Abstract
Introduction
Flavocoxid is a proprietary blend of two flavonoids, baicalin and catechin, and recent evidence has shown that bioflavonoids may exert antiviral activities. The potential antiviral activity of Flavocoxid against hepatitis B virus (HBV) was evaluated. Additionally, it was investigated if Flavocoxid used in combination with Entecavir could potentiate its anti-HBV activity.
Materials and methods
Hepatoma cells replicating HBV were treated with Flavocoxid, or Entecavir alone or in combination for up to 5 days. Viral replicative intermediates, transcripts, and cccDNA levels were evaluated in HBV-replicating cells by real-time PCR, Southern and Northern blotting. Expression profiling was performed using TaqMan low-density arrays.
Results
Flavocoxid treatment induced a reduction of HBV replicative intermediates, the amount of transcripts, and HBsAg levels. Flavocoxid and Entecavir combination therapy further decreased the amount of HBV replicative intermediates, compared to Flavocoxid alone. Importantly, Flavocoxid alone or in combination with Entecavir also induced a reduction of cccDNA. Gene-expression analysis showed that Flavocoxid activates type I IFNs-signaling and dampens the HBV-induced inflammatory response.
Conclusions
Flavocoxid inhibits HBV replication by targeting multiple steps of viral life cycle. These results indicate that the antiviral activity of Entecavir is potentiated by Flavocoxid, suggesting that this medical food might be considered as an adjuvant for anti-HBV therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite the implementation of hepatitis B virus (HBV) vaccination programs, chronic HBV infection continues to represent a global health problem with estimates of 240 million people infected worldwide, and approximately 80,000 dying per year due to HBV-related liver pathologies [1]. HBV infection ranges from very mild and asymptomatic clinical pictures to the most severe liver diseases including fulminant hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) [2]. HBV is one of the smallest viruses infecting humans. Its genome is of approximately 3.2 Kb in length and consists of a partially double-stranded relaxed circular DNA (rcDNA) containing four partially overlapping open-reading frames, which encode for the preS/S envelope proteins, the preC/Core proteins, the viral polymerase (Pol) that possesses reverse transcriptase function and the X regulatory protein. Upon infection of the hepatocytes, the HBV DNA is delivered to the nucleus and converted into covalently closed circular DNA (cccDNA). The cccDNA is of fundamental importance for viral replication by serving as the template for transcription of the pregenomic RNA (pgRNA) and all RNA species encoding the viral proteins [3]. Importantly, the cccDNA is organized into a chromatin-like structure by histone and non-histone proteins, as a mini-chromosome, and plays a crucial role both in viral long-term persistence (even after HBV surface antigen [HBsAg] seroconversion) and in viral relapse after cessation of antiviral therapy [4, 5]. Therefore, the elimination of HBV cccDNA represents the requisite for a therapeutic cure of HBV infection. However, eradication of the infection cannot be achieved with the available drugs licensed for hepatitis B therapy, so far. Currently, there are two different types of antiviral drugs approved for the treatment of chronic hepatitis B (CHB): (a) conventional and pegylated interferon-alpha, which may exert both immunomodulatory and direct antiviral effects, and (b) five oral nucleos(t)ide analogs (NAs) (Lamivudine, Adefovir, Entecavir, Telbivudine and Tenofovir) that efficiently block HBV replication by inhibiting the viral polymerase [6].
Despite the clinical efficacy of these potent antiviral drugs, they do not cure the HBV infection and a sustained virological response in terms of HBsAg loss and HBV surface antibody (anti-HBs) seroconversion is rarely achieved. In particular, therapy with NAs requires life-long administration, has only modest effects on the levels of HBsAg and carries the risk of viral resistance [6, 7]. Therefore, development of novel drugs or supplements to improve therapeutic efficacy for HBV-infected patients is still needed.
Plant-derived bioflavonoids have been shown to have potential effects on a variety of human diseases. Several studies have suggested a protective activity against inflammatory and degenerative diseases including age-related diseases, cancer, and cardiovascular diseases [8,9,10]. Interestingly, recent experimental data have shown that flavonoids possess antiviral activity and display very low toxicity. It has been demonstrated that these compounds may control viral infections by targeting virus entry, reverse transcription or gene expression [11,12,13,14,15]. Flavocoxid is a FDA-regulated medical food containing the naturally occurring flavonoids, such as baicalin extracted from Scutellaria baicalensis, and catechin from Acacia catechu [16].
The aims of this study were to investigate the susceptibility of HBV to Flavocoxid and to evaluate whether the antiviral effect of Entecavir may be potentiated by the medical food. For these purposes, we used a cell-based HBV replication system that recapitulates most of the HBV replication cycle steps, including the nuclear generation of HBV cccDNA molecules [17].
Materials and methods
Cell culture, transfections, and treatments
Both HuH7 and HepG2 well-differentiated human hepatoma cell lines were used in this study. HepG2 cells were obtained from the American Type Culture Collection (ATCC, HEPG2, ATCC ® HB8065 ™); HuH7 cells were a kind gift from Prof. M. Levrero, Laboratory of Gene Expression, La Sapienza University, Italy. Cells were cultured in DMEM with 10% heat-inactivated FBS (Life Technologies, GrandIsland, NY, USA), 1% antibiotic–antimycotic solution, and 1% glutamine (Sigma–Aldrich Srl, Milan, Italy), were incubated at 37 °C with 5% CO2 supplement. Both HuH7 and HepG2 hepatoma cells were transiently transfected with monomeric linear wild-type (WT) HBV genomes of genotype D (HBV-D), or the empty vector (pUC19) as previously described [17] using Lipofectamine transfection reagent (Invitrogen, Milan, Italy), according to the manufacturer’s protocol. Briefly, cells were seeded at a density of 3 × 105 cells in 60-mm diameter Petri dishes and transfected 24 h later with 0.5 µg of SapI-digested HBV DNA [18]. All transfections included 1 µg of reporter plasmid expressing enhanced green fluorescence protein (pGFP) to assess transfection efficiency. All transfection experiments were done at least three times, using independently prepared HBV DNA (Qiagen Maxi Preparation Kit, Qiagen, Milan, Italy).
Treatments with either Flavocoxid (FLAV) or Entecavir (ETV; 5 nM/ml) or their combination started 3 h after transfection and were renewed every other day for 5 days. To determine FLAV half-maximal inhibitory concentrations (IC50), transfected cells were treated with increasing doses of FLAV (25, 75, 100, 125, and 250 µg/ml) or left untreated. At the end of treatments, intracellular encapsidated HBV DNA was extracted and submitted to Southern blot analysis. The IC50 was determined by a 50% decrease in the level of viral DNA detected in treated cells at each time-point compared to untreated cells. The medium was prepared daily with fresh aliquots of the drugs.
FLAV was a kind gift from Primus Pharmaceuticals, Inc (Scottsdale, AZ, USA). ETV was kindly provided by Bristol-Myers Squibb. FLAV and ETV were re-suspended in DMEM, aliquoted, and frozen at −20 °C to avoid repeated freezing and thawing of the drug.
Cytotoxicity assay
Hepatoma cells (both Huh7 and HepG2) transfected with HBV-D or the empty vector (CTRL) were seeded in 96-well plates at a density of 5 × 103 per well and then incubated with different concentrations of the drugs for up to 5 days. Cell viability was determined using the CellTiter 98 AQueous Non-Radioactive Cell Proliferation Assay (Promega Corporation, Madison, WI), according to the manufacturer’s instructions.
Hepatitis B surface antigen in the cell culture medium
Hepatitis B surface antigen (HBsAg) was determined in culture supernatants by ELISA assay using the Murex HBsAg Version 3 kit (DiaSorin, Saluggia, Italy), and quantified using the ARCHITECT HBsAg Assay (Abbott; Abbott Laboratories, Chicago, IL), according to the manufacturers’ instructions.
Purification and analysis of HBV DNA from core particles
To purify HBV DNA from intracellular core particles, HBV-transfected hepatoma cells were lysed in 50 mM Tris–HCl, pH 7.4, 1 mM EDTA, and 1% NP-40. The number of haploid genomes in each cell lysate was then determined by the use of a β-globin gene kit (Light-Cycler β-globin control kit; Roche Diagnostics) for HBV DNA normalization. Lysates from an equal number of cells were then centrifuged at 10,000g for 1 min to pellet nuclei. The supernatant was adjusted to 100 mM MgCl2 and treated with 100 μg of DNase I for 30 min at 37 °C. The reaction was stopped by adding EDTA to a final concentration of 25 mM. Proteins were digested with 0.5 mg/ml proteinase K and 1% SDS for 2 h at 50 °C. DNA was purified by phenol/chloroform (1:1) extraction and ethanol precipitation by adding glycogen and examined by Southern blot analysis, as previously described [19].
Purification and analysis of HBV cccDNA
Nuclear HBV cccDNA was isolated from HBV-transfected cells analyzed by Southern blotting [19]. Briefly, 48 h after transfection with linear HBV monomers, hepatoma cells were harvested, lysed in 500 μL of homogenization buffer (10 mM Tris–HCl, pH 8.0, 10 mM EDTA, 0.5% SDS and 10 mg/ml PK) and incubated overnight at 50 °C. DNA was extracted with phenol/chloroform and precipitated with ethanol in the presence of 10 μg of tRNA.
Aliquots of each extracted DNA were treated for 1 h at 37 °C with 20 U Plasmid-safe DNase (Epicenter, Madison, WI). Twenty microliters of the isolated and Plasmid-safe DNase-treated DNA samples were used for HBV-specific Southern blot analysis [19]. For HBV DNA normalization, evaluation of the number of haploid genomes by the use of Light-Cycler β-globin control kit (Roche DNA control kit; Roche Diagnostics) allowed us to determine the cell number in the homogenate from transfected cell and, consequently, also in the aliquot used for DNA analysis by Southern blotting.
HBV RNAs and cellular mRNA analysis
Total RNA was extracted from HBV-transfected hepatoma cells with the TRIzol reagent (Invitrogen) as recommended by the manufacturer. RNA concentration was measured using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) at 260 nm. RNA quality and quantity were monitored by ethidium bromide staining and by UV absorbance. Northern blot analysis was performed following the standard procedures as previously described [19]. Radioactive probes were prepared by random priming protocol, using either full-length HBV DNA or 18S cDNA templates or 32P α-dCTP (PerkinElmer, Baesweiler, Germany).
For HBV RNA real-time PCR quantification, RNA samples were treated with RQ1 RNase-Free DNase (Promega) for 30 min at 37 °C. RNA quality and quantity were monitored by ethidium bromide staining and by UV absorbance. Two microgram of DNase-treated RNA was then reverse transcribed and amplified by the ThermoScript RT-PCR system (Invitrogen). Two microliters of each cDNA was quantitated by real-time PCR analysis (COBAS TaqMan 48; Roche, Basel, Switzerland) using the following HBV-specific primers and probes: Forward primer 5′-CTC gTg gTg gAC TTC TCT C -3′, reverse primer 5′-CAg CAg gAT gAA gAg gAA-3′, FRET hybridization probes 5′ CAC TCA CCA ACC TCC TGT CCT CCA A-FL-3′, and Red640 5′ TgT CCT ggT TAT CgC Tgg ATg TgT CT-PH-3′. The h-G6PDH housekeeping gene Light Cycler set (Roche Applied Science, Mannhein, Germany) was used to normalize the RNA samples.
For gene expression analysis by TaqMan low-density arrays (TLDAs) (Applied Biosystems, Foster City, CA), total RNA from transfected hepatoma cells was extracted using the RNeasy Mini kit (Qiagen), according to the manufacturer’s instruction. RNA (5 μg) was reverse transcribed using the Transcriptor First Strand cDNA Synthesis kit and 200 ng of each cDNA was loaded in the duplicate on a customized TLDA with 18S RNA used as control. TLDAs were run on an ABI 7900 HT Sequence detection system and, real-time PCR data were collected and analyzed with the SDS2.2 software (Applied Biosystems).
Western blot analysis
Total cellular proteins were extracted from hepatoma cells in a lysis buffer composed by 25 mM Tris–HCl, pH 7.4, 1.0 mM ethylene glycol tetraacetic acid (EGTA), 1.0 mM ethylenediaminetetraacetic acid (EDTA), 0.5 mM phenyl methylsulphonyl fluoride, added with protease and phosphatase inhibitors [100 mM Na3VO4, aprotinin, leupeptin, pepstatin (10 µg/ml each)] to block the endogenous molecules. The cell lysate was centrifuged at 1300×g for 15 min and the supernatant was used for determining protein concentrations by Bio-Rad protein assay (Bio-Rad, Richmond, CA, USA); and then diluted with Laemmli sample buffer (62 mM Tris pH 6.8, 10% glycerol, 2% SDS, 5% β-mercaptoethanol, 0.003% bromophenol blue). Proteins were separated by electrophoresis on SDS (10%) polyacrylamide gel, approximately for 1 h, then the separated proteins were transferred onto a PVDF membrane in a transfer buffer [39 mM glycine, 48 mM Tris–HCl (pH 8.3), 20% methanol] at 200 mA for 1 h. The membranes were stained with Ponceau S (0.005% in 1% acetic acid) to evaluate the amount of loaded proteins, and then blocked with 5% non-fat dry milk in TBS-0.1% Tween-20 for 1 h at room temperature. Membranes were washed three times for 10 min each in TBS-0.1% Tween-20 and incubated with a primary antibody for phospho-NFκB (Cell Signaling, Danvers, MA, USA), iNOS, COX-2 and 5-LOX (Abcam, Cambridge, UK) diluted in TBS-0.1% Tween-20 overnight at 4 °C. After one day, the membranes were washed three times for 10 min in TBS-0.1% Tween-20 and were incubated with a specific peroxidase-conjugated secondary antibody (Pierce, Rockford, IL, USA) for 1 h at room temperature. Following other washings, the membranes were analyzed by the enhanced chemiluminescence system, according to the manufacturer’s protocol (Amersham, Little Chalfont, UK). The protein signals were quantified by scanning densitometry using a bio-image analysis system (C-DiGit, LiCor, Lincoln, NE, USA) and the results were expressed as relative integrated intensity compared to controls. β-Actin (Cell Signaling Technology) was used to confirm equal protein loading and blotting [16].
Measurement of NO2 −
Nitrite levels were determined in the supernatants of hepatoma cells with the Griess colorimetric assay. Supernatants (100 μl) were mixed with 50 μl of sulphanilamide (1%) and 50 μl of naphtyl-ethylendiamine dihydrochloride (0.1%), both dissolved in 2.5% H3PO4, and after 10 min of incubation at room temperature the absorbance was read at 540 nm. Sodium nitrite standards were used as controls. All samples and standards were assayed at least three times in duplicates [16].
Analysis of signal intensity
Autoradiography and PCR images were digitalized and analyzed with a GS-800 Calibrated Densitometer (Bio-Rad Laboratories Segrate MI, Italy). Quantitative analysis of the signal intensity was performed with Quantity One 1-D Analysis Software (BioRad).
Statistical analysis
P-values were determined using the two-tailed Student’s t test. Comparisons between different treatments were analyzed by one-way ANOVA with Tukey post-test for intergroup comparisons. Bar graphs were plotted to show the mean ± standard deviation (SD) of at least three independent experiments. P-values < 0.05 were considered as statistically significant. Statistical analysis was performed with SPSS version 17.0 software package (SPSS Inc, Chicago, IL, USA).
Results
Susceptibility of HBV to FLAV
To evaluate the susceptibility of HBV to FLAV, a cell-based HBV replication system, relying on the generation of transcriptionally active nuclear cccDNA to replicate the virus, was used [17]. Southern blot analysis of intracellular HBV DNA replicative intermediates synthesized in the presence of FLAV in hepatoma cells showed that the most effective concentration was 75 µg/ml following two days of treatment (Fig. 1a). In HBV-replicating cells and in control cells, FLAV showed no inhibitory effect on cell proliferation, as evaluated by MTT assay (Fig. 1b).
Susceptibility of HBV to Flavocoxid (FLAV). a Representative autoradiogram of Southern blot analysis showing in vitro susceptibility profiles of HBV to FLAV. HuH7 cells transiently transfected with monomeric linear full-length HBV genomes (HBV-D) were treated with the indicated increasing concentrations of FLAV, or left untreated (HBV-D NT) for 48 h. Intracellular viral-encapsidated DNA were then isolated and subjected to Southern blot analysis. Migration of a DNA standard is indicated in kilobases (Kb) on the left; position of open circular (OC), double-stranded linear (DS) and single-stranded (SS) DNA is indicated on the right. Right panel: Data plotted in the graph represent the means ± standard deviations (SD) from three different assays, each performed in duplicates, of HBV DNA replicative intermediates. The SS band underneath the linear DS HBV DNA band was used for densitometric quantification. The band corresponding to the DS HBV DNA was not included in the quantitative analysis because this DNA may be partially derived from transfected input DNA. Signal intensity of the SS band was quantified with GS-800 Calibrated Densitometer (Bio-Rad) using the Quantity One 1-D Analysis Software (BioRad). Data are presented as percentage values relative to those of untreated HBV-transfected cells (HBV-D NT), used as control and set at 100. b Influences of FLAV on cell viability in non-transfected cells, empty vector pUC19-transfected cells, and HBV-D transfected cells after 48 h and 120 h (bottom panel) FLAV treatment, as determined by the MTT method. Kb kilobases, NT not treated, OC open circular DNA, DS double-stranded DNA, SS single-stranded DNA
FLAV induces a parallel reduction of cytoplasmic HBV DNA replicative intermediates, nuclear cccDNA, viral transcripts, and HBsAg
To elucidate the antiviral effects of FLAV on an ongoing infection, and to verify its efficacy in comparison with ETV, we treated HBV-D-transfected cells with 75 µg/ml of FLAV or 5 nM/ml of ETV for 5 days.
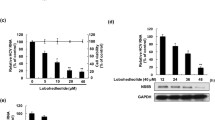
Southern blot experiments revealed that HBV DNA replicative intermediates were reduced by 70 and 50% in FLAV or ETV-treated cells, compared to untreated HBV-replicating cells (Fig. 2a). Of particular interest, a 45% reduction in cccDNA was only observed in FLAV-treated cells, compared to untreated cells (Fig. 2b). Northern blot analysis showed that HBV RNA levels were decreased by 80% after exposure to FLAV, compared to untreated HBV-replicating cells (Fig. 3a). Additionally, the ratio between the 3.5 kb pregenome/C-mRNA and the 2.4–2.1Kb mRNA species (preS/S RNA) did not change during FLAV treatment, indicating that this drug similarly affected all the HBV transcripts. NAs do not target cccDNA transcription, thus ETV did not reduce the expression of viral transcripts in HBV-replicating cells (Fig. 3a). Quantification of total HBV RNA by real-time PCR quantification further confirmed that viral transcripts amounts were significantly reduced in FLAV-treated cells compared to both ETV-treated and untreated cells (Fig. 3b). The strong reduction of HBV transcripts in FLAV-exposed cells was paralleled by a significant decrease (by 60%) in the amount of HBsAg compared to untreated HBV-replicating cells (Fig. 3c). As expected, no reduction in HBsAg concentration was observed in the supernatant of ETV-exposed cells (Fig. 3c). Overall, these results show that FLAV inhibits HBV replication in our system. The natural drug was effective in reducing the levels of all viral replicative intermediates including the HBV cccDNA, in lowering the steady state levels of HBV transcripts, and in reducing the production of HBsAg.
Inhibitory effect of FLAV and ETV on HBV replicative DNA intermediates in HBV-replicating cells. Representative autoradiograms of Southern blotting showing in vitro FLAV and ETV susceptibility profiles of HBV. a HuH7 cells transiently transfected with monomeric linear full-length HBV genomes (HBV-D) were treated with ETV (5 nm/ml), FLAV (75 μg/ml), or left untreated (HBV-D NT) for 120 h. Intracellular capsid-associated HBV DNA was then isolated and subjected to Southern blot analysis. Migration of a DNA standard is indicated in Kb on the left; position of open circular (OC), double-stranded linear (DS) and single-stranded (SS) DNA is indicated on the right. Right panel: data plotted in the graph represent the means ± SD from three different assays, each performed in duplicate, of HBV DNA replicative intermediates. The SS band underneath the linear DS HBV DNA band was used for densitometric quantification. The band corresponding to the DS HBV DNA was not included in the quantitative analysis because this DNA may be partially derived from transfected input DNA. Signal intensity of the SS band was quantified with GS-800 Calibrated Densitometer (Bio-Rad) using the Quantity One 1-D Analysis Software (BioRad). Data are presented as percentage values relative to those of untreated HBV-transfected cells (HBV-D NT), used as control and set at 100. P-values were determined using a student’s t-test comparing FLAV- and ETV-treated HBV-replicating cells to control untreated HBV-replicating cells (HBV-D NT). b HBV cccDNA accumulation in HuH7 cells transiently transfected with monomeric linear full-length HBV genomes (HBV-D) and then treated with ETV (5 nm/ml), FLAV (75 μg/ml), or left untreated (HBV-D NT) for 120 h. After treatment, HBV covalently closed circular DNA (cccDNA) was isolated and subjected to Southern blot analysis. Migration of a DNA standard is indicated in Kb on the left; position of OC and cccDNA is indicated on the right. Right panel: data plotted in the graph represent the means ± SD from three different assays, each performed in duplicates, of HBV cccDNA. Signal intensity of the cccDNA band was quantified with GS-800 Calibrated Densitometer (Bio-Rad) using the Quantity One 1-D Analysis Software (BioRad). Data are presented as percentage values relative to those of untreated HBV-transfected cells (HBV-D NT), used as control and set at 100. P-values were determined using a student’s t-test comparing FLAV- and ETV-treated HBV-replicating cells to control untreated HBV-replicating cells (HBV-D NT). Kb kilobases, OC open circular DNA, DS double-stranded DNA, cccDNA covalently closed circular DNA
Inhibitory effect of FLAV and ETV on HBV transcripts and HBsAg in HBV-replicating cells. a Representative autoradiogram of Northern blot analysis of HBV transcripts isolated from HBV-transfected HuH7 cells treated with ETV (5 nm/ml) or FLAV (75 μg/ml), or left untreated (HBV-D NT) for 120 h. RNA loading was controlled by rehybridization with a 32P-labeled 18S specific probe. Right panels: data plotted in the graph represent the means ± SD from three different assays, each performed in duplicates, of HBV transcripts. Bars indicate SD. Signal intensity of the bands corresponding to pregenomic (pg) (3.5 kb), preS1 (2.4 kb), and preS2/S (2.1 kb) HBV RNAs, respectively, was quantified with GS-800 Calibrated Densitometer (Bio-Rad) using the Quantity One 1-D Analysis Software (BioRad). Data are presented as percentage values relative to those of untreated HBV-transfected cells (HBV-D NT), used as control and set at 100. P-values were determined using a student’s t-test comparing FLAV- and ETV-treated HBV-replicating cells to control untreated HBV-replicating cells (HBV-D NT). b Steady state levels of total HBV RNA levels were analyzed by real-time PCR using selective HBV S region primers and GAPDH primers (for RNA sample normalization). Data plotted in the graph represent the means ± SD from three different assays, each performed in duplicate, of HBV transcripts. Bars indicate SD. Data are expressed as fold induction relative to the untreated HBV-replicating cells (HBV-D NT) using the comparative Ct method. P-values were determined using a student’s t-test comparing FLAV- and ETV-treated HBV-replicating cells to control untreated HBV-replicating cells (HBV-D NT). (C) HBsAg levels in supernatant of HBV-transfected HuH7 cells treated with ETV (5 nm/ml) or FLAV (75 μg/ml), or left untreated (HBV-D NT) for 120 h, as measured by using the ARCHITECT HBsAg Assay (Abbott; Abbott Laboratories, Chicago, IL). Data plotted in the graph represent the mean values ± SD from three different assays, each performed in duplicate, of HBsAg. Bars indicate SD. HBsAg values are expressed as IU/ml. P-values were determined using a student’s t-test comparing FLAV- and ETV-treated HBV-replicating cells to control untreated HBV-replicating cells (HBV-D NT). Kb kilobases, pgRNA pregenomic RNA, HBsAg hepatitis B surface antigen
FLAV potentiates the anti-HBV activity of ETV
To investigate the potential therapeutic use of FLAV in combinational regimen, HBV-replicating cells were treated with FLAV and ETV at fixed concentrations for 5 days. As shown in Fig. 4a, b, FLAV + ETV combination therapy resulted in a significant strong inhibition of HBV replication compared to both untreated cells (HBV-D) and cells treated with ETV alone (HBV-D + ETV). In particular, it is of great importance to note that the combination of FLAV and ETV compared to ETV alone not only significantly reduced the production of HBV DNA replicative intermediates (by about 60%) (Fig. 4a), but also induced a strong and significant reduction of both HBV cccDNA (by 40%) (Fig. 4B) and HBsAg (by 60%) amounts (Fig. 5b). Moreover, treatment with FLAV + ETV also resulted in a significant reduction of the steady state levels of HBV transcripts (by 70%) compared to ETV treatment alone (Fig. 5a). These results indicate that FLAV is able to potentiate the antiviral effect of ETV, and suggest that this medical food might be considered as a promising adjuvant for anti-HBV therapy.
Effect of FLAV, ETV, and ETV plus FLAV combination therapy on HBV replicative DNA intermediates in HBV-replicating cells. Representative autoradiograms of Southern blot analysis showing in vitro susceptibility profiles of HBV to FLAV, ETV, or ETV plus FLAV. a HuH7 cells transiently transfected with monomeric linear full-length HBV genomes (HBV-D) were treated with ETV (5 nm/ml), FLAV (75 μg/ml), or ETV (5 nm/ml) + FLAV (75 μg/ml), or left untreated (HBV-D NT) for 120 h. Intracellular viral-encapsidated DNA were then isolated and subjected to Southern blot analysis. Migration of a DNA standard is indicated in Kb on the left; position of open circular (OC), double-stranded linear (DS) and single-stranded (SS) DNA is indicated on the right. Right panel: Data plotted in the graph represent the means ± standard deviations (SD) from three different assays, each performed in duplicate, of HBV DNA replicative intermediates. The SS band underneath the linear DS HBV DNA band was used for densitometric quantification. The band corresponding to the DS HBV DNA was not included in the quantitative analysis because this DNA may be partially derived from transfected input DNA. Signal intensity of the SS band was quantified with GS-800 Calibrated Densitometer (Bio-Rad) using the Quantity One 1-D Analysis Software (BioRad). Data are presented as percentage values relative to those of untreated HBV-transfected cells (HBV-D NT), used as control and set at 100. P-values were determined using a student’s t-test comparing FLAV- and ETV-treated HBV-replicating cells to control untreated HBV-replicating cells (HBV-D NT) as well as ETV + FLAV- treated HBV-replicating cells to ETV-treated HBV-replicating cells. b HBV cccDNA accumulation in HuH7 cells transiently transfected with monomeric linear full-length HBV genomes (HBV-D) and then treated with ETV (5 nm/ml), FLAV (75 μg/ml), or ETV (5 nm/ml) + FLAV (75 μg/ml), or left untreated (HBV-D NT) for 120 h. After treatment, HBV cccDNA was isolated and subjected to Southern blot analysis. Migration of a DNA standard is indicated in Kb on the left; position of OC and cccDNA is indicated on the right. Right panel: Data plotted in the graph represent the means ± SD from three different assays, each performed in duplicates, of HBV cccDNA. Signal intensity of the cccDNA band was quantified with GS-800 Calibrated Densitometer (Bio-Rad) using the Quantity One 1-D Analysis Software (BioRad). Data are presented as percentage values relative to those of untreated HBV-transfected cells (HBV-D NT), used as control and set at 100. P-values were determined using a student’s t-test comparing FLAV- and ETV-treated HBV-replicating cells to control untreated HBV-replicating cells (HBV-D NT) as well as ETV + FLAV- treated HBV-replicating cells to ETV-treated HBV-replicating cells. Kb kilobases, OC open circular DNA, DS double-stranded DNA, cccDNA covalently closed circular DNA
Effect of FLAV, ETV, and ETV plus FLAV combination therapy on HBV transcripts and HBsAg in HBV-replicating cells. a Representative autoradiogram of Northern blot analysis of HBV transcripts isolated from HBV-transfected HuH7 cells treated with ETV (5 nm/ml), FLAV (75 μg/ml), or ETV (5 nm/ml) + FLAV (75 μg/ml), or left untreated (HBV-D NT) for 120 h. RNA loading was controlled by rehybridization with a 32P-labeled 18S specific probe. Right panels: Data plotted in the graph represent the means ± SD from three different assays, each performed in duplicates, of HBV transcripts. Bars indicate SD. Signal intensity of the bands corresponding to pregenomic (pg) (3.5 kb), preS1 (2.4 kb), and preS2/S (2.1 kb) HBV RNAs, respectively, was quantified with GS-800 Calibrated Densitometer (Bio-Rad) using the Quantity One 1-D Analysis Software (BioRad). Data are presented as percentage values relative to those of untreated HBV-transfected cells (HBV-D NT), used as control and set at 100. P-values were determined using a Student’s T test comparing FLAV- and ETV-treated HBV-replicating cells to control untreated HBV-replicating cells (HBV-D NT) as well as ETV + FLAV- treated HBV-replicating cells to ETV-treated HBV-replicating cells. b HBsAg levels in supernatant of HBV-transfected HuH7 cells treated with ETV (5 nm/ml), FLAV (75 μg/ml), or ETV (5 nm/ml) + FLAV (75 μg/ml), or left untreated (HBV-D NT) for 120 h, as measured by using the ARCHITECT HBsAg Assay (Abbott; Abbott Laboratories). Data plotted in the graph represent the mean values ± SD from three different assays of HBsAg, each performed in duplicate. Bars indicate SD. HBsAg values are expressed as IU/ml. P-values were determined using a Student’s T test comparing FLAV- and ETV-treated HBV-replicating cells to control untreated HBV-replicating cells (HBV-D NT) as well as ETV + FLAV- reated HBV-replicating cells to ETV-treated HBV-replicating cells. Kb kilobases, pgRNA pregenomic RNA, HBsAg hepatitis B surface antigen
Effect of FLAV on IFN-induced signaling
Cells sense virus infection and trigger innate immune response as the first line of defense to control viral replication and dissemination [20]. Innate immune response in HBV-replicating cells treated with FLAV or left untreated was assessed by analyzing the expression levels of selected cytokines and chemokines (such as type I IFNs, IL-1β, IL-6, IL-8) and a number of related genes (namely, signal transducer and activator of transcription [STAT]-1, STAT-2, JAK-1, JAK-2, 2′5′-oligoadenylate synthetase 1 [OAS-1], myxovirus resistance-1 [MX1], IKK-related kinases TANK-binding kinase 1 [TBK1], IFN regulatory factor [IRF]-3, IRF-7, IRF-9, dsRNA-dependent protein kinase R [PKR], PIK3, myeloid differentiation primary response protein 88 [MyD88], IL-1 receptor-associated kinase [IRAK]1, IRAK4, TNF receptor-associated factor 6 [TRAF6], Ik B kinase-ε [IKKε], NF-kB, TNFα, and suppressor of cytokine signaling [SOCS5]) using TLDA.
In cells transfected with an empty plasmid, no change in the expression level of IFNs or other genes was observed following the treatment with FLAV; whereas in HBV-replicating cells treated with FLAV, the expression of IFNα and IFNβ was increased more than threefold and fourfold, respectively, compared to untreated cells (Fig. 6a). By contrast, there was no change in the expression of the several inflammatory genes, as reported in Fig. 6b. Interestingly, the expression of antiviral IFN-inducible gene OAS-1 and of TBK1 and IKKε kinases, which directly activate the IRF3/7 signaling pathways, were significantly increased (6-, 7.5-, and 4-fold, respectively) in FLAV-treated cells (Fig. 6c). In accordance with these results, the expression of SOCS5, an inhibitor of IFN-signaling that allows viruses to evade the host immune response, was significantly reduced in HBV-replicating cells treated with FLAV (Fig. 6d). Remarkably, in these cells the expression level of NF-κB and of the pro-inflammatory cytokines TNF-α, IL-1β, IL-6, and IL-8 were also reduced (Fig. 6d). Altogether, these results indicate that in our HBV replication system FLAV was able to induce an antiviral response by activating type I IFNs-signaling, and to dampen the HBV-induced inflammatory response.
Expression profiling of selected genes in HBV-replicating HepG2 cells treated with FLAV or left untreated. Custom real-time PCR liquid arrays (TLDAs-Applied Biosystems) were loaded with 0.2 μg of cDNA obtained from total RNA extracted from HepG2 cells transfected with 0.5 μg of monomeric linear HBV genomes (HBV-D) or 0.5 μg of an empty pUC19 plasmid and treated with FLAV (75 μg/ml) (HBV-D + FLAV) or left untreated (HBV-D NT). a Relative expression levels of IFNα and IFNβ genes. b Relative expression levels of STAT-1, STAT-2, JAK-1, JAK-2, MX1, IRF-3, IRF-7, IRF-9, PKR, PIK3, MyD88, IRAK1, IRAK4, and TRAF6 genes. c Relative expression levels of OAS-1, TBK1 and IKKε genes. Results are expressed as fold induction over the basal level of expression of empty plasmid transfected cells that was set at 1. All histograms show mean values from three independent experiments, each performed in duplicates; bars indicate SD. *P < 0.05, P-values were determined using a Student’s T test comparing FLAV-treated HBV-replicating cells to control untreated HBV-replicating cells (HBV-D NT)
Synergistic effect of FLAV and ETV on pNF-κB
NF-κB is an early transcription factor that plays a crucial role in the inflammatory pathways [21]. To further confirm gene expression results, we evaluated phosphorylated NF-κB protein levels by Western blot analysis in transfected hepatoma cells treated with ETV (5 nM/ml), FLAV (75 μg/ml) or both treatments for 12 h. Low pNF-κB expression was observed in control cells with and without treatments. On the contrary, pNF-κB was markedly increased in HBV-replicating cells 12 h after transfection (Fig. 7a). Interestingly, ETV or FLAV alone significantly reduced pNF-κB protein compared to untreated transfected cells. Moreover, the simultaneous incubation with FLAV and ETV resulted in an additional decrease of pNF-κB compared to ETV alone (Fig. 7a). These results are in accordance with the blunted NF-κB gene expression in HBV-transfected cells and support our data on the important role of FLAV in potentiating the anti-HBV activity of ETV.
Western blot analysis of pNF-kB (a) and iNOS (b) protein expression in HuH7 transfected (HBV-D) and not transfected (CTRL) cells. Three hours after transfection cells were treated with ETV (5 nm/ml) and FLAV (75 μg/ml) alone or with drugs combination for 12 h. All histograms show mean values from three independent experiments, each performed in duplicates; bars indicate the mean ± SD. *P < 0.05 vs CTRL; #P < 0.05 vs HBV. Nitrite production (C) in HuH7-transfected (HBV-D) and not-transfected (CTRL) cells. Three hours after transfection cells were treated with ETV (5 nm/ml) and FLAV (75 μg/ml) alone or with drugs combination for 12 h. All histograms show mean values from three independent experiments, each performed in duplicates; bars indicate the mean ± SD
FLAV suppression of HBV is independent of its ability to inhibit iNOS
To investigate whether the anti-inflammatory/oxidant ability of FLAV [16, 22] could contribute to the inhibition of HBV replication, we analyzed iNOS by Western blot and nitrite levels. No significant iNOS expression or measurable nitrite levels were observed in the same cell lines after transfection with HBV genomes (Fig. 7b, c). Therefore, these data suggest that FLAV suppression of HBV transcription/replication is independent of its ability to reduce iNOS activation.
Discussion
The general interest in the potential antiviral activity of phytochemicals has increased tremendously in recent years. Indeed, many studies have reported the inhibitory effects of medicinal plant extracts on the replication of several viruses [23]. In particular, there is evidence that a wide variety of active phytochemicals are capable of controlling human immunodeficiency virus, human herpes simplex virus, influenza virus, and hepatitis C virus infections [11, 15]. There is also evidence that several medicinal plants may exhibit anti-HBV activities [23,24,25,26,27]. In fact, flavonoids such as wogonin or ellagic acid appear to be capable of suppressing HBsAg secretion and HBV DNA production, both in HepG2.2.15 cells and in animal models. However, the mechanisms of action, therapeutic effects, and safety profile of these compounds are still largely unknown [28, 29].
FLAV marketed as an FDA-regulated medical food (Limbrel®) for the clinical dietary management of osteoarthritis in the United States contains a proprietary blend of the purified and standardized flavonoids, baicalin and catechin [16]. In vitro and in vivo studies have indicated that FLAV has an optimal preclinical safety profile. Moreover, both clinical trials and a post-marketing surveillance study [30] have confirmed that FLAV has a significant efficacy in the management of OA and a good overall tolerability [31]. FLAV has strong antioxidant and anti-inflammatory activities, it is able to inhibit the peroxidase activities of COX-1 and COX-2, as well as, the activity of 5-LOX enzyme [32], and recent evidence demonstrates that this medical food can also improve survival and restore the histologic architecture of lung and liver of mice subjected to polymicrobial sepsis [22].
The potential effect of FLAV against HBV was tested using a cell-based HBV replication assay relying on the generation of transcriptionally active nuclear cccDNA to replicate HBV. The results showed that FLAV has a strong inhibitory activity on HBV replication, targeting multiple steps of viral life cycle. Indeed, treatment with this medical food was able to affect cccDNA-driven transcription, protein synthesis, and viral replicative intermediate production, leading to a significant reduction of nuclear HBV cccDNA pool without cytotoxicity. Of note, our data also showed that treatment with FLAV resulted in the induction of type I IFNs (IFN-α/β) (known to target the epigenetic control of cccDNA function [33], to impair HBV RNA, capsid, and cccDNA stability [34,35,36]), in the up-regulation of the OAS-1 enzyme (counteracting the early phase of viral infections by degrading viral RNA in combination with RNase L [37]), and in the down-regulation of HBV-induced inflammatory mediators, thus suggesting that FLAV could be capable of promoting both an antiviral state against HBV infection and an anti-inflammatory response against HBV-related liver inflammation. FLAV is also able to reduce the expression of COX-2 and 5-LOX enzymes both in vitro and in vivo models, and to attenuate the expression of iNOS and NO content in macrophages [16, 22]. To investigate whether the downregulation of COX-2, 5-LOX or iNOS by FLAV could contribute to the inhibition of HBV replication, we analyzed COX-2, 5-LOX, and iNOS by Western blot and nitrite levels. COX-2 and 5-LOX were not constitutively expressed and did not show induction after HBV transfection (data not shown), while no changes have been observed in iNOS expression or in nitrite levels. In addition to these possible indirect activities, one might hypothesize a direct antiviral effect of FLAV against HBV, as it has very recently shown for the oligomeric flavonoid proanthocyanidin and its analog oolonghomobisflavanes that are able to directly target the HBV large surface protein, thus inhibiting HBV as well as hepatitis delta virus entry [38]. Noteworthy, the FLAV components baicalin and catechin have been reported to be able to interact with the reverse transcriptase and the glycoprotein gp120 of HIV, respectively, thus blocking virus replication and adsorption [15]. Altogether, these data strongly encourage performing further work to verify whether FLAV might inhibit HBV activities through a direct interaction with viral components.
Very interestingly, the use of FLAV in combination with ETV in our HBV replication system resulted in a potentiation of the antiviral effects of ETV against HBV. Indeed, this combination leads to an effective inhibition of HBV cccDNA transcription, HBsAg synthesis, and—of great importance—reduction of nuclear cccDNA amounts. These findings are of particular interest, considering that current antiviral treatment with NAs efficiently blocks the HBV replication, but fails to affect cccDNA accumulation, and has only modest effects on the circulating HBsAg [7]. As a consequence, monotherapy with NAs often involves long-term commitment, during which durable HBV control is rarely achieved [7]. Indeed, the current research trend is to develop antiviral combination strategies that would lead to finite duration treatment, via the clearance of viral cccDNA and HBsAg seroconversion and the restoration of immune control.
Concern about the use of FLAV in HBV infection may arise from a report suggesting that it may cause liver toxicity [39]. 4 out of 877 cases of suspected drug-induced hepatic toxicity were ascribed to FLAV. They were all women: three were scored as very likely and one as possible due to the combined exposure to several other potential drugs inducing liver injury including pregabalin, duloxetine and tizanidine. However, the report did not include for the three subjects information about the concomitant dietary intake of food containing flavonoids or other natural substances that might influence FLAV pharmacokinetics and also directly concur to liver injury, thus precluding to draw any final conclusion. In addition, a post-marketing surveillance study of FLAV carried out among 284,299 FLAV users by the manufacturing company reported a very low incidence rate of liver adverse effects (0.011%). Finally, FLAV protects against experimental acute liver injury induced in mice by carbon tetrachloride (data on file).
In summary, our data show that in HBV-replicating cells FLAV is able to activate the endogenous IFN system inducing a strong antiviral response and that the combination of FLAV and ETV results in enhanced anti-HBV activity. This suggests that FLAV could be an attractive candidate for the treatment of HBV infection in combination with NAs. Indeed, this combined therapeutic strategy leading to strong viral suppression, induction of antiviral innate immune response, and control of HBV-related liver inflammation could represent a promising immunotherapeutic strategy for the treatment of chronic HBV infection. However, further in vitro and in vivo studies are needed to validate the antiviral activity of FLAV and to better define its mode of action against HBV.
Conclusions
FLAV blunts HBV replication by targeting multiple steps of viral life cycle. Moreover, the use of FLAV in combination with ETV causes a potentiation of the antiviral activity of ETV, suggesting that this medical food might be considered as an adjuvant for anti-HBV therapy.
References
Meireles LC, Marinho RT, Van Damme P. Three decades of hepatitis B control with vaccination. World J Hepatol. 2015;7:2127–32.
Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–45.
Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015;479–480:672–86.
Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. J Hepatol. 2009;51:581–92.
Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C. Structural organization of the hepatitis B virus minichromosome. J Mol Biol. 2001;307:183–96.
EASL.EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167–85.
Zoulim F. Are novel combination therapies needed for chronic hepatitis B? Antiviral Res. 2012;96:256–9.
Middleton E Jr, Kandaswami C, Theoharides TC. The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol Rev. 2000;52:673–751.
Havsteen BH. The biochemistry and medical significance of the flavonoids. Pharmacol Ther. 2002;96:67–202.
Khan N, Mukhtar H. Tea polyphenols for health promotion. Life Sci. 2007;81:519–33.
Friedman M. Overview of antibacterial, antitoxin, antiviral, and antifungal activities of tea flavonoids and teas. Mol Nutr Food Res. 2007;51:116–34.
Calland N, Albecka A, Belouzard S, Wychowski C, Duverlie G, Descamps V. (-)-Epigallocatechin-3-gallate is a new inhibitor of hepatitis C virus entry. Hepatology. 2012;55:720–9.
Song JM, Lee KH, Seong BL. Antiviral effect of catechins in green tea on influenza virus. Antiviral Res. 2005;68:66–74.
Isaacs CE, Wen GY, Xu W, Jia JH, Rohan L, Corbo C. Epigallocatechin gallate inactivates clinical isolates of herpes simplex virus. Antimicrob Agents Chemother. 2008;52:962–70.
De Clercq E. Current lead natural products for the chemotherapy of human immunodeficiency virus (HIV) infection. Med Res Rev. 2000;20:323–49.
Altavilla D, Squadrito F, Bitto A, Polito F, Burnett BP, Di Stefano V, et al. Flavocoxid, a dual inhibitor of cyclooxygenase and 5-lipoxygenase, blunts pro-inflammatory phenotype activation in endotoxin-stimulated macrophages. Br J Pharmacol. 2009;157:1410–8.
Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, et al. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–37.
Pollicino T, Bellinghieri L, Restuccia A, Raffa G, Musolino C, Alibrandi A, et al. Hepatitis B virus (HBV) induces the expression of interleukin-8 that in turn reduces HBV sensitivity to interferon-alpha. Virology. 2013;444:317–28.
Pollicino T, Raffa G, Costantino L, Lisa A, Campello C, Squadrito G, et al. Molecular and functional analysis of occult hepatitis B virus isolates from patients with hepatocellular carcinoma. Hepatology. 2007;45:277–85.
Janeway, CAJr. Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216.
Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–48.
Bitto A, Minutoli L, David A, Irrera N, Rinaldi M, Venuti FS, et al. Flavocoxid, a dual inhibitor of COX-2 and 5-LOX of natural origin, attenuates the inflammatory response and protects mice from sepsis. Crit Care. 2012;16:R32.
Chattopadhyay D, Sarkar MC, Chatterjee T, Sharma Dey R, Bag P, Chakraborti S, et al. Recent advancements for the evaluation of anti-viral activities of natural products. N Biotechnol. 2009;25:347–68.
Xia Y, Luo H, Liu JP, Gluud C. Phyllanthus species versus antiviral drugs for chronic hepatitis B virus infection. Cochrane Database Syst Rev. 2013;4:CD009004.
Qi FH, Wang ZX, Cai PP, Zhao L, Gao JJ, Kokudo N, et al. Traditional Chinese medicine and related active compounds: a review of their role on hepatitis B virus infection. Drug Discov Ther. 2013;7:212–24.
Zhang L, Wang G, Hou W, Li P, Dulin A, Bonkovsky HL. Contemporary clinical research of traditional Chinese medicines for chronic hepatitis B in China: an analytical review. Hepatology. 2010;51:690–8.
Chen Y, Zhu J. Anti-HBV effect of individual traditional Chinese herbal medicine in vitro and in vivo: an analytic review. J Viral Hepat. 2013;20:445–52.
Guo Q, Zhao L, You Q, Yang Y, Gu H, Song G, et al.. Anti-hepatitis B virus activity of wogonin in vitro and in vivo. Antiviral Res. 2007;74:16–24.
Kang EH, Kown TY, Oh GT, Park WF, Park SI, Park SK, et al. The flavonoid ellagic acid from a medicinal herb inhibits host immune tolerance induced by the hepatitis B virus-e antigen. Antiviral Res. 2006;72:100–6.
Pillai L, Burnett BP, Levy RM. Group GSC. GOAL: multicenter, open-label, post-marketing study of flavocoxid, a novel dual pathway inhibitor anti-inflammatory agent of botanical origin. Curr Med Res Opin. 2010;26:1055–63.
Bitto A, Squadrito F, Irrera N, Pizzino G, Pallio G, Mecchio A, et al. Flavocoxid, a nutraceutical approach to blunt inflammatory conditions. Mediators Inflamm. 2014;790851.
Burnett BP, Bitto A, Altavilla D, Squadrito F, Levy RM, Pillai L. Flavocoxid inhibits phospholipase A2, peroxidase moieties of the cyclooxygenases (COX), and 5-lipoxygenase, modifies COX-2 gene expression, and acts as an antioxidant. Mediators Inflamm.2011;385780.
Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T. Pollicino T. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest. 2012;122:529–37.
Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221 – 1128.
Uprichard SL, Wieland SF, Althage A, Chisari FV. Transcriptional and posttranscriptional control of hepatitis B virus gene expression. Proc Natl Acad Sci. 2003;100:1310–5.
Rang A, Gunther S, Will H.. Effect of interferon alpha on hepatitis B virus replication and gene expression in transiently transfected human hepatoma cells. J Hepatol. 1999;31:791–9.
Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809.
Tsukuda S, Watashi K, Hojima T, Isogawa M, Iwamoto M, Omagari K, et al. A new class of hepatitis B and D virus entry inhibitors, proanthocyanidin and its analogs, that directly act on the viral large surface proteins. Hepatology. 2017;65:1104–16.
Chalasani N, Vuppalanchi R, Navarro V, Fontana R, Bonkovsky H, Barnhart H, et al. Acute liver injury due to flavocoxid (Limbrel), a medical food for osteoarthritis: a case series. Ann Intern Med. 2012;156:857–60.
Acknowledgements
Flavocoxid was a kind gift from Primus Pharmaceuticals, Inc (Scottsdale, AZ, USA); Entecavir was kindly provided by Bristol-Myers Squibb. The study has been performed with Departmental funding assigned to Prof. Francesco Squadrito, Prof. Teresa Pollicino, and Prof. Giovanni Raimondo.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors who have taken part in this study declared that they do not have anything to disclose regarding funding or conflict of interest with respect to this manuscript.
Additional information
Responsible Editor: Mauro Teixeira.
Rights and permissions
About this article
Cite this article
Pollicino, T., Musolino, C., Irrera, N. et al. Flavocoxid exerts a potent antiviral effect against hepatitis B virus. Inflamm. Res. 67, 89–103 (2018). https://doi.org/10.1007/s00011-017-1099-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-017-1099-2