Abstract
Objective
This study aimed to test the expression and biological function of miR-140-5p in osteoarthritis (OA), and identify its target gene and explore its mechanism in OA.
Methods
Differential genes were screened and analyzed by gene microarray and WGCNA analysis. The normal human chondrocytes C28/I2 were induced by IL-1β to construct the OA cell model. The expression of miR-140-5p and high mobility group box 1 (HMGB1) was quantified by quantitative real-time PCR (qRT-PCR) in OA tissues and IL-1β-induced chondrocytes. Western blotting was performed to evaluate the expression of HMGB1 and PI3K/AKT pathway activation. The concentrations of tumor necrosis factor (TNF)-α, interleukin (IL)-6, MMP-1 and MMP-3 were determined by ELISA. CCK-8 and flow cytometry were conducted to determine the cellular capabilities of proliferation and cell apoptosis.
Results
Bioinformatics analysis demonstrated that HMGB1 was highly expressed in OA and activated PI3K/AKT pathway. Also, HMGB1 was predicted as a target of miR-140-5p. The levels of miR-140-5p were negatively correlated with HMGB1 in OA tissues and IL-1β-induced chondrocytes. The overexpression of miR-140-5p reduced the expression of HMGB1 protein, p-AKT (Ser473) and p-PI3K in IL-1β-induced chondrocytes. Besides, the expression of p-AKT (Ser473) and p-PI3K was significantly upregulated by employing miR-140-5p inhibitor, but retrieved after treating with LY294002. Furthermore, miR-140-5p inhibited inflammation, matrix metalloprotease expression and apoptosis in IL-1β-induced chondrocytes through regulating HMGB1.
Conclusion
MiR-140-5p was down-regulated while HMGB1 was upregulated in OA. MiR-140-5p could inhibit the PI3K/AKT signaling pathway and suppress the progression of OA through targeting HMGB1.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Osteoarthritis (OA), the most common form of arthritis, is a disease that associated with age-related loss of the cartilage destruction, subchondral bone remodeling and inflammation of the synovial membrane [1]. Chondrocyte death reduced cartilage cellularity in OA, and remaining chondrocytes are activated by growth factors and cytokines to a catabolic and abnormal differentiation that leads to degradation of extracellular matrix (ECM). Matrix metalloproteinases (MMPs) members appear to play key roles in degradation of ECM. MMPs can degrade a variety of components of the ECM, including the fibrillar collagens and cartilage [2]. Compelling evidences has revealed that MMPs play an important role in IL-1β-stimulated human chondrocytes [2]. The high expression of MMP-1, MMP-2, and MMP-9 protein levels has been demonstrated in osteoarthritis [3]. Overexpression of HMGB1 inhibitor reduced IL-1β-induced MMP expression (MMP-1, MMP-3, MMP-9) and the production of inflammatory mediators in human chondrocytes [4]. Over the years, it has become evident that the inflammatory cytokines contribute substantially to the pathogenesis of OA [5]. Increased expression of proinflammatory cytokines (e.g., IL-1β, TNF-α and IL-6) in cartilage, synovial membrane and subchondral bone are linked with the development and progression of structural changes in the OA joint [5]. A number of studies suggested that IL-1β and TNF-α downregulate the synthesis of major ECM components by suppressing anabolic activities of chondrocytes [5]. Besides, highly expressed proinflammatory cytokines in OA chondrocytes can amplify and perpetuate the OA disease process through c-Jun/p38/NF-κB signaling pathway by inducing the production of proinflammatory cytokines such as IL-6 and chemokines [6].
MicroRNAs (miRNAs) are endogenous non-coding RNAs with less than 25 nucleotides [7]. They usually negatively regulate their target genes expression by binding to the 3′-untranslated region (3′-UTR) of target mRNAs [7]. Recent studies have underlined the key role of miRNAs in regulating chondrocyte functions [8]. The comparison between OA and normal cartilage specimens showed different miRNAs expression profiles, highlighting their involvement in the pathogenesis of OA [9]. MiR-140-5p has been considered as a regulator of chondrocyte differentiation, bone development and cartilage homeostasis. It was demonstrated that a reduced expression of miR-140-5p in OA cartilage influenced the expression levels of its target genes MMP-13, ADAMTS-5 and IGFBP-5 [10]. Proctor et al. [11] showed that the down-regulation of miR-140-5p may have either detrimental or protective effects on cartilage, indicating that the role of miR-140-5p is complex. Yin et al. [12] found that miR-140-5p was down-regulated in patients with OA. Tao et al. [13] also suggested that the overexpression of miR-140-5p could enhance cartilage tissue regeneration and prevent OA. Accordingly, miR-140-5p is very likely to have a negative influence in OA joint deterioration.
High mobility group box 1 (HMGB1) is a ubiquitous non-histone DNA-binding protein and plays an important role in inflammation [14]. HMGB1 can activate various inflammatory signaling pathways to produce proinflammatory cytokines and matrix metalloproteinases [14]. High expression of HMGB1 is found in the synovium [15], osteochondral fragments [16], cartilage chondrocytes [17], and the synovial fluid, and it is positively associated with the severity of OA [18]. Though some basic researches have shown that HMGB1 plays important roles in OA, the deeper and newer insights into mechanism of how HMGB1 involves in OA development needs to be further elucidated.
The PI3K/AKT pathway was well recognized as a fundamental intracellular signaling pathway in many diseases [19]. Previous studies revealed that it was involved in the degenerative alterations of articular cartilage of OA, including the degradation of extracellular matrix and the apoptosis of chondrocytes [20]. PRMT5 was found to be able to regulate the production of inflammatory factors and the proliferation of IL-1β-induced chondrocytes through mediating the AKT pathway [21]. The PI3K/AKT pathway was also regulated by HMGB1 in some human diseases, such as prostate cancer [22] and hepatocellular carcinoma [23]. However, the correlation and function of PI3K/AKT pathway and HMGB1 in chondrocyte function is poorly understood.
In this study, we determined the expression levels of miR-140-5p and HMGB1 in OA tissues and IL-1β-induced chondrocytes and investigated their influences on OA development. MiR-140-5p promoted the viability of IL-1β-induced chondrocytes and reduced the synthesis of proinflammatory factor and matrix metalloproteinases through targeting HMGB1. It also modulated the PI3K/AKT pathway in chondrocytes. The results implied that miR-140-5p played a suppressive role in OA, and it might be a potential therapeutic target for this disease.
Materials and methods
Clinical samples
Osteoarthritis articular cartilage samples were obtained from OA patients undergoing total knee arthroplasty (TKA) at the Department of Orthopedics of the Peking Union Medical College Hospital between July 2015 and August 2016 (n = 12, 7 females and 5 males, 68.2 ± 5.6 years). At the time of surgery, the patients had symptomatic disease requiring medical treatment in the form of non-steroidal anti-inflammatory drugs. None had received intra-articular steroid injections within 3 months prior to surgery. While normal articular cartilage samples were obtained from patients who have experienced surgery for fracture with no known history of OA or RA (n = 12, 6 females and 6 males, 26.7 ± 4.3 years). This study had obtained approval from the Institute Review Ethics (IRE) committee of the Peking Union Medical College Hospital and written informed consents from all participants. The articular cartilages of tissue were collected and stored at − 80 °C for further analysis.
Data collection
The genome-wide transcriptomic data sets (Agilent microarrays, GSE117999) from 10 healthy and 10 OA patients for analysis in this study. The data sets were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The differentially expressed genes (DEGs) analysis was performed using “Limma” package in R software with |log2 (fold change)| > 1 and adjust p < 0.05. Next, pairs of DEGs were analyzed by Pearson correlation matrix. Moreover, signal pathway enrichment was determined by KEGG analysis.
OA chondrocytes induced by IL-1β
The normal human chondrocytes C28/I2 purchased from BeNa Culture Collection were induced by IL-1β to develop into IL-1β-induced chondrocytes. C28/I2 cells were maintained in DMEM containing 10% fetal bovine serum (FBS, Gibco, USA) for 24 h at 37 °C in a humidified atmosphere of 5% CO2 and 95% air, and the medium was changed every 2 days. The chondrocytes were plated to 6-well plates and cultured for 24 h. Then, these cells were stimulated with IL-1β (10 ng/mL, Peprotech, US) to produce a cellular OA model [24].
qRT-PCR assay
Total RNA was extracted using the RNeasy Mini kit (Qiagen, Hilden, Germany) and the RNA quantitative analysis was performed using NanoDrop 2000 (Thermo Fisher Scientific. Waltham, MA, USA). 1 µg of total RNA was then reversely transcribed into cDNA by means of the ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan) in line with the manufacturer’s instructions. PCR was performed on MiniOpticon™ qRT-PCR system (Bio-Rad, Hercules, CA, USA) and amplified detection was conducted using SYBR-Green RealMastcrMix (Bio-Rad). For the quantification of the relative expression of each mRNA and miRNA, the threshold cycle (Ct) values were normalized against the endogenous references β-actin and U6. The 2−ΔΔCT method was used. The primer sequences are presented in Table 1.
Cell transfection
The IL-1β-induced chondrocytes were transfected with miR-140-5p mimics, miR-140-5p inhibitor (Applied Biosystems) using Lipofectamine 3000 (Invitrogen, Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s instructions. Nonspecific miR (miR-Control, Dharmacon) was used as a miRNA negative control (NC mimics). For the rescue experiment, pDNA3.1-HMGB1 and small interfering RNA (siRNA) of HMGB1 (siRNA-HMGB1) were synthesized by GenePharma Co. (Shanghai, China). The primer of siRNA-HMGB1 was 5′-GTTGGTTCTAGCGCAGTTT-3′ and the sequence of the siRNA of control was 5′-GCAAACCGTGTATCAGATA-3′.
The IL-1β-induced chondrocytes in logarithmic growth phase were collected 24 h before transfection, digested in trypsin and maintained in medium for re-suspension. After incubation for 24 h in the 6-well plates at a density of 1 × 106 per well, the cell confluence arrived 80–90%. According to the manufacturer’s protocol, all cell transfections were conducted using Lipofectamine 3000 Reagent (Life Technologies Corporation, Carlsbad, CA, USA). The medium was replaced with complete medium after 6-h transfection.
Cell viability assay
Cell viability was determined by Cell Counting Kit-8 assay. After being stimulated by the chondrocyte by IL-1β, the CCK8 solution (10 μl/well) was added to the culture medium. Then, after incubating for another1 h at 37 °C in humidified 95% air and 5% CO2, the optical density (450 nm) was recorded.
Apoptosis assay
To evaluate apoptosis in chondrocytes after IL-1β treatment, we performed flow cytometry analysis with Annexin V-FITC/PI (Beijing Biosea Biotechnology, Beijing, China) staining. In brief, IL-1β-treated chondrocytes (1 × 105 cells/well) were cultured in 6-well plate, then cells were treated for miR-140-5p mimics, inhibitor or pcDNA3.1-HMGB1. After 24 h incubation, cells were washed three times with cold PBS and resuspended in 100 μl binding buffer including 10 μl FITC-Annexin V in the presence of 50 μg/ml RNase A (Sigma-Aldrich, St. Louis, MO, USA), then incubated at room temperature for 15 min in the dark. The samples were then analyzed by flow cytometer (Beckman Coulter, Fullerton, USA) for apoptotic analysis.
Enzyme-linked immunosorbent assay (ELISA)
Culture supernatant was collected and the concentrations of MMP-1, MMP-3, TNF-α and IL-6 were measured by ELISA (R&D Systems, Abingdon, UK) following the manufacturer’s instruction, and were normalized to cell protein concentrations.
Dual-luciferase activity assay
The sequence of HMGB1 3′-UTR containing the predicted miR-140-5p binding site was cloned into pGL2-Basic Vector (E1641, Promega, WI, USA) as the reporter vector HMGB1-wild-type (HMGB1-wt). Mutated HMGB1 3′-UTR was used as a negative control, namely HMGB1-mutated-type (HMGB1-mt). The HMGB1-wt or HMGB1-mut was co-transfected with miR-140-5p mimics or NC mimics into HEK 293T cells (BeNa Culture Collection) by Lipofectamine-mediated gene transfer. The cells were collected 48 h later, and the Firefly and Renilla luciferase activity were measured by using the dual-luciferase reporter system (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Western blot assay
The IL-1β-induced chondrocytes in culture were lysed using the RIPA buffer (Pierce, Rockford, IL, USA) in the presence of a Protease Inhibitor Cocktail (Pierce). Total protein was separated by SDS-PAGE electrophoresis using the Novex NuPAGE system (Invitrogen) and transferred to 0.45 µm PVDF membranes. Membranes were blocked in TBS-T buffer containing 5% nonfat milk for 1 h and incubated overnight at 4 °C with the following primary antibodies: HMGB1 (#ab18256, Abcam, 1:1000), anti-PI3K (#ab151549, 1:1000), anti-PI3K (phospho) (#ab182651, 1:1000), anti-pan-AKT (#ab8805, 1:500), anti-pan-AKT (phospho Ser473) (#4060, Cell signaling, MA, USA, 1:1000), β-actin (#4967, Cell signaling, MA, USA, 1:1000) was the internal control. Then the membranes were washed using TBST and hybridized with the horseradish peroxidase (HRP)-linked antibody goat anti-rabbit IgG (1:2000, Abcam) for 1 h. Signal detection was carried out with an ECL system (Amersham Pharmacia, Piscataway, NJ, USA). The intensity of the bands was quantified using Image Lab™ Software (Bio-Rad, Shanghai, China). The experiment was performed at least in triplicate.
Statistical analysis
Statistical analysis was conducted by means of SPSS 20.0 software (IBM, CA, USA) with all data presented as mean ± standard deviation (SD). All statistical comparisons were performed using unpaired Student’s t test and one-way ANOVA. p < 0.05 was deemed significant difference.
Results
Microarray analysis-identified HMGB1 was upregulated in osteoarthritis chondrocytes
GSE117999 (genome-wide transcriptomic data sets from 10 healthy controls (normal) as well as 10 osteoarthritis (OA)) downloaded from GEO datasets was chosen for analysis in this study. Figure 1a presents the top ten different mRNA between the healthy and OA in the articular cartilage at the screening condition |fold change| > 1 and adjust p value < 0.05. HMGB1 was significantly differential expression was showed between OA and healthy control groups. Furthermore, DEGs were subjected to KEGG enrichment analysis. As shown in Fig. 1b, we observe that PI3K–AKT signal pathway was activated in osteoarthritis. We used the miRNA binding site prediction programs Probability of Interaction by Target Accessibility (PITA) and miRanda to identify candidate HMGB1 targeted miRNAs. In addition, miRNA target pathway prediction program KEGG-miRNA revealed PI3K/AKT signaling pathway associated miRNAs, thus, screen out five miRNAs (Fig. 1c).
Microarray analysis reveals DEG in OA articular cartilage samples from normal patients (healthy) or patients with osteoarthritis. a Heatmaps depict expression profiles of top 10 osteoarthritis-specific mRNA modules’ member genes. b GSEAplot revealed running enrichment score of PI3K–AKT signaling pathway in human osteoarthritis tissues was more than 0, indicating PI3K–AKT signaling pathway was activated. c MiRNAs that targeted HMGB1 were gathered by Venn diagram
The expression of miR-140-5p was inverse correlated with HMGB1 in OA cartilage tissues and IL-1β-induced chondrocytes
To examine the expression of miRNAs in the articular cartilage, qRT-PCR was performed in samples from 12 OA patients and 12 healthy controls. We found that the expression of miR-140-5p was significantly downregulated in the OA group compared with the healthy group. Moreover, the levels of miR-140-5p were negatively correlated with HMGB1 mRNA in OA cartilage tissues (Fig. 2a–g). Similarly, the expression of miR-140-5p and HMGB1 resembled IL-1β-induced chondrocytes compared with OA cartilage tissues (Fig. 2h, i).
The differential expression of miRNAs and HMGB1 in OA cartilage tissues and IL-1β-induced human chondrocytes. a–e The levels of miR190b, miR-543, miR-421, miR-338-3p and miR-140-5p in OA cartilage tissues compared with healthy cartilage tissues by qRT-PCR (*p < 0.05) vs. normal). f, g HMGB1 expression in OA cartilage tissues and normal cartilage tissues was determined in qRT-PCR and the level of HMGB1 was negatively correlated with miR-140-5p (*p < 0.05 vs. normal). h, i The levels of miR-140-5p and HMGB1 mRNA in chondrocytes treated with IL-1β. PBS treatment was used as control (*p < 0.05 vs. control)
The efficiency of miR-140-5p and HMGB1 gene knockdown and overexpression
To investigate the biological function of miR-140-5p and HMGB1, we adopted loss-of-function and gain-of-function strategies. MiR-140-5p expression was significantly decreased in IL-1β-induced chondrocytes upon incubation with miR-140-5p inhibitors when compared to NC mimics (Fig. 3a). Besides, HMGB1 mRNA expression was significantly decreased in IL-1β-induced chondrocytes upon incubation with siRNA when compared to non-targeting siRNA control (Fig. 3b). This result strongly indicates miR-140-5p and HMGB1 were successfully knocked down and overexpressed in a sequence-specific manner.
The overexpression and knockdown efficiency of miR-140-5p and HMGB1. a miR-140-5p mimics and inhibitors were transfected into IL-1β-induced chondrocytes to achieve miR-140-5p overexpression and inhibition. The expression efficiency was verified by using qRT-PCR assays. b Si-HMGB1 or pcDNA-HMGB1 vector were transfected into chondrocytes to achieve HMGB1 overexpression or knockdown. The expression efficiency was verified by RT-qPCR. The data are presented as mean ± SD of three independent experiments (*p < 0.05)
HMGB1 is direct target of miR-140-5p
To explore the underlying molecular mechanism of miR-140-5p regulative role in IL-1β-induced chondrocytes, the prediction of target bioinformatics was employed to analyze the miR-140-5p target. Bioinformatics prediction showed that HMGB1 may be a potential target of miR-140-5p based on the assumed target sequences of the HMGB1 3′-UTR (Fig. 4a). We further performed luciferase assay to determine whether miR-140-5p could target 3′-UTR of HMGB1 directly. The results displayed that miR-140-5p significantly attenuated the luciferase activity of the HMGB1-wt 3′-UTR (*p < 0.05) (Fig. 4b). To summarize, HMGB1 was a targeted gene of miR-140-5p.
HMGB1 was a direct target of miR-140-5p. a The binding sites between miR-140-5p and the 3′UTR region of HMGB1 predicted by PITA/miRanda, b The relative luciferase activity of HMGB1-WT co-transfected with miR-140-5p mimics was decreased, while that of HMGB1-MUT-co-transfected miR-140-5p mimics had no significant change (*p < 0.05 vs. NC mimics)
MIR-140-5p can modulate PI3K/AKT signaling pathway by targeting HMGB1
Previous study have demonstrated that PI3K/AKT was downstream signaling pathway of HMGB1 [22]. Here, we validated whether miR-140-5p/HMGB1 axis participate in PI3K/AKT pathway. By using Western blot assays, we observed that the protein levels of HMGB1, p-PI3K and p-AKT (Ser473) were significantly down-regulated by miR-140-5p mimics. To verify the phosphorylation activation of AKT and PI3K, we used a specific inhibitor of PI3K/AKT pathway: LY294002 [22]. As shown in Fig. 5, LY294002 inhibited AKT and PI3K phosphorylation and miR-140-5p inhibitor combined with LY294002 retrieved AKT and PI3K phosphorylation in IL-1β-induced chondrocytes under our experimental conditions.
MIR-140 suppressed PI3K/AKT signaling pathways activation by binding its target of HMGB1. a The expression of HMGB1, AKT, p-AKT (Ser473), PI3K and p-PI3K in IL-1β-induced human chondrocytes were detected by Western blot. b The relative protein quantification of HMGB1, AKT, p-AKT (Ser473), PI3K and p-PI3K. β-actin was used as the internal control. The quantitation of protein expression was done by densitometry analysis of the protein bands from three independent experiments. Values were normalized against β-actin, and then compared to the control group (*p < 0.05 vs. control; #p < 0.05 vs. IL-1β; ap < 0.05 vs. LY294002)
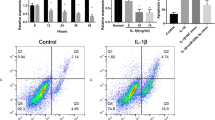
MIR-140-5p inhibit inflammation, matrix metalloprotease expression and apoptosis in IL-1β-induced chondrocytes through regulating HMGB1
To investigate the detailed functions of miR-140-5p/HMGB1 axis in IL-1β-induced chondrocytes, we evaluated the effects of miR-140-5p/HMGB1 axis in IL-1β-induced chondrocytes in vitro. The expression of MMP-1, MMP-3, TNF-α and IL-6 were inhibited after miR-140-5p mimics, while elevated in pcDNA-HMGB1. The co-transfection of pcDNA-HMGB1 and miR-140-5p mimics retrieved the matrix metalloprotease and proinflammatory cytokines expression to the similar level as control in IL-1β-induced chondrocytes (Fig. 6a–d). The chondrocytes viability was inhibited while the cell apoptosis was increased by HMGB1 overexpression, whereas transfected with miR-140-5p mimics showed the favorable influences that cell viability was increased and cell apoptosis was decreased (*p < 0.05) (Fig. 6e, f). Therefore, miR-140-5p overexpression promoted the proliferation and decreased the release of proinflammatory cytokines and matrix metalloprotease in IL-1β-induced chondrocytes.
Effects of miR-140-5p on IL-1β-induced human chondrocytes. With or without IL-1β-induced chondrocytes were transfected NC/miR-140-5p mimics or pcDNA/pcDNA-HMGB1 for 48 h, the level of IL-6 (a), TNF-α (b), MMP-1 (c) and MMP-3 (d) were determined by ELISA. The data are presented as mean ± SD of three independent experiments (*p < 0.05). e, f The overexpression of miR-140-5p promoted chondrocyte proliferation and inhibited apoptosis by targeting HMGB1. The data are presented as mean ± SD of three independent experiments (*p < 0.05)
Discussion
Osteoarthritis (OA), as the most frequent or prevalent chronic joint disease, is characterized by cartilage degradation and musculoskeletal pain, contributing to functional disability and loss of autonomy in the elderly [25]. This study mainly revealed that miR-140-5p could inhibit OA by targeting HMGB1 through PI3K/AKT signaling pathway. MiR-140-5p was down-regulated and restrained the growth in IL-1β-induced chondrocytes, while its target gene HMGB1 displayed an opposite role. The PI3K/AKT pathway was inhibited by miR-140-5p mimics, but activated by miR-140 inhibitor.
MiR-140 had been found to be involved in the development of OA. It has been reported that miR-140 was detected in synovial fluid of both normal knees and OA knees, and its relative expression level was reduced in synovial fluid of OA group compared with normal group [26]. There was significant difference in the expression level of miR-140 between the grade IV subgroup of OA and the grade I–III subgroups as miR-140 expression was negatively related to the severity of OA [27]. The suppressive effect of miR-140 on OA was also revealed by other researchers, which was consistent with our findings. A combined use of miR-140 and miR-29a successfully reversed the destructive effect of IL-1β on the proliferation of chondrocyte [28]. After estrogen treatment, the expression of miR-140 in OA increased, exhibiting an inhibitory effect on IL-1β-induced cartilage matrix degradation [29]. In brief, miR-140 was proved to the suppressor of OA and facilitated its improvement.
HMGB1 can bind to the cell surface receptors (e.g., receptor for advanced glycation end products and Toll-like receptor-2/4/9) leading to the excessive production of IL-1β, IL-6, and TNF-α by activating the NF-kB signaling pathway [30, 31]. OA can suppress the synthesis of proteoglycans and collagens and by enhancing the production of MMPs [32]. Recently, the role of HMGB1 in OA has been highlighted. Stimulation with IL-1β markedly promoted the translocation of HMGB1 from the nucleus to the cytoplasm and increased HMGB1 expression in OA chondrocytes in vitro [33]. Furthermore, HMGB1 could upregulate an increased inflammatory phenotype to induce OA synovial fibroblasts transformed into rheumatoid arthritis synovial fibroblast-like phenotype [34]. These findings supported that HMGB1 participate the pathogenic role in the pathogenesis of OA. The inhibition of HMGB1 shows effective anti-inflammatory effect [35]. Therefore, targeting HMGB1 may provide novel strategies for the treatment of OA.
Nevertheless, limitations in this report are to be taken into consideration. For example, the number of our samples was so small that more samples are needed for further study. In addition, since OA is difficult to cure at present, although our study on OA molecular mechanism allowed us to better understand it, more clinical therapy researches are essential and urgent in future.
In this study, we found the PI3K/AKT signaling pathway was regulated by miR-140-5p. It was activated when the miR-140-5p was down-regulated and vice versa. The PI3K/AKT signaling pathway had been demonstrated to involve in the degradation of extracellular matrix and the death of chondrocytes in OA [20]. The activation of PI3K/AKT signaling pathway could aggravate cartilage degeneration in OA [36]. The Western blot results from our study displayed that miR-140-5p can target HMGB1 to have influence on PI3K/AKT signaling pathway and downstream function of apoptosis, inflammation and matrix degradation. Nevertheless, limitations in this report are to be taken into consideration. For example, our sample number was so small that more samples are needed for further study. In addition, since OA is difficult to cure at present and although our study on OA molecular mechanism allowed us to better understand it, more clinical therapy researches are essential and urgent in future.
In conclusion, miR-140-5p was down-regulated in OA. It prevented the course of OA through reducing apoptosis, increasing proliferation and alleviating inflammation and degradation of ECM. In contrast, its target gene HMGB1 was upregulated, with adverse effect on OA. Down-regulation of miR-140-5p in OA activated the PI3K/AKT signaling pathway through targeting HMGB1. Our current finding suggested miR-140-5p as a potent strategy for miRNA-based OA treatment.
References
Glyn-Jones S, Palmer AJ, Agricola R, Price AJ, Vincent TL, Weinans H, et al. Osteoarthritis. Lancet (London, England). 2015;386(9991):376–87. https://doi.org/10.1016/S0140-6736(14)60802-3.
Kevorkian L, Young DA, Darrah C, Donell ST, Shepstone L, Porter S, et al. Expression profiling of metalloproteinases and their inhibitors in cartilage. Arthritis Rheum. 2004;50(1):131–41. https://doi.org/10.1002/art.11433.
Zeng GQ, Chen AB, Li W, Song JH, Gao CY. High MMP-1, MMP-2, and MMP-9 protein levels in osteoarthritis. Genet Mol Res. 2015;14(4):14811–22. https://doi.org/10.4238/2015.November.18.46.
Fu Y, Lei J, Zhuang Y, Zhang K, Lu D. Overexpression of HMGB1 A-box reduced IL-1beta-induced MMP expression and the production of inflammatory mediators in human chondrocytes. Exp Cell Res. 2016;349(1):184–90. https://doi.org/10.1016/j.yexcr.2016.10.014.
Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 2011;7(1):33–42. https://doi.org/10.1038/nrrheum.2010.196.
Mengshol JA, Vincenti MP, Coon CI, Barchowsky A, Brinckerhoff CE. Interleukin-1 induction of collagenase 3 (matrix metalloproteinase 13) gene expression in chondrocytes requires p38, c-Jun N-terminal kinase, and nuclear factor κB: differential regulation of collagenase 1 and collagenase 3. Arthritis Rheum. 2000;43(4):801–11. https://doi.org/10.1002/1529-0131(200004)43:4%3c801:AID-ANR10%3e3.0.CO;2-4.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. https://doi.org/10.1016/s0092-8674(04)00045-5.
Fioravanti A, Collodel G, Petraglia A, Nerucci F, Moretti E, Galeazzi M. Effect of hydrostatic pressure of various magnitudes on osteoarthritic chondrocytes exposed to IL-1beta. Indian J Med Res. 2010;132:209–17.
Jones SW, Watkins G, Le Good N, Roberts S, Murphy CL, Brockbank SM, et al. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthr Cartil. 2009;17(4):464–72. https://doi.org/10.1016/j.joca.2008.09.012.
Tardif G, Hum D, Pelletier JP, Duval N, Martel-Pelletier J. Regulation of the IGFBP-5 and MMP-13 genes by the microRNAs miR-140 and miR-27a in human osteoarthritic chondrocytes. BMC Musculoskelet Disord. 2009;10:148. https://doi.org/10.1186/1471-2474-10-148.
Proctor CJ, Smith GR. Computer simulation models as a tool to investigate the role of microRNAs in osteoarthritis. PLoS One. 2017;12(11):e0187568. https://doi.org/10.1371/journal.pone.0187568.
Yin X, Wang JQ, Yan SY. Reduced miR26a and miR26b expression contributes to the pathogenesis of osteoarthritis via the promotion of p65 translocation. Mol Med Rep. 2017;15(2):551–8. https://doi.org/10.3892/mmr.2016.6035.
Tao SC, Yuan T, Zhang YL, Yin WJ, Guo SC, Zhang CQ. Exosomes derived from miR-140-5p-overexpressing human synovial mesenchymal stem cells enhance cartilage tissue regeneration and prevent osteoarthritis of the knee in a rat model. Theranostics. 2017;7(1):180–95. https://doi.org/10.7150/thno.17133.
Lu B, Wang C, Wang M, Li W, Chen F, Tracey KJ, et al. Molecular mechanism and therapeutic modulation of high mobility group box 1 release and action: an updated review. Expert Rev Clin Immunol. 2014;10(6):713–27. https://doi.org/10.1586/1744666X.2014.909730.
Garcia-Arnandis I, Guillen MI, Gomar F, Pelletier JP, Martel-Pelletier J, Alcaraz MJ. High mobility group box 1 potentiates the pro-inflammatory effects of interleukin-1β in osteoarthritic synoviocytes. Arthritis Res Ther. 2010;12(4):R165. https://doi.org/10.1186/ar3124.
Ley C, Ekman S, Roneus B, Eloranta ML. Interleukin-6 and high mobility group box protein-1 in synovial membranes and osteochondral fragments in equine osteoarthritis. Res Vet Sci. 2009;86(3):490–7. https://doi.org/10.1016/j.rvsc.2008.10.008.
Heinola T, Kouri VP, Clarijs P, Ciferska H, Sukura A, Salo J, et al. High mobility group box-1 (HMGB-1) in osteoarthritic cartilage. Clin Exp Rheumatol. 2010;28(4):511–8.
Li ZC, Cheng GQ, Hu KZ, Li MQ, Zang WP, Dong YQ, et al. Correlation of synovial fluid HMGB-1 levels with radiographic severity of knee osteoarthritis. Clin Invest Med. 2011;34(5):E298.
Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, Gonzalez-Baron M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30(2):193–204. https://doi.org/10.1016/j.ctrv.2003.07.007.
Chen J, Crawford R, Xiao Y. Vertical inhibition of the PI3K/Akt/mTOR pathway for the treatment of osteoarthritis. J Cell Biochem. 2013;114(2):245–9. https://doi.org/10.1002/jcb.24362.
Chen D, Zeng S, Huang M, Xu H, Liang L, Yang X. Role of protein arginine methyltransferase 5 in inflammation and migration of fibroblast-like synoviocytes in rheumatoid arthritis. J Cell Mol Med. 2017;21(4):781–90. https://doi.org/10.1111/jcmm.13020.
Lv DJ, Song XL, Huang B, Yu YZ, Shu FP, Wang C, et al. HMGB1 promotes prostate cancer development and metastasis by interacting with brahma-related gene 1 and activating the Akt signaling pathway. Theranostics. 2019;9(18):5166–82. https://doi.org/10.7150/thno.33972.
Lu L, Zhang D, Xu Y, Bai G, Lv Y, Liang J. miR-505 enhances doxorubicin-induced cytotoxicity in hepatocellular carcinoma through repressing the Akt pathway by directly targeting HMGB1. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;104:613–21. https://doi.org/10.1016/j.biopha.2018.05.087.
He W, Cheng Y. Inhibition of miR-20 promotes proliferation and autophagy in articular chondrocytes by PI3K/AKT/mTOR signaling pathway. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;97:607–15. https://doi.org/10.1016/j.biopha.2017.10.152.
Salmon JH, Rat AC, Sellam J, Michel M, Eschard JP, Guillemin F, et al. Economic impact of lower-limb osteoarthritis worldwide: a systematic review of cost-of-illness studies. Osteoarthr Cartil. 2016;24(9):1500–8. https://doi.org/10.1016/j.joca.2016.03.012.
Si H, Zeng Y, Zhou Z, Pei F, Lu Y, Cheng J, et al. Expression of miRNA-140 in chondrocytes and synovial fluid of knee joints in patients with osteoarthritis. Chin Med Sci J. 2016;31(4):207–12.
Zhang M, Liu L, Xiao T, Guo W. Detection of the expression level of miR-140 using realtime fluorescent quantitative PCR in knee synovial fluid of osteoarthritis patients. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2012;37(12):1210–4. https://doi.org/10.3969/j.issn.1672-7347.2012.12.005.
Li X, Zhen Z, Tang G, Zheng C, Yang G. MiR-29a and MiR-140 protect chondrocytes against the anti-proliferation and cell matrix signaling changes by IL-1β. Mol Cells. 2016;39(2):103–10. https://doi.org/10.14348/molcells.2016.2179.
Liang Y, Duan L, Xiong J, Zhu W, Liu Q, Wang D, et al. E2 regulates MMP-13 via targeting miR-140 in IL-1beta-induced extracellular matrix degradation in human chondrocytes. Arthritis Res Ther. 2016;18(1):105. https://doi.org/10.1186/s13075-016-0997-y.
Yu M, Wang H, Ding A, Golenbock DT, Latz E, Czura CJ, et al. HMGB1 signals through toll-like receptor (TLR) 4 and TLR2. Shock. 2006;26(2):174–9. https://doi.org/10.1097/01.shk.0000225404.51320.82.
Fiuza C, Bustin M, Talwar S, Tropea M, Gerstenberger E, Shelhamer JH, et al. Inflammation-promoting activity of HMGB1 on human microvascular endothelial cells. Blood. 2003;101(7):2652–60. https://doi.org/10.1182/blood-2002-05-1300.
Lefebvre V, Peeters-Joris C, Vaes G. Modulation by interleukin 1 and tumor necrosis factor alpha of production of collagenase, tissue inhibitor of metalloproteinases and collagen types in differentiated and dedifferentiated articular chondrocytes. Biochim Biophys Acta. 1990;1052(3):366–78. https://doi.org/10.1016/0167-4889(90)90145-4.
Terada C, Yoshida A, Nasu Y, Mori S, Tomono Y, Tanaka M, et al. Gene expression and localization of high-mobility group box chromosomal protein-1 (HMGB-1)in human osteoarthritic cartilage. Acta Med Okayama. 2011;65(6):369–77. https://doi.org/10.18926/AMO/47262.
Wahamaa H, Schierbeck H, Hreggvidsdottir HS, Palmblad K, Aveberger AC, Andersson U, et al. High mobility group box protein 1 in complex with lipopolysaccharide or IL-1 promotes an increased inflammatory phenotype in synovial fibroblasts. Arthritis Res Ther. 2011;13(4):R136. https://doi.org/10.1186/ar3450.
Yuan Z, Luo G, Li X, Chen J, Wu J, Peng Y. PPARgamma inhibits HMGB1 expression through upregulation of miR-142-3p in vitro and in vivo. Cell Signal. 2016;28(3):158–64. https://doi.org/10.1016/j.cellsig.2015.12.013.
Yu F, Zeng H, Lei M, Xiao DM, Li W, Yuan H, et al. Effects of SIRT1 gene knock-out via activation of SREBP2 protein-mediated PI3K/AKT signaling on osteoarthritis in mice. J Huazhong Univ Sci Technol Med Sci. 2016;36(5):683–90. https://doi.org/10.1007/s11596-016-1645-0.
Acknowledgements
The study was supported by Natural Science Foundation of China [Grant Numbers: 81630064; 81871786].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Research involving human participants and/or animals
All procedures performed in studies involving human participants were in accordance with the ethical standards of Peking Union Medical College Hospital and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Responsible Editor: John Di Battista.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, Y., Shen, S., Li, Z. et al. MIR-140-5p affects chondrocyte proliferation, apoptosis, and inflammation by targeting HMGB1 in osteoarthritis. Inflamm. Res. 69, 63–73 (2020). https://doi.org/10.1007/s00011-019-01294-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-019-01294-0