconsistent in Thai [24], Egyptian [25], and Czech [26] populations.
Considering the inconclusive results of the associations between FOXP3, IL13 and IL18 genes polymorphisms and AR risk, we aimed to investigate a precise results by using a meta-analysis.
Methods
Searching strategy
Publications were searched through Pubmed, EMBASE, web of science and China National Knowledge Infrastructure (CNKI) databases up to January, 2020. The procedure followed the Cochrane collaboration definition and PRISMA 2009 guidelines for meta-analysis and systematic review. The keywords: “interleukin 18” or “IL18” and “interleukin 13” or “IL13” and “Forkhead Box Protein 3” or “FOXP3” and “polymorphism” or “variant” or “single nucleotide polymorphism” or “SNP” and “allergic rhinitis” or “AR” were used to retrieve the articles without language restriction. Furthermore, references of all relevant articles were retrieved to identify additional eligible studies.
Inclusion criteria and exclusion criteria
Eligible studies were included if they: (1) were case–control designed studies; (2) had available genotype frequencies in both case and control groups to estimate an odds ratios (OR) and their 95% confidence interval (CI).
Studies were excluded if they: (1) were duplicated data; (2) were case-reports, reviews or abstracts; (3) were lack of genotype frequency data.
Data extraction and quality assessment
The information from eligible studies that according to the inclusion/exclusion criteria were extracted by two independent authors (XQ and XJ). Any disagreement was resolved by discussion. The following information were extracted: Family name of the first author, year, ethnicity, mean ages, gender, genotyping-method, source of controls, number of cases and controls, number of genotypes. All included studies were evaluated using the Newcastle–Ottawa Scale (NOS) [27]. The NOS values arranged from 0 to 8. The studies were included if the NOS values ≥ 6.
Statistical analyses
Statistical analysis for the meta-analysis was conducted by Stata version 12.0 (Stata Corporation, College Station, TX, USA) and Revman 5.2 (Cochrane Collaboration). The ORs and 95% CIs were used to assess the genetic association between the allelic, dominant and recessive models of IL18 (rs187238, rs1946518 and rs360721), FOXP3 (rs2232365 and rs3761548) and IL13 (rs20541 and rs1800925) genes and risk of AR. The significance of the pooled OR was assessed by the Z test, and pZ < 0.05 was considered as statistically significant. The between study heterogeneity was assumed by chi-square-based Q-test and I2-statistic [(I2 = 100% (Q − df)/Q)). A p value > 0.05 or I2 < 50% for the Q test and I2-statistic indicated no heterogeneity among studies, and a fixed model was applied to estimate the pooled ORs. Otherwise the random model was used. Meta regression analysis was undertaken to explore potential sources of heterogeneity across studies when statistical heterogeneity was detected. The stability of the results was assessed using sensitivity analysis by excluding one study each time. Potential publication bias was undertaken by Egger’s test and Begg’ s tests. pEgger or pBegg < 0.05 was considered significant.
Results
Characters of eligible publications
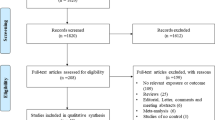
As shown in the Fig. 1, A total of 746 publications were originally obtained, among which 117 irrelevant papers were firstly excluded. Thus, 629 publications were eligible. Then, 122 study that was not case–control designed was eliminated. Finally, 16 publications that met the inclusion criteria were enrolled in the present study. For FOXP3 gene, 6 articles [14,15,16, 28,29,30] encompassing 979 cases and 980 controls were included. Among them, 5 were in Asian population [14, 16, 28,29,30], and 1 was in Caucasian population [15]. For IL13 gene, 5 qualified studies consisted of 879 cases and 684 controls were enrolled [20, 21, 31,32,33]. 4 were in Asian population [20, 31,32,33], and 1 was in Caucasian population [21]. For IL18 gene, 5 publications with 1507 cases and 878 controls were included [23,24,25,26, 34]. And 3 of them were in Caucasian population [23, 25, 26], and others two were in Asian population [24, 34]. The characteristics of these included papers were summarized in Table 1. The articles were published from 2006 to 2017. The NOS scores of all included studies ranged from 7 to 8 stars, indicating that the studies were of high methodological quality (Table s1).
Combined results
The results revealed the frequencies of the allelic, dominant and recessive models of IL18 (rs187238, rs1946518 and rs360721), FOXP3 (rs2232365 and rs3761548), and IL13 rs1800925 were not significantly different in cases and controls (p > 0.05) (Tables 2, 3, 4; Fig. 2, 3, 4). In addition, statistically significant association was found between the IL13 rs20541 and AR risk in the three genetic models (allelic model: p = 0.001, OR 1.32, 95% CI 1.12–1.56, dominant model: p = 0.005, OR 1.43, 95% CI 1.11–1.83; recessive model: p = 0.01, OR 1.64, 95% CI 1.13–2.40) (Table 2; Fig. 4).
Stratification analyses were conducted based on ethnicity. The results indicated that all the genetic models of IL13 rs20541 polymorphism were significantly associated with the increased risk of AR in Asian population (allelic model: p = 0.009, OR 1.36, 95% CI 1.13–1.63, dominant model: p = 0.005, OR 1.43, 95% CI 1.11–1.83; recessive model: p = 0.01, OR 1.64, 95% CI 1.13–2.40). For lack of data, we failed to demonstrate the association between IL13 rs20541 polymorphism and AR risk in Caucasian population. In addition, IL18 rs187238, rs1946518, and rs360721 polymorphisms were not associated with AR risk neither in Asian population nor in Caucasian population (p > 0.05). Furthermore, the FOXP3 rs3761548 polymorphism was found to be significantly associated with AR susceptibility in Caucasian population in three genetic models (allelic model: p = 0.0001, OR 0.45, 95% CI 0.30–0.68; dominant model: p = 0.001, OR 0.04, 95% CI 0.00–0.28; recessive model: p = 0.009, OR 0.44, 95% CI 0.24–0.82) (Tables 2, 3, 4).
Heterogeneity
Significant heterogeneity was found in the recessive model of IL13 rs1800925, the allelic and dominant models of IL18 rs1946518, and the allelic, dominant and recessive models of FOXP3 rs3761548 both in the overall group and Asian subgroup (Tables 2, 3, 4). For IL18 rs1946518, the significant heterogeneity across studies may mainly due to the study conducted by Tungtrongchitr et al. [24]. An I2 = 13% (p = 0.32) were obtained after excluded this study. For FOXP3 rs3761548, the significant heterogeneity across studies may mainly due to the study conducted by Hassannia et al. [29]. An I2 = 10% (p = 0.33) were obtained after excluded this study.
Sensitivity analysis and publication bias
The pooled ORs were not statistically altered when the fixed model changed into random model, and a study was omitted once at a time in each genetic model, which indicated the results of the meta-analysis were stable and trustworthy (Fig. 5). Both Egger’s and Begg’s tests were used to evaluate the publication bias of this meta-analysis. The results revealed that there was no obvious publication bias in overall analysis for IL18 (rs187238, rs1946518 and rs360721), FOXP3 (rs2232365 and rs3761548), and IL13 (rs20541 and rs1800925) polymorphisms (Fig. 6; Table s2).
Discussion
Allergic rhinitis was considered to be caused by dominant differentiation of Th2 cells and over-expression of Th2 cytokines such as IL4 and IL5, which are related to the abnormal balance between Th1 and Th2 cytokine networks [35]. The elevated level of serum total IgE is also an important factor in the pathogenesis of AR [36]. Number of studies have shown that Th2 cytokines including IL-4, IL-5, IL-9, and IL-13 were closely associated with allergic disease, of which IL-13 is currently considered to be one of the most important cytokines [38, 39]. IL-13 is a pluripotent cytokine secreted mainly by Th2 cells of CD40+ [40]. It can directly promote the secretion and improve the activity of B cells, and directly induce the synthesis of excessive IgE by B cells in patients with atopic constitution, thereby increasing the risk of AR [37, 41]. In addition, IL13 can induce T cells to differentiate into Th2 cells, support the expression of Th2 cytokines (IL-4, IL-5 and IL-6), and inhibit the production of IL-12 and INF-y in Th1 cells, thus affecting the occurrence and development of AR [42].
At present, several SNPs of IL13 gene have been found to be associated with the pathogenesis and phenotype of allergic diseases such as asthma [43], atopic dermatitis [44], and AR [31]. The human IL-13 gene is located in region 5q31 and is a highly polymorphic gene. The relative position of 1103C/T (rs20541) within the promoter sequence to the transcription start site is − 1112, and its polymorphism may affect the binding force of nuclear protein and DNA to change the transcription level of IL-13 [45]. Huebner M et al. [18] and Lianes E et al. [19] found that the frequency of TT genotype of the IL13 (rs1800925) significantly reduced in AR group. Wang et al. found the IL-13 rs20541, but not the IL-13 rs1800925 was related to AR susceptibility [20]. There are many possible reasons for the different association between the AR group and IL-13 in different populations. For example: (1) AR is a polygenic disease, and other genes may also be associated with susceptibility to AR; (2) differences in the selection of patients with AR; (3) the effect of IL-13 varies due to changes in dietary habits, geographical environment, human growth status and other environmental factors in different regions. In the previous studies, the results of the genetic associations between the IL13 rs20541 and AR risk was inconsistent in different population, which may due to the limited sample size in individual study. In our meta-analysis, we demonstrated that the IL13 rs20541 was significantly associated with the increased risk of AR in Asian population. For lack of data, we failed to came to the conclusion that IL13 rs20541 was associated with AR in Caucasian population. Thus, large samples in multiple ethnicity and case–control designed studies may shed light on the relationship between IL-13 rs20541 and AR risk.
FOXP3 is a member of the forkhead/winged-helix family of transcription factors [47]. FOXP3 dysfunction in humans can lead to severe systemic immune disorders, such as enteritis [48], autoimmune anemia [49], and type l diabetes [50], as well as severe allergic inflammatory disease [51]. Thus, the normal expression of FOXP3 gene is essential to maintain the function of CD4+CD25+Treg cells and the self-stabilizing state of the whole immune system. A study found that expression of FOXP3 protein decreased in CD4+CD25+Treg cells in asthma patients [52]. Similarly, decreased FOXP3 expression was found in nasal secretions of AR patients [13]. Researchers speculated that SNPs in FOXP3 may reduce the number or influence the function of CD4+CD25+ treg cells, and lead to immune tolerance disorders [53]. Zhang et al. found that the mutation of AC heterozygosity in FOXP3 rs3761548 was correlated with the incidence of AR, while FOXP3 rs3761547 was correlated with AR allergic to dust mites [16]. Our combined results indicated that the FOXP3 rs3761548 was not the susceptible factor for AR. However, because the literature and sample size included in this study were relatively small, the results may be biased to some extent, which still needs to be further studied in the future.
There were limitations in our combined analysis. Firstly, the sample size and the number of included studies were relatively small, especially in Caucasian population, which may partly influence the results of the genetic association between the genes polymorphisms and the risk of AR. To further identify the findings in the present study, more articles with larger number of subjects are necessary. Secondly, both the genetic and environmental factors were determined to play a role in the development of AR. However, we failed to detected the influence of environmental factors and AR risk for lack of sufficient data. Thirdly, some genetic models displayed high heterogeneity, although subgroup analysis was performed to detect the sources of this heterogeneity.
Conclusions
Our findings suggested that the IL13 rs20541 may contribute to the risk of AR in Asian population. To confirm this result, larger number of case–control designed studies with more subjects is necessary in the future.
References
Plaut M, Valentine MD. Allergic Rhinitis. N Engl J Med. 2005;353:1934–44.
Bernstein DI, Schwartz G, Bernstein JA. Allergic rhinitis: mechanisms and treatment. Immunol allergy Clin N Am. 2016;36(2):261–78.
May JR, Dolen WK. Management of allergic rhinitis: a review for the community pharmacist. Clin Ther. 2017;39(12):2410–9.
Sinha B, Vibha X, Singla R, Chowdhury R. Allergic rhinitis: a neglected disease—a community based assessment among adults in Delhi. J Postgrad Med. 2015;61(3):169–75.
Dunlop J, Matsui E, Sharma HP. Allergic rhinitis: environmental determinants. Immunol Allergy Clin N Am. 2016;36(2):367–77.
Lee JH, Koh SH. Genetic role in allergic rhinitis. J Rhinol. 2010;17(1):7–12.
Gu ZW, Wang YX, Cao ZW. Neutralization of interleukin-9 ameliorates symptoms of allergic rhinitis by reducing Th2, Th9, and Th17 responses and increasing the Treg response in a murine model. Oncotarget. 2017;8(9):14314–24.
Xiao C, Li H, Li H, Cheng Z, Qin J, Zhou W, et al. The effect of specific immunotherapy on the regulation of Th1/Th2 cell ratio of the patients with allergic rhinitis in serum. J Clin Otorhinolaryngol. 2010;24(20):924–7.
Yu SQ. Content of CD4+CD25+ regulatory T cells in peripheral blood lymphocytes in patients with allergic rhinitis. J Clin Otorhinolaryngol Head Neck Surg. 2011;25(8):354–9.
Larsson K, Lindstedt M, Lundberg K, Dexlin L, Wingren C, Korsgren M, et al. CD4+ T cells have a key instructive role in educating dendritic cells in allergy. Int Arch Allergy Immunol. 2009;149(1):1–15.
Liu Y, Zeng M, Liu Z. Clinical relevance of Th17 response in allergic rhinitis: more evidence. Clin Exp Allergy. 2015;45(12):1875.
Ciprandi G, Filaci G, Fenoglio D. Th17 cells and allergic rhinitis: is there clinical relevance? Otolaryngol Head Neck Surg. 2010;143(4):604–5.
Lee SM, Gao BX, Dahl M, Calhoun K, Fang D. Decreased FOXP3 gene expression in the nasal secretions from patients with allergic rhinitis. Otolaryngol Head Neck Surg. 2009;140(2):197–201.
Zhang L, Zhang Y, Desrosiers M, Wang C, Zhao Y, Han D. Genetic association study of FOXP3 polymorphisms in allergic rhinitis in a Chinese population. Human Immunol. 2009;70(11):934.
Fodor E, Garaczi E, Hilda P, et al. The rs3761548 polymorphism of FOXP3 is a protective genetic factor against allergic rhinitis in the Hungarian female population. Human Immunol. 2011;72(10):930–4.
Zhang Y, Duan S, Wei X, Zhao Y, Zhao L, Zhang L. Association between polymorphisms in FOXP3 and EBI3 genes and the risk for development of allergic rhinitis in Chinese subjects. Hum Immunol. 2012;73(9):939–45.
Miller AL. The etiologies, pathophysiology, and alternative/complementary treatment of asthma. Altern Med Rev. 2001;6(1):20–47.
Huebner M, Kim DY, Ewart S, Karmaus W, Sadeghnejad A, Arshad SH. Patterns of GATA3 and IL13 gene polymorphisms associated with childhood rhinitis and atopy in a birth cohort. J Allergy Clin Immunol. 2008;121(2):408–14.
Llanes E, Quiralte J, López E. Analysis of polymorphisms in olive pollen allergy: IL13, IL4RA, IL5 and ADRB2 genes. Int Arch Allergy Immunol. 2009;148(3):228–38.
Wang M, Xing ZM, Lu C, Ma YX, Yu DL, Yan Z. A common IL-13 Arg130Gln single nucleotide polymorphism among Chinese atopy patients with allergic rhinitis. Hum Genet. 2003;113(5):387–90.
Bottema RWB, Nolte IM, Howard TD. Interleukin 13 and interleukin 4 receptor-α polymorphisms in rhinitis and asthma. Int Arch Allergy Immunol. 2010;153(3):259–67.
Wong CK, Ho CY, Ko FW, Chan CH, Ho AS, Hui DS, et al. Proinflammatory cytokines (IL-17, IL-6, IL-18 and IL-12) and Th cytokines (IFN-gamma, IL-4, IL-10 and IL-13) in patients with allergic asthma. Clin Exp Immunol. 2001;125(2):177–83.
Sebelova S, Izakovicova-Holla L, Stejskalova A, Schüller M, Znojil V, Vasku A. Interleukin-18 and its three gene polymorphisms relating to allergic rhinitis. J Hum Genet. 2007;52(2):152–8.
Tungtrongchitr A, Jumpasri J, Sookrung N, Visitsunthorn N, Tantilipikorn P, Piboonpocanan O, et al. Alteration of -656(G/T) and -607(C/A) polymorphisms in interleukin-18 (IL-18) gene in house dust mite-sensitive allergic rhinitis patients in Thailand. Genet Mol Res. 2017;16:3.
Ibrahim GH, Eltabbakh MT, Gomaa AHA, Mohamed EA. Interleukin-18 gene polymorphisms in Egyptian patients with allergic diseases. Am J Rhinol Allergy. 2012;26(5):385–9.
Holla LI, Hrdliková B, Schüller M, Buckova D, Kindlova D, Izakovic V, et al. Haplotype analysis of the interleukin-18 gene in Czech patients with allergic disorders. Human Immunol. 2010;71(6):597.
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Euro J Epidemiol. 2010;25(9):603–5.
Li ZL, Li YG, Lu JP, Chen SQ. Study on FOXP3 -924 polymorphisms in patients with allergic rhinitis. Anat Res. 2014;36(2):124–7.
Hassannia H, Abediankenari S, Ghaffari J. FOXP3 and TGFβgene polymorphisms in allergic rhinitis. Iran J Immunol. 2011;8(4):218–25.
Song QL. Study on the relationship between FOXP3 gene polymorphism and allergic rhinitis. Int J Immunol. 2016;39(2):139–41.
Kim JJ, Min JY, Lee JH. Polymorphisms in the IL-13 and IL-4 receptor alpha genes and allergic rhinitis. Eur Arch Otorhinolaryngol. 2007;264(4):395–9.
Micheal S, Minhas K, Ishaque M, Ahmed F, Ahmed A. IL-4 gene polymorphisms and their association with atopic asthma and allergic rhinitis in Pakistani patients. J Investig Allergol Clin Immunol. 2013;23(2):107–11.
Li YG, Liu ZW, Lu JP. Investigation into the polymorphism of interleukin-13 1112 C%3eT in allergic rhinitis patients. China Trop Med. 2012;12(1):67–9.
Lee HM, Park SA, Chung SW, Woo JS, Chae SW, Lee SH, et al. Interleukin-18/-607 gene polymorphism in allergic rhinitis. Int J Pediatr Otorhinolaryngol. 2006;70(6):1085–8.
KleinJan A, Willart M, Van Nimwegen M, Leman K, Hoogsteden HC, Hendriks RW, et al. United airways: circulating Th2 effector cells in an allergic rhinitis model are responsible for promoting lower airways inflammation. Clin Exp Allergy. 2010;40(3):494–504.
Ciprandi G. Symptom severity and allergen-specific IgE in allergic rhinitis. Iran J Allergy Asthma Immunol. 2017;16(1):79–81.
Devos FC, Pollaris L, Cremer J, Seys S, Hoshino T, Ceuppens J, et al. IL-13 is a central mediator of chemical-induced airway hyperreactivity in mice. PLoS ONE. 2017;12(7):e0180690.
Barlow JL, Bellosi A, Hardman CS, Drynan LF, Wong SH, Cruickshank JP, et al. Innate IL-13-producing nuocytes arise during allergic lung inflammation and contribute to airways hyperreactivity. J Allergy Clin Immunol. 2011;129(1):191.e1–4–8.e1–4.
Yosri H, Elkashef WF, Said E, Gameil NM. Crocin modulates IL-4/IL-13 signaling and ameliorates experimentally induced allergic airway asthma in a murine model. Int Immunopharmacol. 2017;50:305–12.
Guo LY, Li JH, Paul WE. Probabilistic regulation in TH2 Cells accounts for monoallelic expression of IL-4 and IL-13. Immunity. 2005;23(1):89–99.
Defrance T. Interleukin 13 is a B cell stimulating factor. J Exp Med. 1994;179(1):135–43.
Ngoc PL, Ngoc LP, Gold DR, Tzianabos AO, Weiss ST, Celedón JC. Cytokines, allergy, and asthma. Curr Opin Allergy Clin Immunol. 2005;5(2):161–6.
Alasandagutti ML, Ansari MSS, Sagurthi SR, Valluri V, Gaddam S. Role of IL-13 genetic variants in signalling of asthma. Inflammation. 2017;40(2):1–12.
Gleń J, Trzeciak M, Sobjanek M, Bandurski T, Wilkowska A, Nedoszytko B, et al. Interleukin-13 promoter gene polymorphism -1112 C/T is associated with Atopic dermatitis in Polish patients. Acta Dermatovenerol Croat. 2012;20(4):231–8.
Hummelshoj T, Bodtger U, Datta P, Malling HJ, Oturai A, Poulsen LK, et al. Association between an interleukin-13 promoter polymorphism and atopy. Int J Immunogenet. 2003;30(5):355–9.
Ying XJ, Zhao SW, Wang GL, Tzianabos AO, Weiss ST, Celedón JC. Association of interleukin-13 SNP rs20541 with allergic rhinitis risk: a meta-analysis. Gene. 2013;521(2):222–6.
Ziegler SF. FOXP3: of mice and men. Annu Rev Immunol. 2006;24(1):209–26.
Wang K, Zhu TJ, Wang HJ, Yang JX, Du SS, Dong GY, et al. Adoptive transfers of CD4+ CD25+ Tregs raise FOXP3 expression and alleviate mouse enteritis. Biomed Res Int. 2018;2018(2):1–9.
Solomou EE, Rezvani K, Mielke S, Malide D, Visconte V, Keyvanfer K, et al. FOXP3-positive regulatory T-cells in acquired aplastic anemia. Blood. 2006;108(11):2248.
Cui G, Zhang Y, Gong Z, Zhang JZ, Zang YQ. Induction of CD4+CD25+FOXP3+ regulatory T cell response by glatiramer acetate in type 1 diabetes. Cell Res. 2009;19(5):574–83.
Choi JM, Shin JH, Sohn MH. Cell-permeable FOXP3 protein alleviates autoimmune disease associated with inflammatory bowel disease and allergic airway inflammation. Proc Natl Acad Sci USA. 2010;107(50):21943–21943.
Lee JH, Yu HH, Wang LC, Yang YH, Lin YT, Chiang BL. The levels of CD4+CD25+ regulatory T cells in paediatric patients with allergic rhinitis and bronchial asthma. Clin Exp Immunol. 2007;148(1):53–63.
Ban Y, Tozaki T, Tobe T, Ban Y, Jacobson EM, Concepcion ES, et al. The regulatory T cell gene FOXP3 and genetic susceptibility to thyroid autoimmunity: an association analysis in Caucasian and Japanese cohorts. J Autoimmun. 2007;28(4):201–7.
Acknowledgements
The present study was funded by the National Natural Science Foundation of China (Grant nos. 81873780, 61702054); Hunan Natural Science Foundation Youth Program (2019JJ50697, 2018JJ3568); The Changsha Outstanding Innovative Young People Training Scheme (kq1905047, kq1905045); The Foundation of the Education Department of Hunan Province (16A027, 19A058); The Foundation of Health and Family Planning Commission of Hunan Province (20201918); The Application Characteristic Discipline of Hunan Province; The Hunan Key Laboratory Cultivation Base of the Research and Development of Novel Pharmaceutical Preparations (No. 2016TP1029); The clinical research center of neurodegenerative diseases in Hunan province (2018SK4002); The Hunan Provincial Innovation Platform and Talents Program (No. 2018RS3105); The Hunan provincial science and technology department and Hunan provincial health and family planning commission (Grant No.[2018]85); The Natural science foundation of Hunan province (Grant No.[2017]1); The Key project of Hunan provincial commission of health and family planning (Grant No.[2017]144); The Hunan province science and technology major project (Grant No.[2016]158); Hunan Provincial Science and Technology Department Clinical Medical Technology Innovation Guide Project(2018SK51711).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Additional information
Responsible Editor: John Di Battista.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tang, L., Chen, Y., Xiang, Q. et al. The association between IL18, FOXP3 and IL13 genes polymorphisms and risk of allergic rhinitis: a meta-analysis. Inflamm. Res. 69, 911–923 (2020). https://doi.org/10.1007/s00011-020-01368-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-020-01368-4