Abstract
Background
The recent development of effective immunotherapies with immune checkpoint inhibitors for the treatment of cancer has rekindled the interest for the immune system and its activation for an anti-cancer response. At the same time, it has become evident that not all types of cancers respond equally to these treatments, and even within the same tumor type only a subset of patients derive clinical benefit. Biomarkers predictive of response to immunotherapy have been sought and in certain occasions incorporated in the indication for treatment. These include expression of PD-L1 and defects in DNA mismatch repair (MMR).
Objective
Tumor mutation burden (TMB) has been associated with response to immune checkpoint inhibitors. The current investigation examines TMB as a biomarker of response to immunotherapy in breast cancer.
Patients and Methods
Publicly available data from the breast cancer study of The Cancer Genome Atlas (TCGA) and the METABRIC study were analyzed. Parameters examined included the TMB and specific mutations that may impact on TMB. In addition, correlations with breast cancer sub-types were investigated.
Results
The percentage of breast cancers with high TMB (more than 192 mutations per sample) was low (3.5–4.6%) in luminal and triple-negative cancers and higher (14.1%) in the HER2-positive subset. Almost all cancers with high TMB had defects in MMR proteins or the replicative polymerases POLE and POLD1.
Conclusions
Small sub-sets of breast cancers with high TMB exist and may present an opportunity for effective immunotherapeutic targeting.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Breast cancer is not very sensitive to targeted immunotherapy with immune checkpoint inhibitors due to its low immunogenicity. |
Subsets of breast cancer may be more immunogenic due to higher tumor mutation burden. |
Tumor mutation burden and other biomarkers that may correlate with sensitivity to immunotherapy in breast cancer will help the successful development of immunotherapy for this disease. |
1 Introduction
Recent successes with anti-cancer immunotherapy using immune checkpoint inhibitors targeting PD-1 or its ligand PD-L1 and CTLA-4 have created a need for the characterization of cancers that would be more probable to respond to these treatments through the discovery of predictive factors. Lead predictive factors characterized to date include the expression of PD-L1 in the tumor micro-environment and the characterization of the number of mutations that a tumor contains, either directly through measurement of the tumor mutation burden (TMB) or indirectly by determining the microsatellite status of the cancer, given that the great majority of microsatellite unstable cancers (MSI-H) contain a high TMB and respond to immunotherapy [1,2,3]. In fact, one of the immune checkpoint inhibitors, pembrolizumab, has received a tumor agnostic indication in any cancer with MSI-H status, based on the high response rate in these cancers [4]. Another immune checkpoint inhibitor, nivolumab, is indicated in combination with low-dose ipilimumb for MSI-H or mismatch repair-deficient (dMMR) colorectal cancer [5].
Tumor non-synonymous mutation burden and neo-antigen load is a predictor of response to ipilimumab in melanoma patients and to pembrolizumab in non-small-cell lung cancer patients [6,7,8]. In other tumor types, tumor mutation burden is also a predictor of response and progression-free survival [9]. An evaluation of these studies suggested that a critical cut-off of tumor mutation burden beyond which the response to therapy increases significantly exists and is situated at around 192 non-synonymous mutations [10].
Another parameter identified as critical for immunotherapy response to immune checkpoint inhibitors is the amount of immune cells infiltrating the tumor micro-environment and specifically tumor-infiltrating lymphocytes (TILs). T lymphocytes are the effectors of the immune attack and thus their absence from the tumor would make even tumors with a high tumor mutation burden less responsive or unresponsive to immune checkpoint inhibitors [11]. TILs are more frequently present in triple-negative and HER2+ breast cancers and are independently predictive of better outcomes, especially in triple-negative tumors [12].
Identification and validation of biomarkers of response to immune checkpoint inhibitors may pave the way for introducing these treatments in cancers that are not good candidates for treatment as a whole due to their low immunogenicity, but that may contain sub-sets with better prospects to response related to specific molecular lesions. As an example, colorectal cancers with MSI could be a group of responsive colorectal cancers among the majority of non-responsive colorectal cancers that are microsatellite stable [2]. Breast cancer is also not considered a good candidate for immune checkpoint therapy and overall trial results have been sobering. However, specific subsets that could respond better to such drugs may exist and remain to be better characterized [13]. The focus of clinical trials so far has been in the non-luminal subtypes with encouraging albeit variable results [14, 15]. The success story so far is with the PD-L1 inhibitor atezolizumab, which has recently gained US Food and Drug Administration (FDA) approval in combination with nanoparticle albumin-bound paclitaxel for patients with PD-L1 positive triple-negative metastatic breast cancer [14]. This article investigates characteristics and molecular correlates of breast cancers with high tumor mutation burden based on a series with publicly available data in order to identify clinical cases that could be the best candidates for immunotherapeutic interventions.
2 Methods
Cases included in the breast cancer genomic study of The Cancer Genome Atlas (TCGA) [16] were analyzed in the cBioCancer Genomics Portal (cBioportal, http://www.cbioportal.org) platform [17, 18] for number of mutations and other clinical and genomic characteristics of interest. The METABRIC breast cancer study [19] also included in the cBioportal was further considered in some analyses. The cBioportal platform allows for interrogation of each contained study for genetic lesions in any gene of interest. Clinical characteristics of cases with higher TMB and underlying mutations in critical genes (MSI-related, polymerases epsilon and delta, and others) possibly related to the pathophysiology of increased mutation burden were recorded in order to determine possible clinico-pathologic associations. Status of PD-L1 expression was investigated and molecular sub-type profiles of cases with increased expression were compared with those of normal expression.
Mutations of the MSI-associated genes and POLE and POLD1 genes associated with cases with high TMB were assessed using the mutation assessor server (mutationassessor.org), which assigns a prediction score of functional significance to each mutation derived from a multiple sequence alignment (msa) algorithm [20]. Additionally, information from the OncoKB database was integrated in mutation evaluation [21].
Survival of patients with high PD-L1 mRNA expression versus those with low PD-L1 mRNA expression was compared using the online tool Kaplan–Meier Plotter (kmplot.com) [22].
In analyses that could not be directly performed in the cBioportal platform, the primary clinical data of the breast cancer TCGA study were transferred to an Excel sheet (Microsoft Corp., Redmond, WA) for further analysis and calculations. Categorical and continuous data were compared using Fisher’s exact test or the χ2 test and t test, respectively. All statistical comparisons were considered significant with p < 0.05.
3 Results
In the TCGA breast cancer study, samples that contained more than 192 mutations per sample were sought for analysis. This cut-off was selected based on a previous study that had identified tumors with more than 192 mutations to be more responsive to immune inhibitors [10]. Among 1009 patient samples with data available in the TCGA breast cancer study, 47 patients (4.7%) had tumors with more than 192 mutations. Among sub-types, tumors categorized as HER2-positive had the higher percentage of cases with more than 192 mutations (14.1%, 11 cases of total of 78) (Fig. 1). The three other subtypes had significantly lower percentages of cases with more than 192 mutations. These percentages were 4.6% (nine of 197 samples) for luminal B breast cancers, 3.6% (18 of 499 samples) for luminal A cancers, and surprisingly an equally low percentage of 3.5% (six of 171 samples) for basal cancers (Fig. 1). These differences were statistically significant (χ2p < 0.001). Mean number of mutations was 72.2 in luminal A cancers (n = 499), 75.1 in luminal B cancers, 163.6 in HER2-positive cancers, and 96.5 in basal cancers. The mean number of mutations in cancers with ductal histology was 82.9 and in lobular carcinomas it was 86.6 (t test p = 0.8). The percentage of cases with more than 192 mutations was 4.1% and 6% in ductal and lobular carcinomas, respectively (Fisher’s exact test p = 0.25).
The METABRIC study investigators analyzed a total of 173 genes for mutations in breast cancer [19]. They also proposed a new classification that was based on a genomic analysis of copy number alterations and classified breast cancers in 11 integrative clusters (from 1 to 10 and the cluster 4 was subdivided into an ER + and an ER- sub-group). The integrative clusters corresponded very imperfectly to the five known intrinsic subtypes, with cluster 10 containing most triple-negative cases and cluster 5 containing several of the HER2-positive cases, with the rest of the clusters being a mix of luminal types mainly and some cases of other subtypes. The mean number of total mutations in the 173 genes examined in the 11 integrative clusters of METABRIC did not differ significantly, ranging from 5.1 to 6.1 with only cluster 6 having a somewhat lower mean number of mutations of 4.6. The percentage of cases with 10 or more mutations in the 172 genes in the 11 integrative clusters ranged from 10 to 13% in all clusters except for clusters 2 and 6, which had a lower number (5.6% and 3.5% of cases with 10 or more mutations; Fig. 2).
Next, breast cancers with the higher number of mutations (more than 1000 mutations per case) were evaluated for specific underlying mutations that could be associated with hypermutability. Seven cases in the TCGA breast cancer series (less than 1%) contained more than 1000 mutations each (Table 1). Six of these cases had mutations in one or more of the mismatch repair (MMR)-associated genes (MSH2, MSH6, MLH1, and PMS2) or the polymerases epsilon or delta (POLE and POLD1), which are associated with polymerase proofreading-associated polyposis (PPAP) [23]. The seventh case contained a homodeletion of MLH1. Among the seven cases, all sub-types, both ductal and lobular histologies, and a wide range of patient age were represented (Table 1). In five of the six cases with mutations, these mutations were considered as likely oncogenic by the oncoKB database. The sixth case contained a POLE mutation that was classified by oncoKB as having unknown significance.
An additional nine cases in the TCGA breast cancer study contained 500–1000 mutations. Among these, five cases had mutations in the MMR-associated genes or POLE and POLD1 (Table 2). All but one of these mutations are classified as of unknown oncogenic potential by the oncoKB database, and one is classified as likely oncogenic. However, three of them are classified by mutation assessor as having a high probability of functional interference with the normal respective protein function. The remaining four cases with 500–1000 mutations in the TCGA breast cancer study contained no MMR-associated genes or POLE and POLD1 mutations, but had mutations in other repair genes such as PARP1, BRCA2, PALB2, ERCC4, APOBEC3B, FANCA, and MUTYH (Table 3). In addition, mutations in the tumor suppressors TP53 and PTEN and the oncogene PIK3CA were present in two cases each.
Further evaluation of each MSI- and PPAP-associated polymerases genes as well as of the additional repair genes mentioned above and identified as mutated in some cases with high mutation burden was performed in order to characterize the patients’ mutational landscape and association with cases of different TMB in breast cancer. MSH2 was mutated in eight patients and these cases presented a wide range of total mutation burden from 32 to 4261, with a median of 155 (Table 4). Three of eight MSH2 mutated cases (37.25%) had a total number of mutations above 192. Table 5 summarizes these data and presents similar data for other repair genes. Table 5 also includes data for other genes that are part of the genetic panel of rare breast-cancer predisposing genes such as the Peutz-Jeghers syndrome predisposing gene STK11 (also known as LKB1), the polyposis coli gene APC, and the kinase CHEK2 gene, which senses DNA damage and activates the p53 pathway. Cases with mutations in all genes examined present with a wide range of total mutation burden (Table 5). A wide range of patient age is also observed and all subtypes are represented. Interestingly, among the 11 PARP1-mutated cases there were no luminal A cancers, while HER2 + and basal subtypes were over-represented with four and three cases, respectively. BRCA1 mutations, the most common mutations associated with genetic breast cancer predisposition, are commonly observed in the basal sub-type, which represented 48.1% of cases with BRCA1 mutations. BRCA1 mutations were observed in 7.6% of basal breast cancers, while 1.8%, 1.5%, and 2.5% of luminal A, luminal B, and HER2-positive cancers, respectively, harbored such mutations. Cases with mutations in BRCA1 were not associated with a particularly elevated total mutation burden (Table 5).
Several of the cases with the highest mutation burden possess several mutations in repair genes. Although the presence of these multiple mutations may be a chance effect due to the high number of total mutations that would increase the probability that any gene would be mutated, it may contribute to propagation of increased mutation burden by synergistically impeding DNA repair.
The wide range of total mutation number in cases with MSI- or PAPP-associated polymerases could be the result of differences in functional repercussions of each mutation due to their position in the protein product of the affected gene and due to the production of a truncated protein (insertions or deletions or nonsense mutations) versus a replacement of a single amino acid (missense mutations). To illustrate this point, among the eight breast cancer cases with MSH6 mutations, the two cases that contained nonsense mutations had a mutation burden of over 4000 mutations, while the remaining six cases with missense mutations in MSH6 had between 33 and 689 mutations (Table 6). Similarly, among the 15 cases in the TCGA breast cancer study with POLE mutations, the two cases with mutations producing frame-shifts had over 4000 mutations each, while all the rest with missense POLE mutations, but one, had a much lower number of total mutations, between 13 and 797 (Table 7). The single case with a missense POLE mutation and a high mutation burden of 5397 had the hot-spot P286R mutation, in addition to mutations in other repair genes, including two nonsense mutations in the APC gene deemed likely oncogenic in the OncoKB database. APC disabling mutations may activate the β-catenin pathway. Activation of β-catenin promotes an immune-suppressive tumor micro-environment and may contribute to immune evasion of tumors with such high tumor mutation burden.
The association of known oncogenes (PIK3CA) or tumor suppressors (TP53, CDH1, PTEN) commonly mutated in breast cancer with tumor mutation burden was investigated next. Among the 46 cases with more than 192 mutations in the TCGA breast cancer study, the number of cases with mutations in CDH1, TP53, PTEN, and PIK3CA genes were 16 (34.8%), 15 (32.6%), 13 (28.3%), and 25 (54.3%), respectively. These represented 12.5% (16 of 128 total CDH1 mutated cases), 4.4% (15 of 344 total TP53 mutated cases), 23.2% (13 of 56 total PTEN mutated cases), and 7.2% (25 of 345 total PIK3CA mutated cases) of the total cases with mutations in each of these commonly mutated genes in breast cancer (Fig. 3). The respective percentages for cases with more than 192 mutations in patients that were wild-type for each of these genes were 3.5% (30 of 868) for CDH1, 4.8% (31 of 652) for TP53, 3.5% (33 of 940) for PTEN, and 3.2% (21 of 651) for PIK3CA. These data suggest that mutations of CDH1, PTEN, and PIK3CA favor further mutation accumulation while TP53 mutation status does not affect the total mutation burden. However, only a minority of cases with mutations in the three other genes have a total mutation burden above the proposed critical level of 192 mutations (Fig. 3). Mutations of TP53 were observed in 89.5% of basal sub-type cancers, in 70.5% of HER2-positive cancers, 36% of luminal B sub-type, and 10.6% of luminal A cancers. This variability of distribution corroborates with the fact that TP53 mutation status does not affect the total mutation burden as luminal cancers and basal cancers that have similar burdens have quite different TP53 mutations prevalence.
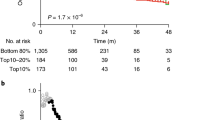
The PD-L1 gene was altered in 55 cases (6%) in the TCGA breast cancer study. Most lesions were amplifications (13 cases) or mRNA over-expressions (35 cases), while deletions and mutations of PD-L1 were very rare. The distribution of PD-L1 amplifications/over-expressions according to sub-type was six in luminal A cases (1.2% of total luminal A cases in the series), five in luminal B cases (2.5% of total luminal B cases), six in HER2-positive sub-type (7.7% of total HER2-positive cases), and 29 in basal sub-type (17% of total basal cases, χ2p < 0.001). PD-L1 lesions in the METABRIC study were present in 7% of samples. The respective percentages of PD-L1 amplifications and mRNA over-expression according to breast cancer sub-type in the METABRIC study were 2% for luminal A cancers, 3.2% for luminal B, 5.8% for HER2-positive cancers, 12% for basal sub-type and 13.8% for claudin-low cancers (χ2p < 0.001). Patients with higher mRNA expression of PD-L1 across breast cancer sub-types had a significant better relapse-free survival (RFS) than patients with lower mRNA expression of the immune ligand (p < 0.000, Fig. 4). This was also true when ER-positive/HER2-negative cancers and triple-negative cancers were analyzed separately (Figs. 5, 6). Although HER2-positive cancers with higher PD-L1 expression had a better RFS than counterparts with lower PD-L1 expression, this did not reach statistical significance, possibly due to lower numbers of analyzed tumors with this sub-type (Fig. 7).
Relapse-free survival (RFS) of HER2-positive breast cancer cases with higher than the mean or lower than the mean mRNA expression of PD-L1 (probe 227458_at). Cancers with high PD-L1 had a better RFS than cases with lower PD-L1, but the difference did not reach statistical significance (HR 0.71, LogRank p = 0.22)
4 Discussion
Mutations present in the genome of a cancer lead to the creation of neo-antigens that are a pre-requisite for recognition of a tumor cell by the host immune system as foreign. Thus, the higher the total mutation burden that a tumor possesses in general, the higher are the probabilities that neo-antigens will be created for possible presentation and recognition by the immune system. Indeed, TMB was shown in melanoma, non-small-cell lung cancer, and other cancers to be a predictor of response to immune checkpoint inhibitors [6,7,8,9]. In contrast, clinical correlates such as patient age, site of the primary, number of previous therapies, and LDH level were not associated with ipilimumab response [6]. Mutations in genes whose protein products are implicated in correction of DNA-replication errors such as in mismatch repair (MMR) or the replication polymerases epsilon and delta lead to increased mutation loads. Due to their high mutation burden, MMR or tumors with high microsatellite instability (MSI-H) have been the subject of a first approval of an anti-cancer drug agnostic of site of the tumor [4].
Immunotherapy has not been widely used in breast cancer, but several studies over the last few years show encouraging results [24]. Recently, the PD-L1 inhibitor atezolizumab in combination with nanoparticle albumin-bound (nab) paclitaxel was shown in a phase III trial to prolong progression-free survival (PFS) compared with nab-paclitaxel monotherapy in triple-negative metastatic breast cancer [14]. In the sub-group of patients expressing PD-L1, median overall survival (OS) was 25.0 months in the group of patients that received combination therapy and 15.5 months in the group of patients who received nab-paclitaxel. In a small phase II study, the PD-1 inhibitor nivolumab was also found to be more effective in metastatic triple-negative breast cancer when a short induction of doxorubicin or cisplatin chemotherapy preceded its administration [25]. Chemotherapy led to up-regulation of PD-L1 expression and inflammation-related genes. The PD-1 inhibitor pembrolizumab was studied as monotherapy in a phase Ib study and in different cohorts of a phase II study in triple-negative breast cancer in the first-line setting and as second or later line [26,27,28]. Median PFS was about 2 months in all three publications and median OS was 18 months in the first-line patients and 9–11 months in the later-line patients. The first-line phase II study included only PD-L1 positive patients [26]. The later-line phase II study allowed both positive (1% or higher) and negative for PD-L1 patients, and PFS, OS, and response rates were similar in the positive and negative cohorts [27]. One-year survival was 61.7% in the first line and 39.8% in second or later line, and several patients in each study had prolonged responses.
In breast cancer, the mutation burden is lower than in other cancers that are associated with mutagen exposures and are more responsive to immunotherapy, such as melanoma and non-small-cell lung cancer [29, 30]. In addition, MSI or MMR defects are uncommon in breast cancer [31], and this is confirmed in the TCGA breast cancer cohort. The current investigation used published genomic data to characterize the clinical and mutation landscape of hypermutated breast cancers, and attempted to define genetic lesions with hyper-mutability associations, beyond the known MSI-related and PPAP syndrome genes. Several conclusions can be drawn from the data presented. First, the rare breast cancers with very high mutation burden do have MSI-related and PPAP syndrome gene mutations. Second, most breast cancers with moderately high mutation burden level above the proposed critical of 192 mutations per tumor still have either mutations MSI/PPAP genes or other DNA repair genes. Third, the sub-type with the higher percentage of cases above this critical level is the HER2-positive groups. In contrast, triple-negative cancers have the same percentage of cases above the proposed critical level as luminal cancers. This concurs with a small study of 53 patients in which HER2-positive tumors had a higher TMB, while TMB was not significantly different in ER-positive tumors versus ER-negative ones and PR-positive tumors versus PR-negative tumors [32]. However, other investigations suggest that triple-negative cancers have the highest mutation burden among all sub-types, followed by HER2-positive sub-type [33].
Another conclusion of the current study is that p53 mutations do not correlate with increased mutations (above 192) compared to tumors with a wild-type p53. In contrast, mutations in the three other commonly mutated genes in breast cancer, CDH1, PTEN, and PIK3CA show an increase in the percentage of tumors with higher mutation load, which is more significant in the case of PTEN. Two scenarios may account for this association of mutations of these three genes (and any other gene) with an increased percentage of high tumor mutation burden tumors: They may be passenger mutations and be more commonly mutated randomly by chance in tumors with higher total number of mutations (i.e. a consequence of higher mutation number due to other causes) or contributors to this increase (i.e. having a causal relationship for example by being involved in DNA repair dysfunction similarly to MSI-associated genes) [34]. Although the second scenario is probable given the multiple mechanistic associations of oncogenes and tumor suppressors with DNA repair, the first scenario is also possible, especially if mutations in these genes are of unknown significance. For example, the only gene more commonly mutated than PIK3CA in patients with more than 192 mutations is the TTN gene encoding for the striated muscle protein titin, which is mutated in 36 out of 46 of those patients. Several cases have multiple alterations in TTN gene and all mutations are listed in oncoKB database as having unknown significance. Additional suggestions that these mutations are passenger and not contributing causally to mutation load despite their increasing frequency in cases with high TMB include the fact that the protein is not expressed in breast epithelium, the gene is very big, encoding for a protein of over 34,000 amino acids, and mutations are common in cases with less than 192 mutations (151 of these cases in the TCGA breast cancer study have one or more TTN mutations).
High PD-L1 expression and improved outcomes in breast cancer has been reported before and is associated with increased tumor infiltration by TILs [35]. Thus, PD-L1, besides being the target of inhibitors of the PD-L1/PD-1 pair and a predictor of response to these drugs at least in some settings, is also a prognostic factor associated with an inflamed tumor micro-environment [36]. This association may also be equally important for its value as a predictive factor for PD-L1/PD-1 inhibitors, denoting tumors with TILs ready to be activated.
The broad range of total mutation number in cases with lesions in the same causative MSI or PPAP polymerase gene suggests that the specific sites of the mutation or type of mutations (insertion/deletions, nonsense mutations versus missense mutations) have different repercussions. The variable immunogenicity of different mutation types has been described and may contribute to high responses of renal cell carcinomas, which possess a higher percentage of insertion/deletions, to immune checkpoint inhibitors, despite the lower TMB in these cancers [37].
A higher number of cases with the critical number of mutations in HER2-positive cancers may suggest a higher probability of response to immune checkpoint inhibitors in these cancers. This hypothesis has been tested in the recently presented PANACEA study [15]. This was a multicenter, non-randomized, phase Ib study with a phase II expansion examining treatment with trastuzumab in combination with pembrolizumab in patients with HER2-positive metastatic breast cancer that had progressed after at least one previous line of anti-HER2 therapy containing trastuzumab. Most patients (40 of the 52 in the expansion cohort) were positive for PD-L1. A modest response rate (RR) of 15% and disease control rate of 39% in PD-L1+ patients was observed [15]. Interestingly, this RR mirrors the percentage of HER+ patients with a tumor mutation burden above 192 in the TCGA breast cancer study.
Despite their interest as a predictive marker of immune checkpoint inhibitors, neither TMB nor MSI are perfect markers. Several reasons could account for a tumor with high TMB or MSI-H being resistant or less responsive to these drugs. In the case of MSI, some defects in the genes that are involved in MMR may not lead to MSI tumors. This phenomenon has been observed in ER-positive breast cancers with defects of the MutL complex (MLH1/3 and PMS1/2) that lead to endocrine resistance but not a hypermutated phenotype [38]. In addition, even if a high TMB exists neo-antigens need to be presented by the cell antigen presentation machinery in order to elicit an immune response [39, 40]. Cancers with a high TMB, independently of cause, may possess mutations in the antigen-processing and presentation machinery that renders neo-antigens invisible to the incoming immune cells [41, 42]. Tumors with a high TMB may also develop resistance to immune checkpoint inhibitors by mechanisms preventing immune cell infiltration or prevention of activation of TILs by creation of an immunosuppressive tumor micro-environment. Association of presence of neo-antigens with immunosuppressive tumor micro-environment as witnessed by the expression of CTLA-4, PD-1, and LAG-3 immune checkpoint molecules and indoleamine 2,3-dioxygenase 1 (IDO1) as well as immunosuppressive cytokine IL-10 has been documented in renal cell carcinomas and other cancers [43, 44]. Specific tumor signaling pathways such as the Wnt/β-catenin pathway and TGF-β pathway are associated with immune exclusion from the tumor micro-environment leading to so-called cold tumors that are unresponsive to immune checkpoint inhibitors [45, 46]. This may be relevant, for example, in breast cancers with CDH1 mutations such as lobular carcinomas, where E-cadherin absence favors β-catenin nuclear localization and activation of transcription [47]. Overall, the very existence of tumors with high TMB is interconnected with failed immunoediting either due to failure of neo-antigen presentation or paralysis/exhaustion of the immune system in the tumor micro-environment, both possible mechanisms of resistance to immune checkpoint inhibitor therapy. An additional piece of evidence to consider in the field of TMB as an immunotherapy predictive marker is that its value may vary depending on the specific immune checkpoint inhibitor considered. In patients treated with a combination of PD-1 inhibitors with CTLA-4 inhibitors, survival outcomes were better than outcomes of PD-1 inhibitors monotherapy independently of TMB, whence high TMB was predictive of better outcomes in monotherapy-treated patients [8]. These results suggest that high TMB is less critical for combination CTLA-4/anti-PD-1 response, at least in some patients, possibly because the more robustly activated immune system can attack the tumor effectively even in the presence of fewer neo-antigens.
As a conclusion, the TMB presents a valuable opportunity for clinical use as a predictive marker in breast cancer and high burdens are associated with breast cancers of all sub-types, often due to MMR or PPAP polymerases defects. A combined marker taking into consideration TMB, TILs presence, and immune receptors, such as PD-L1, expression may be the optimal predictor of immunotherapy response and should be prioritized in further studies [48].
References
Lee V, Murphy A, Le DT, Diaz LA Jr. Mismatch repair deficiency and response to immune checkpoint blockade. Oncologist. 2016;21:1200–11.
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–22.
Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res. 2016;22:813–20.
Lemery S, Keegan P, Pazdur R. First FDA approval agnostic of cancer site-when a biomarker defines the indication. N Engl J Med. 2017;377:1409–12.
Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36:773–9.
Van Allen E, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11.
Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99.
Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8.
Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–608.
Colli LM, Machiela MJ, Myers TA, et al. Burden of nonsynonymous mutations among TCGA cancers and candidate immune checkpoint inhibitor responses. Cancer Res. 2016;76:3767–72.
Maleki Vareki S. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer. 2018;6:157.
Stanton SE, Disis ML. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J Immunother Cancer. 2016;4:59.
Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34.
Schmid P, Adams S, Rugo HS, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379:2108–21.
Loi S, Giobbie-Hurder A, Gombos A, et al. Pembrolizumab plus trastuzumab in trastuzumab-resistant, advanced, HER2-positive breast cancer (PANACEA): a single-arm, multicentre, phase 1b-2 trial. Lancet Oncol. 2019;20:371–82.
Network Cancer Genome Atlas. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:269.
Pereira B, Chin SF, Rueda OM, et al. The somatic mutation profiles of 2433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479.
Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118.
Chakravarty D, Gao J, Phillips S, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017. https://doi.org/10.1200/po.17.00011.
Szász AM, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1065 patients. Oncotarget. 2016;7:49322–33.
Palles C, Cazier JB, Howarth KM, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2012;45:136–44.
Voutsadakis IA. Immune blockade inhibition in breast cancer. Anticancer Res. 2016;36:5607–22.
Voorwerk L, Slagter M, Horlings HM, et al. Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: the TONIC trial. Nat Med. 2019;25(6):920–8.
Adams S, Loi S, Toppmeyer D, et al. Pembrolizumab monotherapy for previously untreated, PD-L1-positive, metastatic triple-negative breast cancer: cohort B of the phase II KEYNOTE-086 study. Ann Oncol. 2019;30:405–11.
Adams S, Schmid P, Rugo HS, et al. Pembrolizumab monotherapy for previously treated metastatic triple-negative breast cancer: cohort A of the phase II KEYNOTE-086 study. Ann Oncol. 2019;30:397–404.
Nanda R, Chow LQM, Dees EC, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 study. J Clin Oncol. 2016;34:2460–7.
Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9.
Hayward NK, Wilmott JS, Waddell N, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–80.
Davies H, Morganella S, Purdie CA, et al. Whole-genome sequencing reveals breast cancers with mismatch repair deficiency. Cancer Res. 2017;77:4755–62.
Xu J, Guo X, Jing M, Sun T. Prediction of tumor mutation burden in breast cancer based on the expression of ER, PR, HER-2 and Ki-67. Oncotarget Ther. 2018;11:2269–75.
Narang P, Chen M, Sharma AA, et al. The neoepitope landscape of breast cancer: implications of immunotherapy. BMC Cancer. 2019;19:200.
Van de Haar J, Canisius S, Yu MK, et al. Identifying epistasis in cancer genomes: a delicate affair. Cell. 2019;177:1375–83.
Schalper KA, Velcheti V, Carvajal D, et al. In situ tumor PD-L1 mRNA expression is associated with increased TILs and better outcome in breast carcinomas. Clin Cancer Res. 2014;20:2773–82.
Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51.
Turajlic S, Litchfield K, Xu H, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 2017;18:1009–21.
Haricharan S, Punturi N, Singh P, et al. Loss of MutL disrupts CHK2-dependent cell-cycle control through CDK4/6 to promote intrinsic endocrine therapy resistance in primary breast cancer. Cancer Discov. 2017;7:1168–83.
Voutsadakis IA. Proteasome expression and activity in cancer and cancer stem cells. Tumour Biol. 2017;39:1010428317692248.
Stratikos E. Modulating antigen processing for cancer immunotherapy. Oncoimmunology. 2014;3:e27568.
Voutsadakis IA. Polymerase epsilon mutations and concomitant β2-microglobulin mutations in cancer. Gene. 2018;647:31–8.
Grasso CS, Giannakis M, Wells DK, et al. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov. 2018;8:730–49.
Matsushita H, Sato Y, Karasaki T, et al. Neoantigen load, antigen presentation machinery, and immune signatures determine prognosis in clear cell renal cell carcinoma. Cancer Immunol Res. 2016;4:463–71.
Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61.
Luke JJ, Bao R, Sweis RF, et al. WNT/β-catenin pathway activation correlates with immune exclusion across human cancers. Clin Cancer Res. 2019;25:3074–83.
Mariathasan S, Turley SJ, Nickles D, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–8.
Voutsadakis IA. Pathogenesis of colorectal carcinoma and therapeutic implications: the roles of the ubiquitin-proteasome system and Cox-2. J Cell Mol Med. 2007;11:252–85.
Vonderheide RH, Domchek SM, Clark AS. Immunotherapy for breast cancer: what are we missing? Clin Cancer Res. 2017;23:2640–6.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No external funding was used in the preparation of this article.
Conflict of interest
Ioannis A. Voutsadakis declares that he has no conflicts of interest that might be relevant to the contents of this article.
Rights and permissions
About this article
Cite this article
Voutsadakis, I.A. High Tumor Mutation Burden and Other Immunotherapy Response Predictors in Breast Cancers: Associations and Therapeutic Opportunities. Targ Oncol 15, 127–138 (2020). https://doi.org/10.1007/s11523-019-00689-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11523-019-00689-7