Abstract
Silicon (Si) confers several benefits to many plant species when absorbed as silicic acid through nodulin 26-like intrinsic proteins (NIPs). The NIPs belong to major intrinsic protein (MIP) family, members of which form channels with high selectivity to control transport of water and different solutes. Here, comparative genomic analysis of the MIPs was performed to investigate the presence of Si transporter MIPs in soybean. Thorough analysis of phylogeny, gene organization, transcriptome profiling and protein modeling was performed to characterize MIPs in rice, Arabidopsis and soybean. Based on several attributes, two putative Si transporter genes, GmNIP2-1 and GmNIP2-2, were identified, characterized and cloned from soybean. Expression of both genes was detected in shoot and root tissues, and decreased as Si increased. The protein encoded by GmNIP2-2 showed functionality for Si transport when expressed in Xenopus oocytes, thus confirming the genetic capability of soybean to absorb the element. Comparative analysis of MIPs in plants provides opportunities to decipher gene evolution, functionality and selectivity of nutrient uptake mechanisms. Exploitation of this strategy has helped to uncover unique features of MIPs in soybean. The identification and functional characterization of Si transporters can be exploited to optimize the benefits that plants can derive from Si absorption.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Silicon (Si), a major constituent of the earth’s crust, is the second most abundant element in the soil (Epstein 1994; Richmond and Sussman 2003). Long considered non-essential for plant growth, it is now gaining more attention by plant biologists because of its reported dynamic roles in alleviating biotic and abiotic stresses (Fauteux et al. 2005; Guntzer et al. 2012). Plants absorb Si in the form of silicic acid and accumulate from 0.1 to 10 % of the dry mass depending on the plant species (Epstein 1994). On the basis of percent Si accumulation, plant species have been categorized as accumulators (>1 %) or excluders (Guntzer et al. 2012). In general, monocots and primitive plant species are considered good accumulators in contrast to dicots that are mostly excluders (Montpetit et al. 2012; Guntzer et al. 2012). The Si influx into plants is mediated through nodulin 26-like intrinsic proteins (NIPs), which are family members of major intrinsic proteins (MIPs; Ma et al. 2006).
MIPs play an important role in many organisms since they form transmembrane channels that facilitate and discriminate transport of different solutes across the membrane. Most plant MIPs have water transport ability and are therefore commonly referred as aquaporins (Maurel et al. 2008). MIPs have been classified in five major classes including NIPs, which constitute a diverse subfamily possessing very high selectivity for different solutes. Back in 1987, a study conducted to analyze proteins associated with symbiotic nitrogen fixation in soybean led to the identification of the first NIP gene (Fortin et al. 1987). This discovery served as the basis for uncovering several other NIPs in different plant species. The NIPs can be classified into three distinct groups on the basis of similarity at the aromatic/arginine (Ar/R) constriction and selective filters since they dictate solute specificity (Mitani et al. 2008). The NIP functional classes, namely NIP1, NIP2 and NIP3, have a very diverse range of substrate specificities. NIP1 was found to be more permeable to water and glycerol (Dean et al. 1999), whereas NIP3 shows transport activity of relatively larger molecules like urea and formamide (Wallace and Roberts 2005). The NIP2 subgroup can transport Si, a rare feat among living organisms since Si transporters had been reported previously only in diatoms (Ma and Yamaji 2006). In addition, the NIP2 subgroup appears to be specific to high Si absorbing plant species. For instance, most of the NIP2 genes characterized for Si absorption have been cloned from monocots or lower plant species (Ma and Yamaji 2006; Montpetit et al. 2012; Grégoire et al. 2012). To date, among dicots, only members of the Cucurbitaceae family have been found to have a Si transporting NIP2 gene (Mitani-Ueno et al. 2011). Therefore, our information about the functional, structural and evolutionary dynamics of NIP2s is lacking in dicots in general, and more particularly in leguminous plant species such as soybean where Si absorption has recently been shown to provide great benefits (Shen et al. 2010; Arsenault-Labrecque et al. 2012; Table S1).
The three-dimensional (3D) structure of proteins is very important to understand their molecular mechanisms. As a matter of fact, the 3D structure is often more highly conserved among homologs than the amino acid sequence (Kaczanowski and Zielenkiewicz 2010). In this regard, the 3D structures of several MIPs modeled at high resolution have great importance (Gonen et al. 2004; Harries et al. 2004; Lee et al. 2005; Newby et al. 2008). The available 3D structures of MIPs have revealed several unique features that are conserved. All MIPs have a structure resembling an hour glass formed by six transmembrane (TM) helices (H1 to H6) separated by five loops (A to E), two conserved NPA domains present in half-helices, and an Ar/R selectivity filter. Both the NPA domains and the Ar/R filter form the constriction that will dictate the selectivity of the MIP. The Ar/R selectivity filter is composed of four amino acids present in each of the transmembrane domains H2 and H5, and two more in loop E (LE) (Lee et al. 2005; Newby et al. 2008). The information about MIP attributes can be used strategically to identify novel homologs in diverse plant species.
In some plant species, a wide range of genes coding for MIPs have been identified, amounting to 35 in Arabidopsis, 33 in rice and 28 in grapes (Quigley et al. 2002; Sakurai et al. 2005; Fouquet et al. 2008). In a recent study, Zhang et al. (2013) have identified 66 MIPs, including 13 NIPs, in the soybean genome although their analysis did not identify putative Si transporters. The MIP information available in rice and Arabidopsis can be extremely valuable to identify specific features required for the Si permeability since these are model species described, respectively, as a strong accumulator and excluder of Si (Ma and Yamaji 2006; Montpetit et al. 2012). Thus, it is possible to perform comparative genomics among these species in order to identify whether soybean has the genetic ability to uptake Si.
In this study, we have characterized and compared MIP genes identified in rice, Arabidopsis and soybean. The detailed analysis has led to the identification of new MIP genes in soybean, and revealed several unique features inherent to soybean MIPs including the presence of Si transporters and their functionality, the first such report for a leguminous species.
Materials and methods
Identification of major intrinsic proteins in soybean
The Glyma1.0 chromosome-scale annotated assembly of the soybean genome was retrieved from the phytozome database (www.phytozome.net, Schmutz et al. 2010). A local database of transcript and protein sequences of soybean genes was created using NCBI command-line BLAST utilities in BioEdit (Version 7.0.9.0; Hall 1999). The putative MIP genes were identified in the local database with BLASTp using 70 MIP genes as query sequences (Table S2). These included 35 rice and 33 Arabidopsis aquaporins, which represent most of the MIP genes characterized to date in monocots and dicots, respectively (Quigley et al. 2002; Sakurai et al. 2005). In addition, two XIP aquaporins, one each cloned from lotus and tomato were also used as query sequences (Bienert et al. 2011; Giovannetti et al. 2012). An e-value of 10−5 was used as an initial cut-off to claim significant matches. Then, the BLAST output was tabulated and top hits on the basis of bit scores were selected. BLAST hits with less than a 100 bit-score were removed.
Evaluation of NPA motifs and transmembrane domains
The NPA motifs were identified in protein sequences using NCBI’s Conserved Domain Database (CDD, www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Transmembrane domains in the genes identified by the BLAST search were detected using TMHMM, SOSUI and TOPCONS software tools (www.cbs.dtu.dk; http://bp.nuap.nagoya-u.ac.jp; Bernsel et al. 2009). The results were then manually examined for altered and/or missing transmembrane domains.
Classification and phylogeny of soybean MIPs
Multiple alignments of MIP sequences were performed using CLUSTALW as implemented in MEGA5 (Kumar et al. 2008). The phylogenetic tree was constructed using the neighbor-joining method and the stability of branch nodes was measured by performing 1,000 bootstraps. The subgroups PIP, TIP, NIP, and SIP formed in the phylogenetic tree were classified in accordance with the nomenclature used for the Arabidopsis and rice MIPs (Quigley et al. 2002; Sakurai et al. 2005). XIPs were assigned on the basis of sequence similarity with lotus and tomato XIPs (Bienert et al. 2011; Giovannetti et al. 2012). A phylogenetic tree of Arabidopsis, rice, and soybean MIPs was also constructed.
Homology modeling of soybean MIPs
All the soybean MIP sequences were submitted to the Phyre2 protein-modeling server (www.sbg.bio.ic.ac.uk/~phyre2). Model quality was assessed based on the z-score calculated using the Prosa server (Wiederstein and Sippl 2007).
Conserved motif identification
Motifs conserved in the MIPs were identified using the ‘Multiple EM for Motif Elicitation’ (MEME) program (Bailey et al. 2006). The default settings (minimum width 6 and maximum width 50 amino acid motifs) were used. The final output of MEME was manually evaluated.
Expression profiling of MIPs using Illumina transcriptome sequence and microarray data
Transcriptome sequence data (Illumina Genome Analyzer II) generated by Severin et al. (2010) for 14 different tissues including leaf, flower, pod, two stages of podshell, root, nodule and seven stages of seed development were used to analyze expression profiles of soybean MIP genes. Hierarchical clustering of expression data was performed using Cluster 3.0 (de Hoon et al. 2004). Average linkage method provided in Cluster 3.0 was used to cluster gene and tissue types.
Affymetrix GeneChip expression data available in the Genevestigator database were also used for expression profiling of MIPs in Arabidopsis, rice, and soybean (www.genevestigator.com). The gene expression data available at Genevestigator were retrieved using phytozome Ids for soybean, TIGR Ids for rice and TAIR Ids for Arabidopsis.
Plant material
Soybean plants were grown in a greenhouse maintained at a constant temperature (25 ± 2 °C) and light for a photoperiod of 16 h a day. Seeds of soybean cultivars Jack, Williams 82 and Hikmoksorip were surface sterilized using 2 % sodium hypochloride treatment for 5 min followed by three subsequent washes with distilled water. Plants were grown in hydroponic conditions in Hoagland solution supplemented with or without 1.7 mM Si (pH 7.0) in the form of potassium silicate (Kasil #6, 23.6 % SiO2; National Silicates, Quebec, QC, Canada).
Silicon quantification in plant tissue
Leaf samples of 1-month-old plants were harvested and then dried at 65 °C for 24 h. Dried samples were ground to a fine powder using a mixer mill and compressed into pellets of 5-mm thickness and 13-mm diameter. Pellets were used for Si measurement using a Niton XL3t900 GOLDD XRF Analyzer (Thermo Scientific NITON, Billerica, MA, USA). The material was prepared as previously described by Reidinger et al. (2012). For calibration of the instrument, a synthetic methyl cellulose spike (Sigma-Aldrich Canada Ltd. Oakville, ON, Canada) was used. A matrix of spiked methyl cellulose powder with different percentages of silica powder (Fisher Scientific, Montréal, QC, Canada) was prepared and used to draw a standard curve for calibration. Signal intensity in kilo counts per second (kcps) was obtained for each sample in three replicates and converted into percentage using the calibration curve.
RNA extraction and quantitative RT-PCR
Total RNA was extracted from soybean root and shoot tissues using a combined Trizol/Qiagen RNeasy mini kit (Qiagen, Mississauga, ON, Canada). First strand cDNA from 2 μg of total RNA was synthesized by using Superscript III reverse transcriptase (Invitrogen Canada Inc., Burlington, ON, Canada) and an Oligo(dT) primer. Primers for quantitative reverse transcription PCR (qPCR) were designed using Primer3 (Table S3; http://frodo.wi.mit.edu). For expression analysis, quantitative RT-PCR was performed using standard cycler conditions:initial step of 94 °C for 2 min, followed by 30 cycles of 94 °C for 30 s, 65 °C for 30 s, 72 °C for 30 s and a final extension at 72 °C for 5 min. A soybean actin gene (XP_003547582) was used as reference to normalize expression levels across the tissues.
Cloning of soybean NIP2 gene subfamily
The coding sequences (CDS) of the soybean GmNIP2-1 and GmNIP2-2 genes were amplified by Phusion Taq polymerase (New England Biolabs, Whitby, ON, Canada) using root cDNA as a template. Amplified CDS were first cloned in pUC18 plasmid vector. Then, the clones were sequenced and confirmed for accuracy of reading frame. To facilitate the heterologous expression of GmNIP2-1 in Xenopus laevis oocytes, the CDS was further cloned in the Pol1 plasmid vector, which was derived from pGEMT and contains the T7 promoter, untranslated regions (UTR) of the X. laevis globin gene and a poly(A) sequence (Caron et al. 2000). For directional cloning of the CDS, forward and reverse primers with unique restriction sites (underlined) at their 5′ ends were designed (GmNIP2-2-EcoRI-F AGTGGAATTCATGGAGGGGACCACCAGCC and GmNIP2-2-XbaI-R AGTTCTAGACCTATCACACCAAGCATCTTTG). The resulting amplicons were ligated with linearized Pol1 vector and cloned in Escherichia coli TOP10 (Invitrogen, Carlsbad, CA, USA).
Heterologous expression assay in Xenopus laevis oocytes
Pol1-GmNIP2-2 plasmid DNA was extracted from freshly grown Escherichia coli cultures using a QIAprep Spin Miniprep kit (Qiagen, Mississauga, ON, Canada). Plasmid was linearized with NheI (Roche Applied Science, Laval, PQ, Canada) and 1 μg of plasmid DNA was used to synthesize complementary RNA (cRNA). In vitro transcription was performed using the mMESSAGEmMACHINE T7 ULTRA kit (Applied Biosystems/Ambion, Austin, TX). Resultant cRNA was purified and dissolved in ultrapure DNase-/RNase-free distilled water.
Oocytes at stages V or VI were extracted from adult female X. laevis frogs. The defolliculated oocytes were injected with 25 nl of 500 ng/nlcRNA or equal volume of H20 as negative control. Then oocytes were incubated at 18 °C in Barth’s (MBS) medium (88 mM NaCl, 1 mM KCI, 2.4 mM NaHCO3, 0.82 mM MgSO4, 0.33 mM Ca(NO3)2·4H20, 0.41 mM CaCl2, 15 mM HEPES, pH 7.6) supplemented with 100 μM each of penicillin and streptomycin. After 3 days of incubation, pools of 10 oocytes for each condition were exposed for 1 h to MBS solution containing 1.7 mM Si. After exposure, oocytes were rinsed in solution containing 0.32 M sucrose and 5.0 mM HEPES (pH 7.4). For quantification of intracellular silicon, 25 μl of concentrated nitric acid were added into each pool of 10 oocytes then dried for 2 h at 82 °C. To dissolve dried oocyte samples, 100 μl of plasma grade water were added and samples were placed at room temperature for 60 min. Thereafter, the samples were briefly mixed and centrifuged at 13,000g for 5 min. Using 10 μl of the resultant supernatant, Si measurement was performed with a Zeeman atomic spectrometer AA240Z (Varian, Palo Alto, CA, USA) equipped with a GTA120 Zeeman graphite tube atomizer. Analysis of the spectrometer observance data was performed using JMP 9.0.2 (SAS Institute Inc.).
Results
Major intrinsic proteins (MIPs) in the soybean genome
Initially, 84 MIP genes were identified in the soybean genome (Table S4). Functional annotation based on similarity with known proteins available at Pfam also confirmed the genes as MIPs (Table S5). The phylogenetic tree of these genes revealed five distinct subgroups that were named according to their similarity with Arabidopsis and rice genes (Fig. S1, Table S4). Of the 84 soybean MIP genes, only 53 showed the typical presence of six transmembrane domains considered as a required attribute of aquaporins (Table S6). Incidentally, only 58 out of the 70 MIPs reported in Arabidopsis, rice, tomato and lotus bore six transmembrane domains (Table S7).
Some of the MIP genes initially identified in soybean were found to code for truncated or altered protein sequences. Therefore, to remove defective genes, multiple sequence alignments of individual subfamilies of MIPs were manually analyzed. In the end, 72 MIPs including 22 plasma membrane intrinsic proteins (PIPs), 23 tonoplast intrinsic proteins (TIPs), eight small intrinsic proteins (SIPs), two uncharacterized intrinsic proteins (XIPs) and 17 NIPs were sorted on the basis of multiple alignment (Table 1). The number of genes in most of the sub-families was comparable to that recently reported by Zhang et al. 2013, with the notable exception of a higher number of NIPs (17 vs. 13) and SIPs (8 vs. 6) observed in the present study (Fig. 1, Fig. S2).
Phylogenetic tree representing different groups of major intrinsic proteins (MIPs) identified in the soybean genome, compared with rice and Arabidopsis MIPs. The genes from Arabidopsis, rice, and soybean are indicated with the prefixes At, Os, and Gm, respectively. In the XIP subfamily, genes with prefixes Lj and Sl are from lotus and tomato, respectively
Phylogenetic analysis of the soybean PIP genes along with Arabidopsis and rice PIPs revealed two major subgroups (Fig. 1). The gene structure of PIPs was found to be conserved across Arabidopsis, rice, and soybean (Fig.S3). All the PIPs have two conserved NPA domains and an Ar/R selective filter (FHTR). The amino acids forming the Ar/R selective filter in PIPs are highly hydrophilic, which suggests its suitability for water transport (Fig. S4). In the case of TIPs, 23 genes separated in five distinct subgroups similar to rice and Arabidopsis were found in soybean (Fig. 1). Unlike PIPs, variation at the Ar/R filter positions exists among the TIP subfamilies (Fig. S5, Table 1). The Ar/R filter amino acids are more hydrophobic in TIP1s as compared to the rest of the TIP subgroups.The SIPs formed two distinct groups, SIP1 and SIP2, whereas XIPs were grouped in a single cluster (Fig. 1). It seems that soybean SIP and XIP genes present a wider range of gene length than their homologs in rice and Arabidopsis (Fig. S3b). In SIP1s, the NPA domain in loop B was comparatively less conserved than the NPA in loop E (Fig. S6a, Table 1). In SIP1s, the residues forming the Ar/R filter were hydrophobic, whereas in SIP2s they were hydrophilic. The XIPs also have hydrophobic amino acids composing the Ar/R filter and deformed NPA domains as described above in SIPs (Fig. S6b, Table 1).
Nodulin 26-like intrinsic protein (NIP)
The 17 NIP genes present in the soybean genome clustered into five distinct groups (Fig. 1). The NIP1 subgroup contains the largest number of genes (nine), followed by NIP3 (three), NIP2 and NIP4 (with two each) and NIP5 (one gene). The NIP genes contain a greater number of exons compared to other MIPs (Fig. S3b). Sequence alignment of soybean NIPs revealed a high level of sequence variation in the loops and terminal regions (Fig. S7). All the NIP subfamilies have conserved NPA motif in loops A and B except for the NIP3s, which have a NPV motif instead of NPA in loop B. NIP subgroups 1, 2, and 4 have more hydrophobic amino acids at Ar/R filter position, whereas NIP3s and NIP5 have hydrophilic amino acids at Ar/R filter position.
Conserved motifs, phosphorylation sites, and sub-cellular localization of soybean MIPs
A total of eight conserved motifs were identified in MIPs from Arabidopsis, rice, and soybean using the MEME search tool (Fig. S3c, Fig. S8, http://meme.nbcr.net). The two most significant motifs possess NPA domains that bear characteristic features of MIPs. Among these, the motif with NPA domain present in loop E was found in 136 out of 142 MIPs analyzed (Fig. S8). The second motif with NPA, present in loop B, is less conserved and was observed in 126 MIPs. The longest motifs (3 and 4) were almost exclusive to PIPs. Exceptionally, motifs 3 and 4 were also observed in a few TIPs (Fig. S3c). The relatively small motif 8 was exclusively present in PIPs and has four highly conserved glycine residues.
Motifs 2, 3 and 4 have very highly conserved serine residues. These were predicted as probable sites for phosphorylation (Table S8). The greatest number of serine potential phosphorylation sites was predicted in the NIP subfamily followed by PIPs, with an average of 8.5 and 8.1 sites per gene, respectively. TIPs and SIPs have an average of 4.3 and 2.7 serine potential phosphorylation sites per gene, which is considerably less compared to PIPs and NIPs.
A wide range of sub-cellular localizations for soybean MIPs was predicted (Table S9). All soybean PIPs were predicted to be located in the plasma membrane (Table S9). By contrast, only five out of 23 soybean TIPs were predicted to be localized in the plasma membrane. The majority of TIPs (nine) were targeted to the cytoplasm and vacuoles (six). The SIPs were located either in the plasma membrane or vacuoles, and XIPs in the cytoplasm. In the case of NIPs, most of the genes appeared to be associated with the plasma membrane.
Expression profile of soybean MIPs
Analysis of Illumina transcriptome data revealed diverse patterns of MIP gene expression in different soybean tissues (Fig. 2). None of the tissue types exhibited an exclusive expression of a particular MIP subfamily. For instance, in developing seeds, expression of genes belonging to almost all MIP subfamilies was observed. On the other hand, expression of some of the subgroups was highly specific, such as TIP3s, which were specifically expressed in developing seeds (Fig. 2). Interestingly, a large switch in gene expression was observed between different stages of seed development. The expression profiles of genes during early and late stages of seed development were found to be clustered in separate groups (Fig. 2).
As a complement, microarray expression data from hundreds of independent studies were also exploited. Hierarchical clustering of microarray expression data generated by Genevestigator (www.genevestigator.com) revealed typical expression patterns for MIP genes across different soybean tissues (Fig. S9). As with the Illumina analysis, seed-specific expression of the soybean TIP3 subfamily was also observed in microarray data. Similarly, seed-specific expression of TIP3 was also observed in rice and Arabidopsis (Fig. S10, Fig. S11). For the most part, expression levels of tandem-repeated genes and genes with a high level of similarity were found to be quite comparable in the various tissues.
GmNIP2-1 and GmNIP2-2 have the characteristics of a bona fide silicon transporter
The predicted protein sequences revealed several features of GmNIP2-1 and GmNIP2-2 that suggested they might code for Si transport (Fig. 3). To supplement in silico predictions, the coding sequences of GmNIP2-1 and GmNIP2-2 were cloned and revealed open reading frames of 882 and 888 bp, respectively. The predicted protein sequences exhibited 93 % homology and most of the variation was observed in the terminal region (Fig. 3). Homology modeling of GmNIP2-1 and GmNIP2-2 proteins formed a highly similar 3D structure (Fig. 4a). Two constrictions were predicted to form at the center of the pore, one involving the two NPA domains and the other Ar/R selective filter formed by four amino acids (GSGR). Zhang et al. (2013) have also observed the GSGR selective filter in the NIP2 proteins.
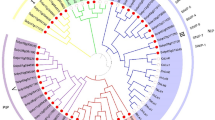
Characteristic features and experimental validation for silicon (Si) permeability of NIP2s in soybean. a Similar type of homology based on 3-D structure of GmNIP2-1 and GMNIP2-2 genes showing position of G-S-G-R selective filter amino acids and superimposition of both structures showing no variation in pore. b The Si permeability of GmNIP2-2 gene revealed in Xenopus oocyte assay. c Accumulation of Si observed in leaves of soybean plants grown with and without Si. d Expression profile of GmNIP2-1 and GmNIP2-2 genes revealed by quantitative RT-PCR in plants grown with and without Si. e Phylogenetic tree showing evolutionary relation of soybean NIP2s with known Si transporter genes from different plant species
Protein sequence alignment of soybean NIP2s with previously identified Si-competent NIPs revealed very little sequence conservation in the C- and N-terminal regions (Fig. 3). However, not a single gap was detected in the aligned sequences within the transmembrane regions and loops present between the transmembrane regions. The amino acid sequences in both NPA domains and Ar/R selective filter are highly conserved. Another important characteristic sequence known as Froger’s residues (P1–P5) was also conserved among NIP2s (Froger et al. 1998).
Silicon permeability, accumulation and expression of GmNIP2-1 and GmNIP2-2
In order to determine the functionality of GmNIP2, GmNIP2-2 was expressed in X. laevis oocytes and these were tested for their permeability to Si. GmNIP2-2 expression resulted in a significant increase in Si absorption in X. laevis oocytes, confirming the function of GmNIP2-2 as a Si transporter as predicted in silico (Fig. 4b). In addition, soybean plants grown in hydroponic conditions with Si supply accumulated 2.5 times more Si in leaf tissue than the control plants grown without Si (Fig. 4c). To verify if Si availability influenced the expression of soybean NIP2s, qRT-PCR was performed on roots of soybean plants grown with or without Si supply. The data revealed a significant decrease in gene expression for GmNIP2-1 and GmNIP2-2 (43 and 34 %, respectively) in plants grown in presence of Si (Fig. 4d). The soybean NIP2s forms a separate group from the known Si transporters (NIP2s) of non-legume dicot and monocots (Fig. 4e).
Sequence conservation and uniform expression of NIP2s in soybean
The coding DNA sequences of GmNIP2-1 and GmNIP2-2 were found to be conserved among three different soybean cultivars, Jack, Hikmoksorip, and Williams 82. However, when grown in the presence of Si, these cultivars accumulated varying concentrations of Si with Hikmoksorip absorbing as much as six times more Si than Williams 82 (Table 2). This differential property did not appear to be linked with a differential activity of NIP2s since both GmNIP2-1 and GmNIP2-2 had similar expression levels across all three cultivars (Fig. S12).
Discussion
Si absorption in plants was poorly understood for many years until the discovery that it was under the control of specific influx and efflux transporters (Ma et al. 2006, 2007). The influx transporters represent the first barrier to Si uptake and determine if a given plant has the ability to absorb Si. Soybean has received limited attention for its Si accumulating potential despite the fact that several recent reports have highlighted the benefits it derives from Si uptake (Shen et al. 2010; Arsenault-Labrecque et al. 2012; Table S1). Given that all Si influx transporters identified to date belong to the large family of MIPs, we report here a characterization of soybean Si transporters identified through genome mining and comparative analysis of MIPs in soybean along with rice and Arabidopsis data. Our results corroborate closely those of Zhang et al. (2013) who found 66 MIPs in soybean. The disparity lies mostly in our identification of 17 NIPs compared to 13 in the previous study. Incidentally, we were mostly interested in soybean NIPs as a way to determine if soybean contained Si transporters, based on homology with previously reported NIP2s, the only known plant aquaporins that display substrate selectivity for Si in superior plants (Ma et al. 2006; Ma and Yamaji 2006). We were thus able to identify two genes, GmNIP2-1 and GmNIP2-2, characterized as Si transporters, the first such report for a leguminous species.
Comparison among Arabidopsis, rice, and soybean MIPs
As in the work by Zhang et al. (2013), we found more than twice as many MIPs in soybean compared to rice or Arabidopsis. This is most likely because of the very recent genome duplication event in the evolution of soybean compared to rice and Arabidopsis (Roulin et al. 2013 ). Moreover, duplicated gene pairs are found to be expressed in all three species (Fig. S9–S11). This is mostly because of the interdependency of MIPs for their functionality, which also leads to an increased number of family members. Recently, such interdependency of PIP1 and PIP2 has been reported in rice (Matsumoto et al. 2009). The rice PIP1 and PIP2 genes have been found to result in membrane permeability only when expressed together in Xenopus oocyte experiments (Matsumoto et al. 2009). However, some of the MIP subgroups show very limited duplication events that might have either recent origin or vanished due to dose effects. It can be correlated with the fact that the higher absorption of some minerals is toxic to plant and MIPs permeable for such mineral might be lethal. For instance NIP genes are known to transport boric acid, arsenic, germanium and Si and NIPs have very little duplication compared to other subfamilies.
Patterns of MIP distribution observed in the present study are fairly similar to previous reports in rice and Arabidopsis (Quigley et al. 2002; Sakurai et al. 2005). The formation of comparable subgroups in the subfamilies PIP, TIP, NIP and SIP leads one to conclude that the common ancestor of monocots and dicots also contained these four subfamilies. Since XIPs are missing in rice and Arabidopsis, it suggests that they have evolved later in some dicot plant families including legumes. In the case of NIP subfamilies, NIP2s and NIP4s were found to be missing in Arabidopsis and rice, respectively, probably being lost after the initial duplications in a common ancestor. As a result, the nomenclature based on Arabidopsis may be limiting and should rely more on phylogenetic clustering to accommodate additional NIP groups that may arise as more plant genomes are being sequenced (Sonah et al. 2011).
Complex arbitrary pattern of MIP expression
Most of the MIPs showed ubiquitous expression in soybean. Genes of the same MIP subfamily were found to be expressed in different organs and did not exhibit specific expression patterns. This suggests that MIP organization may have specificity at the cellular or tissue level rather than at the organ level. However, some genes from the TIP3 subgroup were found to be expressed specifically in developing seeds (Fig. 2). Such specificity for different organs was also observed in some of the TIPs in rice and Arabidopsis (Fig. S10, S11). The TIP3s have also been reported to be specific to seed maturation and the early phases of germination in Arabidopsis (Gattolin et al. 2011).
Silicon absorption in soybean
The identification of two Si influx transporters in soybean in this study confirm the ability of the species to absorb the element and bring a scientific rationale to the previously reported beneficial effects of Si amendment in disease resistance and abiotic stress in soybean (Shen et al. 2010; Arsenault-Labrecque et al. 2012; Table S1). By using a comparative approach with a Si-accumulating (rice) and non-accumulating (Arabidopsis) species, it was possible to rely on in silico predictions to identify GmNIP2-1 and GmNIP2-2 genes. The characteristic features of soybean Si transporters include two conserved NPA domains, G-S-G-R selective residues forming an Ar/R filter, and Froger’s residues (L-T-A-Y-F), which are common to all Si transporters identified in rice and other monocots (Fig. 3; Froger et al. 1998). Quite surprisingly, they differ from the only other dicot transport gene found in pumpkin by having a NPA motif instead of NPV in helix E (HE) (Mitani-Ueno et al. 2011). This indicates that the NPV in HE is not a feature specific to dicots. The permeability of GmNIP2-2 to Si was confirmed using a Xenopus oocyte bioassay, a heterologous expression system that has proven quite reliable for the purpose of testing Si transporters (Ma et al. 2006; Montpetit et al. 2012; Mitani-Ueno et al. 2011). By using Si directly as a substrate instead of germanium, we have also eliminated any possible bias associated with a surrogate substrate.
Genotypic difference in soybean for silicon accumulation
Based on the identification of GmNIP2-1 and GmNIP2-2 genes and subsequent demonstration of their functionality, Si absorption in three soybean genotypes did confirm that soybean should indeed be considered as a Si accumulator. However, the differential level of absorption among the three cultivars and the unexpectedly high value of 2.4 % Si observed in cv. Hikmoksorip raise important questions. While plenty of data are available for inter-species differences, very little work has addressed the notion of intra-species variation in terms of Si absorption (Hodson et al. 2005). In rice, genotypic variations have amounted to about 35 % between the lowest and highest accumulators, a difference that was attributed to the level of gene expression of Si transporters (Deren 2001; Ma et al. 2007). In pumpkin, the difference between a low- and a high-absorbing cultivar was attributed to a change in the protein sequence rendering the transporter defective (Mitani-Ueno et al. 2011). In our work, the 600 % difference (0.4 vs. 2.4 %) between cvs Hikmoksorip and Williams 82 represents an exceptionally large difference that prompted further investigations. As a first approach, comparison of GmNIP2 sequences revealed a perfect match among all three genotypes, thus eliminating the possibility of a divergent protein structure. Similarly, expression levels were identical and could not account for the differential absorption. As no differences in GmNIP2 gene structure and expression were observed, it is tempting to speculate that differences in a Si efflux transporter (Lsi2) could possibly explain the disparity in Si accumulation between soybean cultivars. However, Lsi2 is not an aquaporin and was not examined in this study. Considering that Hikmoksorip displayed a significantly increased level of resistance against soybean rust when supplied with Si (Arsenault-Labrecque et al. 2012), this suggests that increasing Si absorption in soybean could yield important benefits and that a better understanding of the mechanisms underlying expression of this trait would prove useful.
In conclusion, through a genome wide analysis of MIPS, we have identified two Si influx transporters in soybean and confirmed their role as the first determinant allowing Si uptake in this species. Our results also highlight important differences between cultivars in the quantitative Si concentrations found in soybean tissues. This suggests that other factors, including efflux transporters, act synergistically with influx transporters to regulate Si accumulation in soybean.
References
Arsenault-Labrecque G, Menzies JG, Bélanger RR (2012) Effect of silicon absorption on soybean resistance to Phakopsora pachyrhizi in different cultivars. Plant Dis 96(1):37–42
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34(Suppl 2):W369–W373
Bernsel A, Viklund H, Hennerdal A, Elofsson A (2009) TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res 37(Suppl 2):W465–W468
Bienert GP, Bienert MD, Jahn TP, Boutry M, Chaumont F (2011) Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J 66(2):306–317
Caron L, Rousseau F, Gagnon E, Isenring P (2000) Cloning and functional characterization of a cation Cl− cotransporter-interacting protein. J Biol Chem 275(41):32027–32036
de Hoon MJ, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20(9):1453–1454
Dean RM, Rivers RL, Zeidel ML, Roberts DM (1999) Purification and functional reconstitution of soybean nodulin 26. An aquaporin with water and glycerol transport properties. Biochemistry 38(1):347–353
Deren CW (2001) Plant genotype, silicon concentration, and silicon-related responses. Stud Plant Sci 8:149–158
Epstein E (1994) The anomaly of silicon in plant biology. Proc Natl Acad Sci USA 91:11–17
Fauteux F, Rémus-Borel W, Menzies JG, Bélanger RR (2005) Silicon and plant disease resistance against pathogenic fungi. FEMS Microbiol Lett 249(1):1–6
Fortin MG, Morrison NA, Verma DP (1987) Nodulin 26, a peribacteroid membrane nodulin is expressed independently of the development of the peribacteroid compartment. Nucleic Acids Res 15(2):813–824
Fouquet R, Léon C, Ollat N, Barrieu F (2008) Identification of grapevine aquaporins and expression analysis in developing berries. Plant Cell Rep 27(9):1541–1550
Froger A, Thomas D, Delamarche C, Tallur B (1998) Prediction of functional residues in water channels and related proteins. Protein Sci 7(6):1458–1468
Gattolin S, Sorieul M, Frigerio L (2011) Mapping of tonoplast intrinsic proteins in maturing and germinating Arabidopsis seeds reveals dual localization of embryonic TIPs to the tonoplast and plasma membrane. Mol Plant 4(1):180–189
Giovannetti M, Balestrini R, Volpe V, Guether M, Straub D, Costa A, Ludewig U, Bonfante P (2012) Two putative-aquaporin genes are differentially expressed during arbuscular mycorrhizal symbiosis in Lotus japonicus. BMC Plant Biol 12(1):186
Gonen T, Sliz P, Kistler J, Cheng Y, Walz T (2004) Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature 429(6988):193–197
Grégoire C, Rémus-Borel W, Vivancos J, Labbé C, Belzile F, Bélanger RR (2012) Discovery of a multigene family of aquaporin silicon transporters in the primitive plant Equisetum arvense. Plant J72(2):320–330
Guntzer F, Keller C, Meunier JD (2012) Benefits of plant silicon for crops: a review. Agron Sustain Dev 32(1):201–213
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Harries WEC, Akhavan D, Miercke LJW, Khademi S, Stroud RM (2004) The channel architecture of aquaporin 0 at a 2.2-A resolution. Proc Natl Acad Sci USA 101:14045–14050
Hodson MJ, White PJ, Mead A, Broadley MR (2005) Phylogenetic variation in the silicon composition of plants. Ann Bot 96(6):1027–1046
Kaczanowski S, Zielenkiewicz P (2010) Why similar protein sequences encode similar three-dimensional structures? Theor Chem Acc Theory Comput Model (Theoretica Chimica Acta) 125:643–650
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9(4):299–306
Lee JK, Kozono D, Remis J, Kitagawa Y, Agre P, Stroud RM (2005) Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A. Proc Natl Acad Sci USA 102(52):18932–18937
Ma JF, Yamaji N (2006) Silicon uptake and accumulation in higher plants. Trends Plant Sci 11(8):392–397
Ma JF, Tamai K, Yamaji N, Mitani N, Konishi S, Katsuhara M, Ishiguro M, Murata Y, Yano M (2006) A silicon transporter in rice. Nature 440(7084):688–691
Ma JF, Yamaji N, Mitani N, Tamai K, Konishi S, Fujiwara T, Katsuhara M, Yano M (2007) An efflux transporter of silicon in rice. Nature 448(7150):209–212
Matsumoto T, Lian HL, Su WA, Tanaka D, Liu C, Iwasaki I, Kitagawa Y (2009) Role of the aquaporin PIP1 subfamily in the chilling tolerance of rice. Plant Cell Physiol 50(2):216–229
Maurel C, Verdoucq L, Luu DT, Santoni V (2008) Plant aquaporins: membrane channels with multiple integrated functions. Ann Rev Plant Biol 59:595–624
Mitani N, Yamaji N, Ma JF (2008) Characterization of substrate specificity of a rice silicon transporter, Lsi1. Pflügers Archiv Eur J Physiol 456(4):679–686
Mitani-Ueno N, Yamaji N, Zhao FJ, Ma JF (2011) The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J Exp Bot 62(12):4391–4398
Montpetit J, Vivancos J, Mitani-Ueno N, Yamaji N, Rémus-Borel W, Belzile F, Ma FJ, Bélanger RR (2012) Cloning, functional characterization and heterologous expression of TaLsi1, a wheat silicon transporter gene. Plant Mol Biol 79(1–2):35–46
Newby ZE, O’Connell LJ, Robles-Colmenares Y, Khademi S, Miercke LJW, Stroud RM (2008) Crystal structure of the aquaglyceroporin PfAQP from the malarial parasite Plasmodium falciparum. Nat Struct Mol Biol 15(6):619–625
Quigley F, Rosenberg JM, Shachar-Hill Y, Bohnert HJ (2002) From genome to function: the Arabidopsis aquaporins. Genome Biol 3(1):1–17
Reidinger S, Ramsey M, Hartley SE (2012) Rapid and accurate analyses of silicon and phosphorus in plants using a portable X-ray fluorescence spectrometer. New Phytol 195(3):699–706
Richmond KE, Sussman M (2003) Got silicon? The non-essential beneficial plant nutrient. Curr Opin Plant Biol 6(3):268–272
Roulin A, Auer PL, Libault M, Schlueter J, Farmer A, May G, Stacey G, Doerge RW, Jackson SA (2013) The fate of duplicated genes in a polyploid plant genome. Plant J73(1):143–153
Sakurai J, Ishikawa F, Yamaguchi T, Uemura M, Maeshima M (2005) Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol 46(9):1568–1577
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J et al (2010) Genome sequence of the palaeopolyploid soybean. Nature 463(7278):178–183
Severin AJ, Woody JL, Bolon YT, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE et al (2010) RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol 10(10):160
Shen X, Li X, Li Z, Li J, Duan L, Eneji AE (2010) Growth, physiological attributes and antioxidant enzyme activities in soybean seedlings treated with or without silicon under UV-B radiation stress. J Agron Crop Sci 196(6):431–439
Sonah H, Deshmukh RK, Singh VP, Gupta DK, Singh NK, Sharma TR (2011) Genomic resources in horticultural crops: status, utility and challenges. Biotech Adv 29(2):199–209
Wallace IS, Roberts DM (2005) Distinct transport selectivity of two structural subclasses of the nodulin-like intrinsic protein family of plant aquaglyceroporin channels. Biochemistry 44(51):16826–16834
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35(Suppl 2):W407–W410
Zhang DY, Ali Z, Wang CB, Xu L, Yi JX, Xu ZL, Liu XQ, He XL, Huang YH, Khan IA et al (2013) Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.). PLoS ONE 8(2):e56312
Acknowledgments
The project was funded by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) in collaboration with Syngenta Biotechnology and the Canada Research Chairs Program to RRB. The authors would like to thank the group of Dr. P. Isenring from the Nephrology Research Group of the CHUQ-L’Hôtel-Dieu de Québec Institution for help with the oocyte assays.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Deshmukh, R.K., Vivancos, J., Guérin, V. et al. Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant Mol Biol 83, 303–315 (2013). https://doi.org/10.1007/s11103-013-0087-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-013-0087-3