Abstract
G protein-coupled receptors (GPCR) are important drug discovery targets. Despite progress, many GPCR structures have not yet been solved. For these targets, comparative modeling is used in virtual ligand screening to prioritize experimental efforts. However, the structure of extracellular loop 2 (ECL2) is often poorly predicted. This is significant due to involvement of ECL2 in ligand binding for many Class A GPCR. Here we examine the performance of loop modeling protocols available in the Rosetta (cyclic coordinate descent [CCD], KIC with fragments [KICF] and next generation KIC [NGK]) and Molecular Operating Environment (MOE) software suites (de novo search). ECL2 from GPCR crystal structures served as the structure prediction targets and were divided into four sets depending on loop length. Results suggest that KICF and NGK sampled and scored more loop models with sub-angstrom and near-atomic accuracy than CCD or de novo search for loops of 24 or fewer residues. None of the methods were able to sample loop conformations with near-atomic accuracy for the longest targets ranging from 25 to 32 residues based on 1000 models generated. For these long loop targets, increased conformational sampling is necessary. The strongly conserved disulfide bond between Cys3.25 and Cys45.50 in ECL2 proved an effective filter. Setting an upper limit of 5.1 Å on the S–S distance improved the lowest RMSD model included in the top 10 scored structures in Groups 1–4 on average between 0.33 and 1.27 Å. Disulfide bond formation and geometry optimization of ECL2 provided an additional incremental benefit in structure quality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
G protein-coupled receptors (GPCR) comprise one of the largest families of integral membrane proteins in eukaryotes. GPCR serve central roles in amplifying and regulating a wide range of intracellular responses to extracellular stimuli. In response to ligand activation, GPCR undergo conformational changes that influence coupling with intracellular partners, including heterotrimeric G proteins (Gα, Gβ, Gγ subunits), β-arrestins, G protein-coupled receptor kinases (GRK), and other effectors [1,2,3]. It is estimated that between 27–50% of FDA-approved drugs interact with GPCR targets including classes of drugs such as beta-blockers, antihistamines, and antipsychotics [4, 5].
All GPCR share a common topology featuring a core bundle of seven transmembrane (TM) α-helices with the N-termini and C-termini located on the extracellular and intracellular sides of the cell membrane, respectively. The extracellular and intracellular loops (ECL and ICL) are the protein segments that connect adjacent TM domains (ECL1–3, ICL1–3). The ECL and ICL of GPCR have lower sequence and structural conservation than the TM domains. With respect to the available known GPCR structures (mostly Class A GPCR), ECL2 is generally the longest and most diverse in terms of amino acid identity and three-dimensional structure (Fig. 1). Despite low sequence conservation, an overwhelming majority of GPCR contain a disulfide bond between highly conserved cysteine residues in ECL2 and the extracellular end of TM3. Based on the analysis of 367 GPCR sequences representing members from Class A, B1, B2, C, and F that were downloaded using the GPCRdb alignment tools [6, 7], 89% (327 out of 367) of sequences contain the conserved cysteine residues in ECL2/TM3. Also, the disulfide bond between the cysteine sidechains is observable in 94% (47 out of 50) of representative crystal structures of unique GPCR (as of May 2018). Three lipid receptors (LPAR1, S1PR1, CB1) that have known crystal structures lack the conserved disulfide bond between ECL2/TM3. Instead, they contain an intra-loop disulfide bond that constrains the loop conformation.
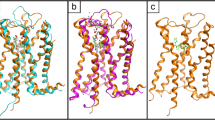
Extracellular loop comparison of seventeen superposed GPCR crystal structures. The PDB IDs of the superposed GPCR crystal structures above are as follows: Bovine RHO (1GZM, green), B2AR (2RH1, orange), B1AR(2VT4, magenta), Squid RHO (2Z73, gold), AA2AR (3EML, cyan), DRD3 (3PBL, pink), H1R (3RZE, maroon), ACM2 (3UON, light blue), CXCR4 (3ODU, brown), S1PR1 (3V2Y, dark green), PAR1 (3VW7, yellow), NTR1 (4GRV, blue), ACM3 (4DAJ, light brown), OPRK (4DJH, purple), OPRM (4DKL, dark blue), NOP (4EA3, salmon), OPRD (4EJ4, dark brown)
For many GPCR, ECL2 plays important roles in GPCR activation, orthosteric ligand binding, and allosteric ligand interactions [8, 9]. Mutagenesis experiments on ECL2 of the complement C5a receptor and thrombin receptor resulted in constitutively active GPCR [10, 11]. These findings suggest that in some GPCR, ECL2 functions as a negative regulator that dampens signaling by restricting the transition to active receptor states in the absence of endogenous ligand. However, ECL2 mutagenesis doesn’t uniformly confer constitutive GPCR activity. For example, the A204E mutation in ECL2 of the ghrelin receptor resulted in diminished constitutive activity [12]. Thus, it is generally understood that ECL2 plays a role in GPCR function, but many of the details are receptor-dependent.
Given that the binding pocket features of closely-related GPCR are relatively similar, there must be other structural aspects that give rise to observed differences in receptor-ligand specificity. Indeed, the diversity among ECL2 amino acid sequences and structural features even between closely-related GPCR contributes to receptor-specific ligand interactions [13]. Table 1 shows examples of GPCR crystal structures that display a range of receptor-ligand interactions with a variable number of ECL2 residues.
Template-based modeling methods rely on an underlying structural similarity which is often lacking when comparing ECL2 between members of the GPCR family. GPCR loop segments display low sequence conservation relative to the TM domains and tend to exhibit variable lengths which inevitably introduces gaps in GPCR sequence alignments. Furthermore, there are instances of loops with low structural similarities despite having relatively high sequence identities by homology modeling standards. For reliable homology-based prediction of ligand-receptor interactions, the pairwise percentage identity of target-template sequence identity is suggested to be around 35–40% based on the GPCR Dock assessments from 2008 to 2010 [30, 31]. For example, the dopamine receptor D2 (DRD2) homology modeling target shares 66.9% and 36.8% sequence identity overall with the GPCR templates, DRD3 (PDB: 3PBL) and 5HT2C (PDB: 6BQH), respectively. While these templates certainly meet or exceed the suggested 35–40% sequence identity threshold for reliable homology modeling, the DRD3 and 5HT2C crystal structures would be poor templates for the ECL2 segment of the DRD2 target (Fig. 2).
Although not a comprehensive list, Table 2 shows several GPCR with more than two crystal structures available at the beginning of this study. The dynamic nature of these loops is apparent in these sets of superposed crystal structures. The ECL2 Cα atoms in different structures of a single protein have root mean square deviation (RMSD) values ranging from 0.3 to 1.5 Å. Traditionally the gold standard of structure prediction is achieving top-ranking models with sub-angstrom accuracy (Cα RMSD under 1.0 Å) to the reference “native” structure. However, ECL2 experimental variability in different crystal structures of the same GPCR can exceed this value. For the ECL2 targets in this benchmark, it is more reasonable to consider methods that produce top-scoring models with near-atomic accuracy (Cα RMSD within 2.5 Å) as the threshold for success [32]. Often, models with near-atomic accuracy are sufficient for applications downstream of modeling [33].
Ab initio loop modeling can be described as a mini-protein folding problem with success largely depending on two general components: sampling and scoring. An extensive search to sample loop conformational space is implemented with the target sequence. Also, a method to evaluate or score the loop model conformations in order to select near-native structures (close to reference structure) is necessary. The prediction involves generating model loop structures and ranking them based on some criteria (i.e. energy functions as in Rosetta and MOE) that desirably correlate with experimentally determined structures. Our primary research question at the start of this benchmark study was, which of the available modeling software methods can accurately sample and score near-native loop models?
Methods
The reference GPCR crystal structures used in this benchmark for ECL2 modeling are listed in Table 3. Crystal structures with poorly resolved or completely missing residues in the ECL2 segment were excluded from the benchmark. The highest resolution structures were chosen as references for individual GPCR that had multiple crystal structures available. The preferred chain of each benchmark target PDB file was extracted using the GPCRdb structure browser application for further preparation.
The molecular operating environment (MOE version 2016.08) software package was used for GPCR structure preparation and visualization. The Rosetta software suite (Rosetta version 3.8) and MOE were used for modeling the ECL2 segment of the benchmark reference structures. For each of the GPCR in the benchmark, the Robetta fragment server was used to generate the 9mer and 3mer fragment library files that are necessary for Rosetta CCD and KICF loop modeling methods [124]. Fragment generation for each reference GPCR excluded any PDB data from the GPCR crystal structure itself, as verified by manual inspection of PDB ID codes in each fragment file.
Structure preparation for GPCR targets using MOE
The benchmark GPCR structures listed in Table 3 were downloaded from the PDB [125] and prepared for loop modeling with MOE. Ligands and water molecules were deleted from the structure files. For each GPCR structure, the “QuickPrep” process was used to streamline the structure preparation process. QuickPrep corrects any structural issues (i.e. residues with alternate locations, missing atoms, chain breaks, etc.) that often accompany structural data, adds explicit hydrogens and partial charges with “Protonate 3D,” and performs a tethered-receptor energy minimization using the Amber12EHT [126,127,128] forcefield (RMS gradient of 0.1 kcal/mol/Å). The final energy minimization step was performed to improve any inaccurate geometries derived from the crystallographic data. During the minimization process, the receptor atoms were tethered to ensure that changes to the initial positions are modest.
Description of loop modeling software
Rosetta loop modeling protocols can be used for loop refinement or loop reconstruction. This study only implements modeling in the context of loop reconstruction—ab initio/de novo prediction of the “native” loop conformation based on amino acid sequence, but the initial backbone and sidechain conformations were discarded prior to modeling. Loop refinement, on the other hand, is utilized in finding lower energy conformations starting from a given loop conformation that is potentially close to the “native” structure.
The sampling process was implemented in two stages with iterations of Monte Carlo simulated annealing: An initial low-resolution/coarse-grained stage where the sidechain atoms were represented as “centroids” and a high-resolution/full-atom stage where the sidechain atoms were explicitly represented. This process was coupled with one of several scoring functions. Figure 3 shows a schematic overview of the general Rosetta loop modeling process. The available loop modeling algorithms in Rosetta differ in conformational search (sampling) strategy and solutions to the loop closure problem.
The cyclic coordinate descent (CCD) algorithm proceeds by optimizing the dihedral angles through consecutive loop residues from the N- to C-terminus where the goal is to minimize the distance between the free C-terminus end of the loop and the fixed anchor position [129, 130]. The CCD algorithm in Rosetta uses experimentally-derived fragment libraries to guide the conformational search during loop modeling. The fragment libraries contain the coupled phi/psi dihedrals of peptide segments with 9 and 3 residues (9mers, 3mers) from the PDB.
The kinematic closure (KIC) method selects three pivot atoms (remaining loop backbone atoms are designated non-pivot) and divides the loop into two segments for conformational sampling of the non-pivot phi, psi dihedral angles. Subsequently, the pivot dihedral angles (six phi, psi angles for three pivots) were analytically solved to position each rigid segment for loop closure [131]. The standard KIC protocol for loop modeling has subsequently been replaced by the next generation KIC (NGK) and KIC with fragments (KICF) methods.
The next generation KIC (NGK) algorithm employs intensification strategies during non-pivot conformational sampling in both low- and high-resolution stages of the loop modeling process [132]. In the high-resolution stage, NGK implements additional annealing strategies that modulate the energy function to overcome large energy barriers. The intensification strategies involve (1) using neighbor-dependent Ramachandran distributions (Rama2b term) to select phi/psi dihedral combinations during sampling and (2) independently sampling ω angles based on observations in high-resolution crystal structures. Traditionally, the planar character of the peptide bond restricts the ω dihedral angle to either 180° for the common trans-configuration or 0° for the less common cis-configuration. However, analyses of high-resolution protein structures concluded that trans peptide ω values can vary by more than 25° from planarity in some cases, and that the non-planar character of peptide bonds are more common than previously known [133]. During the NGK method, ω sampling is performed independently of the phi/psi dihedrals from a Gaussian around the observed mean of 179.1° ± 6.3° [132]. The annealing strategies implemented in the NGK method involve ramping the weights of (1) the repulsive component of the Lennard-Jones potential and (2) the Rama score (distinct from Rama2b term used in intensification strategy), which is the likelihood of a phi/psi combination occurring given an amino acid type. While the intensification strategies (Rama2b and ω sampling) are applied in both low- and high-resolution stages of loop modeling, the annealing strategies are only implemented in the high-resolution stage. Overall, these intensification and annealing strategies were found to greatly improve loop modeling accuracy compared to the standard KIC method in a previous benchmark [132].
The KIC with fragments (KICF) method combines the fragment library sampling strategy from the CCD method with the KIC loop closure method. The main difference between this method and the NGK method is the way in which loop backbone conformations are sampled. The fragment-based sampling of phi/psi/omega dihedral angles consists of four major steps: (1) one of the given fragment libraries is selected at random and searched for alignment frames where fragments overlap with subsegments of the loop; (2) one of the alignment frames and fragments within that frame is selected at random; (3) the phi/psi/omega dihedral angles of that fragment are applied to the loop subsegment; and finally (4) kinematic closure (KIC) calculations are performed to achieve loop closure.
The Rosetta all-atom energy function to evaluate/score biomolecular structures and models has evolved over many versions (Score12, Talaris2013, Talaris2014), with the Rosetta Energy Function 2015 (REF2015) becoming the default scoring function as of July 2017 [134,135,136,137]. However, the most recently available scoring function (REF2015) and loop modeling algorithm, (KICF), had not been tested on the 12-residue, or 14–17 residue loop modeling benchmark sets used by Rosetta developers (as of May 2018).
The MOE loop modeler application has a de novo search method and a PDB search method for sampling potential loop backbone conformations. For this study, only the de novo search method was used to model the ECL2 of the benchmark GPCR. MOE loop modeling protocols also consisted of distinct low-/high-resolution stages for loop modeling. The initial de novo search stage only deals with the loop backbone atoms, generating potential loop conformations that were ranked by an initial coarse scoring function before advancing to the full-atom stage. Figure 4 shows a schematic overview of the general MOE loop modeling process.
MOE loop modeling uses an extension of the CCD algorithm, full CCD (FCCD) [138]. This method differs from CCD by solely operating on the Cα backbone atoms with pseudo bond angles and dihedral angles, optimizing both terms to achieve loop closure. Probability densities calculated from high-resolution PDB structures resulted in specific profiles for the Cα pseudo bond and dihedral angles. These profiles were used for random sampling of loop Cα conformational space during the de novo search stage, followed by FCCD loop closure.
After loop closure by FCCD, it is necessary to optimize the Cα backbone atoms. This is accomplished by using a component of the protein chain reconstruction algorithm (PULCHRA) method which employs a steepest descent gradient minimization and a simple harmonic potential to optimize the Cα positions before full backbone reconstruction [139]. To reconstruct backbone atom positions from the Cα loop traces generated, MOE uses the backbone building from quadrilaterals (BBQ) method which is based on proximal distance geometries for sets of four sequential Cα atoms in the loop. Additionally, MOE backbone packer performs a minimization to relieve any strained backbone geometries and atom clashes. This step is followed by a final geometry and duplicate check to ensure that non-redundant backbone conformations with reasonable bond and dihedral angles are being evaluated by the coarse scoring function. The top-ranking loop backbone conformations are advanced to the full-atom stage.
In the full-atom stage, sidechain atoms were added to the loop and optimized with respect to the sidechain orientations. The entire loop segment was energy minimized through multiple steps before the final scoring step. The full-atom loop conformations generated were scored using generalized-born volume integral (GBVI). The potential energy of the system using GBVI has been shown to recover loop conformations close to the native from the Jacobson Loop Decoy Dataset [140].
Rosetta and MOE loop modeling methods
A total of 1000 ECL2 models were generated for the GPCR benchmark targets after structure preparation using each loop modeling method. All 28 reference structures in the benchmark were reconstructed with the Rosetta methods discussed previously, but only 7 of the 28 reference structures were modeled with MOE. This was due to non-competitive performance on the shorter ECL2 targets which will be discussed in the "Results" section.
The following descriptions are the main Rosetta command line options used to perform NGK, KICF, and CCD loop modeling methods in low-resolution and full-atom stages. The remodel stage was the term for the initial coarse-grained modeling step. The remodel stage samples loop backbone conformations using a reduced representation of amino acid sidechains and a Rosetta low-resolution scoring function. This was initiated by the following options associated with the ‘-loops:remodel’ command: ‘perturb_kic’, ‘perturb_kic_with_fragments’, ‘perturb_ccd’. A loop definition file was separately generated for each reference GPCR to define the residues of the loop (ECL2) to be remodeled. By enabling the “extend loop” field in the loop files, the target loop segment’s bond lengths, angles, and omega torsions were idealized, and all phi/psi values were replaced randomly from Ramachandran space to give an initial closed conformation at the start of remodel stage. This was to ensure that loop reconstruction wasn’t influenced by the initial loop conformation. Subsequently, the loop phi/psi dihedrals were sampled using the fragment data described previously, followed by KIC or CCD calculations to achieve loop closure. Finally, the loop underwent minimization using the Broyden–Fletcher–Goldfarb–Shanno (BFGS) algorithm and a Metropolis criterion acceptance test using the Rosetta low-resolution scoring function, score4L. The number of Monte Carlo steps in both stages of loop modeling was determined by the number of outer and inner cycles, (outer_cycles * inner_cycles). The default number of outer (5) and inner cycles (evaluated by: min(1000, number_of_loop_residues * 20)) were used for all Rosetta loop modeling jobs in this benchmark study. Loop poses were set to the lowest energy conformation evaluated at the end of each outer cycle. The temperature decreased exponentially from 1.5 to 0.5 KT from the first step to the last. The refine stage was the term for the all-atom loop modeling step and was activated by the following options associated with the ‘-loops:refine’ command: ‘refine_kic’, ‘refine_kic_with_fragments’, ‘refine_ccd’. This stage implements a similar scheme to the perturb stage, with major differences in the all-atom treatment of the loop during conformational sampling and the scoring function used to guide and evaluate loop conformations during model production. Out of the three Rosetta loop modeling algorithms used in this benchmark, NGK was the quickest among the more accurate methods at generating a given set of loop models. Therefore, if subsequent sets of ECL2 models of benchmark targets were produced, loop modeling was performed initially with the NGK algorithm.
The following MOE loop modeling options were used during the de novo search stage, full-atom model generation, and final model scoring. The loop sequence for each GPCR ECL2 was selected in the sequence editor window and the following options were provided in the main MOE Loop Modeler window: only the de novo search method was enabled, the default RMSD limit of 0.50 was decreased to 0.25 Å, the max iterations and energy window were set to 1000 and 10, respectively. The number of de novo search runs and final models built were set to 1000 total. Subsequently, SVL batch files were created to run the loop modeler jobs on a high-performance computing cluster. Due to the molecular database (.mdb files) storage limitations, 10 sets of 100 final models or 20 sets of 50 final models were generated.
Filtering and optimization
The majority of GPCR share a conserved disulfide bond between Cys45.50 in ECL2 and Cys3.25 at the top of the third transmembrane domain. In the 28-receptor benchmark set used in this study, 25 of the receptors share this structural constraint. We assessed the utility of using the S–S distance as a filter for improving ECL2 models. Distance-based filtering used a cut-off of 5.1 Å. This distance is approximately double the van der Waals contact distance for sulfer and was selected to avoid the possibility of selecting loop models in which atoms occur between the two sulfur atoms expected to form a disulfide bond. Disulfide bonds were constructed in the top 10 scored models meeting this distance cutoff followed by geometry optimization of the loop using the MOE software.
Results and discussion
Loop modeling performance throughout this benchmark study was assessed by comparing de novo models of ECL2 for proteins from Table 3 to the crystallographic reference structures indicated in the same table. Superpositions were performed for residues not modeled de novo before calculation of (Cα) RMSD values. The metrics reported throughout this section include: lowest RMSD model (LRM), top scored model (T1), lowest RMSD model in the top 10 scored (T10), and lowest RMSD model in the top 25 scored (T25).
Rosetta scoring function comparison
To decide which scoring function to use with Rosetta loop modeling, the NGK loop modeling algorithm was used to sample ECL2 conformations for the first four targets in Group 1 (Table 3) using both the Rosetta Energy Function 2015 (REF2015) and its predecessor, Talaris 2014. The LRM results show that models meeting the near-atomic accuracy metric of 2.5 Å were sampled in each run (Fig. 5). However, when modeling loops of unknown structure, the T1 is more relevant than the LRM. In 3 out of 4 cases, REF2015 improved reference structure prediction accuracy (lower RMSD) for the top scored models. These data suggest that the REF2015 scoring function is more suitable for identifying models closer to the reference ECL2 structures in the benchmark. Furthermore, the most recent energy function has been parametrized to estimate energies in units of kilocalories per mole, whereas all previous Rosetta energy functions used arbitrary units [137]. Therefore, REF2015 was used in the Rosetta loop modeling protocols for all benchmark targets and method comparisons.
Energy function comparison with Rosetta NGK loop modeling. Comparisons of the energy function influence on loop modeling performance is shown for the shortest GPCR ECL2 targets in the benchmark. The lowest RMSD models (LRM), top scored models (T1), lowest RMSD models in the top 10 scored (T10), and lowest RMSD models in the top 25 scored (T25) are shown from 1000 models generated for each ECL2 target using NGK loop modeling. The number in parentheses represents the ECL2 length
Group 1 loop modeling results
Loop modeling was performed using three algorithms in Rosetta and one algorithm in MOE for the Group 1 ECL2 targets from Table 3. The results from Group 1 in Fig. 6a show that the NGK and KICF methods were able to sample models with better accuracy than either CCD or MOE based on the RMSD of the LRM to the reference ECL2 structures. Notably, ECL2 of the muscarinic acetylcholine receptor M4 (ACM4, Fig. 7) and dopamine receptor D3 (DRD3) targets were modeled with sub-angstrom accuracy using both the NGK (LRM = 0.34, 0.71 Å, respectively) and KICF (LRM = 0.35, 0.76 Å, respectively) algorithms. For the ECL2 of CXCR4 (Fig. 8), loop modeling using KICF also displayed sub-angstrom accuracy for the LRM (0.50 Å). However, none of the targets were modeled with sub-angstrom accuracy using the CCD algorithm or MOE. Six out of the seven ECL2 targets in Group 1 were modeled with near-atomic accuracy (RMSD ≤ 2.5 Å) using the NGK and CCD algorithms. Additionally, all seven of the targets were modeled with near-atomic accuracy using the KICF algorithm. From the Group 1 targets, there were only two cases where the LRM from MOE had RMSD values below 2.5 Å to the reference structure. The average LRM values for the NGK, KICF, and CCD algorithms were 1.66, 1.09, and 2.07 Å, respectively. In comparison, the average LRM using the MOE loop modeling algorithm was much higher, 3.37 Å.
Group 1 loop modeling benchmark set. a The RMSD values of the LRM and T1 are shown out of 1000 models generated for Group 1 ECL2 targets using NGK, KICF, CCD, and MOE loop modeling algorithms. In total, the LRM had sub-atomic accuracy in two and three cases when using the NGK and KICF algorithms, respectively. Additionally, the LRM had near-atomic accuracy in seven cases when using the KICF algorithm and six cases when using the NGK or CCD algorithms. b The RMSD values of the T1 is compared to the T10 and T25 using NGK, KICF, CCD, and MOE
ECL2 models of ACM4 superposed with reference structure. a The LRM (orange) and T1 (magenta) out of 1000 total models generated using NGK loop modeling had RMSD values of 0.34 Å and 0.40 Å to the reference structure (green). b The LRM and T1 out of 1000 total models generated using KICF loop modeling had RMSD values of 0.35 Å and 0.62 Å to the reference structure
ECL2 models of CXCR4 superposed with reference structure. a The LRM (orange) and T1 (magenta) out of 1000 total models generated using NGK loop modeling had RMSD values of 3.08 Å and 7.02 Å to the reference structure (green). b The LRM and T1 out of 1000 total models generated using KICF loop modeling had RMSD values of 0.50 Å and 4.19 Å to the reference structure
While generating ECL2 models with sub-angstrom or near-atomic accuracy overall is desirable, sampling loop conformations is only one aspect of structure prediction when the target structure is unknown. Loop models with low RMSD values to the target must also be scored or ranked favorably so they can be distinguished from the rest of the generated models. To evaluate the scoring component of the loop modeling protocols, the lowest RMSD within the top 1, 10, and 25 scored models (T1, T10, T25) compared to the reference ECL2 structure is illustrated for Group 1 in Fig. 6b. Overall, the MOE loop modeling algorithm had a substantially higher average RMSD value for the T1 (7.96 Å) compared to the algorithms used within Rosetta. The NGK, KICF, and CCD algorithms had average T1 values of 3.61, 3.40, and 4.56 Å, respectively. The T1 produced by CCD or MOE loop modeling algorithms had RMSD values outside of the near-atomic accuracy threshold for all seven Group 1 ECL2 targets. The NGK and KICF algorithms were both able to produce T1 with RMSD values below the near-atomic accuracy threshold for two of the targets. For a majority of the Group 1 targets, selecting higher-quality loop models than the T1 was necessary to reliably study receptor-ligand interactions through docking experiments. Therefore, we sought to establish selection guidelines for the number of final loop models to retain from the top scored models. Specifically, we wanted to assess the accuracy of final models within subsets of the top 10 or 25 scored models produced. For every ECL2 target in Group 1, the RMSD values for T10 and T25 are lower than the RMSD value for the T1, regardless of sampling methodology. However, expanding the number of retained ECL2 models from the top 10 to the top 25 scored did not consistently result in substantially lower RMSD models. In Group 1, there were only 3 of 28 total cases in which the T10 had an RMSD value above the 2.5 Å threshold, but the corresponding T25 had an RMSD below that threshold. Models with near-atomic accuracy were generated and scored within the top 25 for the targets CRFR1 (using KICF), ACM4 (using CCD), and US28 (using CCD) that were not scored within the top 10. However, for most of the sampling algorithm and target combinations, there was not a significant advantage in retaining the top 25 scored models rather than the top 10 out of 1000 models total.
Group 2 loop modeling results
All Group 2 ECL2 targets from Table 3 were modeled using the three loop modeling algorithms in Rosetta. Loop modeling results for Group 2 targets (Fig. 9) show that the NGK and KICF methods were able to sample loop conformations of the cannabinoid receptor type 1 (CB1) ECL2 with sub-angstrom accuracy overall (NGK/KICF LRM = 0.85/0.82 Å, Fig. 10). Loop modeling with the KICF method also achieved sub-angstrom accuracy with the longest loop in Group 2, CCR5 (LRM = 0.75 Å). In terms of models with near-atomic accuracy (including models with sub-angstrom accuracy), there were three and four cases where the NGK and KICF algorithms sampled loop conformations with RMSD values ≤ 2.5 Å, respectively. There were three cases where the CCD algorithm sampled loop conformations with near-atomic accuracy, but no models with sub-angstrom accuracy were generated.
Group 2 loop modeling benchmark set. a The RMSD values of the LRM and T1 are shown out of 1000 models generated for Group 2 ECL2 targets using NGK, KICF, and CCD algorithms. In total, the LRM had sub-atomic accuracy in one and two cases when using the NGK and KICF algorithms, respectively. Additionally, the LRM had near-atomic accuracy in four cases when using the KICF algorithm and in three cases when using the NGK or CCD algorithms. b The RMSD values of the T1 is compared to the T10 and T25 using NGK, KICF, and CCD. The upper and lower dotted lines represent the near-atomic and sub-angstrom accuracy thresholds, respectively
ECL2 models of CB1 superposed with reference structure. a The LRM (orange) and T1 (magenta) out of 1000 models total using NGK loop modeling had RMSD values of 0.85 Å and 4.32 Å to the reference structure (green). b The LRM and T1 when KICF loop modeling was used had RMSD values of 0.82 Å and 1.93 Å to the reference structure
The T1 for a majority of Group 2 targets had a substantially larger RMSD value than the LRM which is consistent with the results from Group 1. However, for the CB1 ECL2 target the T1 using KICF displayed near-atomic accuracy to the reference structure with an RMSD of 1.93 Å and the LRM was scored within the top 10 models (Figs. 7, 8b). On the other hand, the T1 found using NGK had an RMSD of 4.32 Å to the reference structure and the LRM was not scored within the top 10 or 25 scored models. The lowest RMSD model found in the top 10 scored models using the NGK method displayed near-atomic accuracy to the reference structure with an RMSD of 1.39 Å. For the CCR5 ECL2 target, both methods that use fragment assembly (KICF and CCD) outperformed the NGK method in all four metrics. Notably, the LRM and T10 values from the KICF method with CCR5 were 0.75 and 0.81 Å.
In Group 2, the smoothened receptor (SMO) ECL2 was the most troublesome target for all three Rosetta loop modeling algorithms. The KICF algorithm yielded the most accurate LRM with an RMSD of 1.58 Å to the reference structure. The LRM found using the NGK and CCD algorithms had RMSD values of 6.22 and 5.84, respectively. However, the top scored model from this method had an RMSD of ~ 20 Å. While the ECL2 is situated just above the center of the TM bundle in the reference structure, SMO (Class F GPCR) differs from the other benchmark structures in many ways. Particularly, SMO has a longer ECL1 than other known GPCR structures (mostly Class A) and an extracellular domain (ECD) linker region that essentially form a lid over ECL2 and the TM bundle center (Fig. 11). The long ECL1 and ECD linker regions might sterically hinder a search for ECL2 loop conformations that are close to the reference structure. In other words, loop models that position ECL2 away from the TM bundle center, ECL1, and the ECD linker regions may be scored better. Since the SMO reference structure contained a co-crystallized ligand with contacts to the ECLs (ECL2 contacts shown in Table 1), it is also plausible that the conformation of the reference ECL2 is not as energetically favorable when the ligand is absent. A second set of ECL2 models (n = 1000) was generated for SMO using the NGK algorithm, but the ECD linker domain on the N-terminus was deleted prior to loop modeling. Out of the second set, the T1 and LRM were the same and had an RMSD value of 3.30 Å to the reference ECL2. This is a significant improvement over the T1 and LRM RMSD values from the initial set of models produced by NGK which were 17.0 Å and 6.22 Å, respectively. This demonstrates that the steric hindrance provided by the ECD linker domain is one impediment to sampling loop conformations similar to the reference SMO structure.
ECL2 models of SMO superposed with reference structure. a Side view of SMO crystal structure (PDB:4JKV) highlighting the native ECL2 (green) buried underneath ECL1 (gray) and the ECD linker (salmon). b Top view of the extracellular side of the SMO crystal structure. c The LRM (orange), T1 (magenta), and T10 (cyan) using the NGK algorithm had RMSD values of 6.22 Å, 17.0 Å, and 12.7 Å to the reference structure, respectively. d The LRM, T1, and T10 using the KICF algorithm had RMSD values of 1.58 Å, 19.9 Å, and 6.89 Å to the reference structure, respectively. The ECD linker and ECL1 regions were hidden in panels C and D to visualize ECL2 and models clearly
Group 3 loop modeling results
Group 3 targets from Table 3 were modeled using the Rosetta loop modeling algorithms. The results from modeling the ECL2 of Group 3 targets (Fig. 12) show that KICF was the only algorithm capable of sampling ECL2 models with sub-angstrom accuracy relative to the reference structures (2 of 7 targets). The LRM generated by KICF for the P2Y1R (Fig. 13b) and AT2R ECL2 targets had sub-angstrom accuracy with RMSD values of 0.63 and 0.54 Å, respectively. The LRM of P2Y1R had near-atomic accuracy (RMSD ≤ 2.5 Å) to the reference structure for all three loop modeling algorithms, suggesting it is a relatively ‘easy’ case.
Group 3 loop modeling benchmark set. a The RMSD values of the LRM and T1 are shown out of 1000 models generated for Group 3 ECL2 targets using the NGK, KICF, and CCD algorithms. In total, the LRM had sub-atomic accuracy in two cases when using the KICF algorithm. Additionally, the LRM had near-atomic accuracy in three cases when using the KICF algorithm and in single cases when using the NGK or CCD algorithms. b The RMSD values of the T1 is compared to the T10 and T25 using NGK, KICF, and CCD. The upper and lower dotted lines represent the near-atomic and sub-angstrom accuracy thresholds, respectively
ECL2 models of P2YR1 superposed with reference structure. a The LRM (orange), T1 (magenta), and T10 (cyan) out of 1000 models generated using NGK loop modeling had RMSD values of 2.44 Å, 6.53 Å, and 2.55 Å to the reference structure (green), respectively. b In this case, the LRM was also the T1 (magenta) when KICF loop modeling was used (LRM = T1 = 0.63 Å RMSD to reference structure)
In the case of the P2Y1R ECL2 target, the LRM produced by the KICF algorithm was also evaluated as the T1. This was not the case for the AT2R ECL2 target, but the top scored model produced by the KICF algorithm still had sub-angstrom accuracy (RMSD = 0.81 Å) when compared to the reference structure. On the other hand, the KICF and NGK algorithms produced top scored models with high RMSD values (~ 18 Å for both methods) compared to the ECL2 reference structure of β2AR (Fig. 14). For the top 25 scored β2AR ECL2 models, none of the methods were able to generate loop models with RMSD values below 4 Å. Since this target had one of the longer loops in the benchmark, it is possible that increased sampling was necessary to produce models closer to the reference structure. To determine if increased sampling would substantially improve models, a second set of 4000 ECL2 models was generated for β2AR using the NGK loop modeling algorithm. From the larger set of models, the LRM had an RMSD of 2.93 Å to the reference target. While this is only a slight improvement from the LRM from the initial set of 1000 models, the T1 from the set of 4000 models had a RMSD value of 5.46 Å which is substantially lower than the T1 from the initial set (18.1 Å). Out of the 4000-model set, the T10 and T25 both had an RMSD of 3.67 Å to the reference structure which was also lower relative to the T10 and T25 from the initial set (6.81 and 5.83 Å).
ECL2 models of B2AR superposed with reference structure. a The LRM (orange), T1 (magenta), and T10 (cyan) out of 1000 models generated using NGK loop modeling had RMSD values of 3.84 Å, 18.1 Å, and 6.81 Å to the reference structure (green), respectively. b The LRM, T1, and T10 generated using KICF loop modeling had RMSD values of 2.60 Å, 18.6 Å, and 5.04 Å to the reference structure, respectively
Group 4 loop modeling results
Group 4 targets from Table 3 were also modeled using the Rosetta loop modeling algorithms (Fig. 15). The average LRM values resulting from modeling with the NGK, KICF, and CCD algorithms were 4.55, 4.98, and 5.38 Å, respectively. None of the loop modeling methods used was able to generate models with sub-angstrom or near-atomic accuracy to the reference structures. Based on these results, an increase in conformational sampling is likely necessary. Potentially due to inadequate sampling of conformational space, deficiencies in the scoring function to distinguish accurate models were evident for the longer loops in this benchmark set. The average ECL2 RMSD values for the T1 using the NGK, KICF and CCD algorithms were 13.18, 11.01, and 11.70 Å, respectively. A decrease in the average ECL2 RMSD values was observed for the T10 using the same three algorithms (7.65, 7.37, and 7.72 Å), but were still substantially higher than the 2.5 Å threshold for useful models.
Group 4 loop modeling benchmark set. a The RMSD values of the LRM and T1 are shown out of 1000 models generated for Group 4 ECL2 targets using the NGK, KICF, and CCD algorithms. The T10 and T25 are shown out of 1000 models generated for Group 4 ECL2 targets using NGK, KICF, and CCD. b The RMSD values of the T1 is compared to the T10 and T25 using NGK, KICF, and CCD. The upper and lower dotted lines represent the near-atomic and sub-angstrom accuracy thresholds, respectively
Filtering and optimization results
Many GPCR structures characterized to date share a conserved disulfide bond between Cys3.25 at the top of TM3 and Cys45.50 in ECL2. The models generated using the KICF algorithm for the 25 of 28 receptors studied here were therefore filtered based on the distance between the sulfur atoms in these residues to see if such filtering selected an improved set of ten models. Additionally, formation of the disulfide bond followed by geometry optimization of ECL2 was examined as a potential loop optimization strategy. Table 4 compares T10 results in the absence of filtering/optimization (T10), after filtering (T10SSdist), and after filtering/disulfide formation/optimization (T10SSbond) across all four groups of loop modeling targets.
The results in Table 4 demonstrate that the use of the Cys3.25–Cys45.50 S–S distance as a filter produced modest improvements for most receptors in the lowest RMSD found within the top 10 scoring models. The average improvement in the group 1, 2, 3, and 4 targets was 0.33, 1.71, 0.55 and 1.27 Å, respectively. Formation of the disulfide bond followed by ECL2 geometry optimization provided slightly larger improvements of 0.37, 2.11, 0.86, and 1.41 Å across the same groups. A few targets showed more substantial changes, including PAR2 for which T10 dropped from 15.49 to 4.68 Å upon filtering and AA2AR for which T10 changed in the opposite direction from 7.05 to 9.88 Å upon filtering. Figure 16 compares the PAR2 structures selected solely based on score (panel a) and those selected based on score after filtering based on S–S distance (panel c) with the reference crystal structure (panel b). This figure illustrates that the ten top scored loop models consistently show interactions between ECL2 and the transmembrane domain, with ECL2 intruding into the lipid bilayer, which was absent during loop modeling. Use of the S–S distance filter eliminated many of the models in which ECL2 occupies space that should be reserved for the lipid bilayer, producing the > 10 Å improvement in the lowest RMSD found within the top 10 scored structures (Table 4). It is likely that the models in Fig. 16a scored well due to burial of hydrophobic sidechains, an important driver for folding of soluble proteins that have been used in parameterizing typical energy functions. Alternative filters (such as setting a minimum distance between atoms in ECL2 and those in the membrane-embedded sidechains of the TM segments) or hybrid scoring functions that appropriately treat the membrane-embedded region of transmembrane proteins could also effectively filter out unwanted structures like those in Fig. 16a. In contrast to the substantial benefit filtering provided in selecting better models of PAR2 relative to scores alone, filtering lowered the quality of the best model in the top 10 scored structures for AA2AR. As shown in Fig. 17, the majority of the top 10 AA2AR ECL2 models did not exhibit an ECL2 position overlapping with the position of the lipid bilayer. In this case and for others in the Group 3 target set, ECL2 models with the desired accuracy (< 2.5 Å RMSD were not present in the 1000 models initially sampled, and more sampling would be required in order to obtain improved models).
PAR2 ECL2 models (KICF) and reference structure. a Top 10 scored PAR2 ECL2 models produced within 1000 structures generated using KICF in Rosetta. b PAR2 crystallographic reference structure. c Top 10 scored PAR2 ECL2 models after filtering 1000 structures generated using KICF in Rosetta for Cys3.25–Cys45.50 S–S distances 5.1 ≤ Å
AA2AR ECL2 models (KICF) and reference structure. a Top 10 scored AA2AR ECL2 models produced within 1000 structures generated using KICF in Rosetta. b AA2AR crystallographic reference structure. c Top 10 scored AA2AR ECL2 models after filtering 1000 structures generated using KICF in Rosetta for Cys3.25–Cys45.50 S–S distances 5.1 ≤ Å
Comparison to other GPCR structure prediction benchmarks
Advances in GPCR structure prediction have been assessed in the community-wide GPCR DOCK experiments in 2008, 2010, 2013 [30, 31, 141]. In these experiments, researchers were tasked with modeling a target GPCR with a bound ligand prior to the publication of the target crystal structures. In GPCR DOCK 2010, two of the targets utilized were the crystal structures of the DRD3/eticlopride (PDB: 3PBL) and CXCR4/IT1t (PDB: 3ODU) receptor-ligand complexes [31]. Overall, there were no models submitted for either target where the ECL2 had a backbone RMSD within 2.5 Å in comparison to the crystal structure. While the best DRD3 models had ECL2 RMSD values of 2.69 Å, CXCR4 was a more difficult modelling target where the two best models had ECL2 RMSD values of 4.32 and 6.61 Å. Based on all submitted models of DRD3 and CXCR4, the median RMSD values for ECL2 were 4.11 and 9.19 Å, respectively. While the RMSD values presented in this benchmark study show significant improvement in modeling the ECL2 of DRD3 and CXCR4 in terms of sampling, it is remarkable that the LRM obtained from KICF loop modeling with CXCR4 and DRD3 were ranked within the top 10 scored models after Cys3.25–Cys45.50 S–S distance filtering and had RMSD values of 0.52 and 0.86 Å, respectively. However, it should be noted that there is a significant advantage in modeling loops starting with a GPCR crystal structure versus a homology model. In general, a template-based GPCR model (without additional refinement) has an equivalent backbone structure to the aligned segments of the template. However, GPCR structures tend to diverge at the extracellular ends, and thus homology models matched to a template structure may not reflect real structural differences between the target and template structure. Arora et al. showed that variations in loop anchor positions can have significant influence on modeling accuracy for GPCR loops [142]. Therefore, the results obtained from this benchmark study represent a best-case scenario for modeling GPCR loops where the starting structure contained no errors in the anchor positions or in the structures of the geometrically proximal ECL1 or ECL3.
Conclusion
Overall, the results from modeling Group 1 and 2 ECL2 targets showed that KICF sampled the most loop conformations with sub-angstrom and near-atomic accuracy to the reference structure. The NGK algorithm followed just behind KICF in terms of building loop models with sub-angstrom accuracy, but both algorithms had the same number of cases of modeling loops with near-atomic accuracy. Although the CCD algorithm was not able to build any loop models with sub-angstrom accuracy for the targets in Group 1 and 2, CCD produced models with near-atomic accuracy for 9 of the 14 targets. The results from modeling Group 3 ECL2 targets showed that only the KICF algorithm was able to sample loop models with sub-angstrom accuracy. Overall, these data suggest that the KIC with fragments and next generation KIC methods within Rosetta perform better than the cyclic coordinate descent method or the de novo search method within MOE loop modeler for loops with up to 21 residues (Groups 1 and 2). For the longer loops in Group 3 (20–24 residues), KIC with Fragments outperforms all the other methods. Out of all 28 GPCR loops modeled, KICF generated the most models under 2.5 Å out of 1000 produced total. For loop targets analogous to those in Group 4 (25–32 residues) where no models were within 2.5 Å RMSD regardless of loop modeling method, it is recommended that a greater number of models be produced (i.e. > 4000) to increase sampling of ECL2 conformational space. Application of loop modeling to generate unknown structures requires that models from the sampled set be selected. Regardless of loop length, the RMSD of the T1 was much higher than the LRM in most cases. This was also observed in another benchmark study targeting 13 GPCR ECL2 using the CABS modeling software [143]. Scores alone were less effective than the combination of scores and use of the conserved disulfide bond between Cys3.25 in TM3 and Cys45.50 in ECL2 as a filter. Notably, selection of ten structures provided substantial improvement in model quality within the set over selection of only one structure, but inclusion of additional structures up to 25 did not provide significant additional gains. Overall, 11 of the 21 targets in Groups 1–3 included at least one model with an RMSD ≤ 2.5 Å within the top 10 scored models meeting the S–S distance filter. Four targets in Groups 1–3 sampled a model with RMSD was ≤ 2.5 Å that did not score in the top ten models after S–S distance filtering. In these cases, adjustments to the scoring function, refinement methods, or loop structure environment would be needed improve loop structure prediction.
References
Ribas C, Penela P, Murga C, Salcedo A, García-Hoz C, Jurado-Pueyo M, Aymerich I, Mayor F (2007) The G protein-coupled receptor kinase (GRK) interactome: role of GRKs in GPCR regulation and signaling. Biochim Biophys Acta BBA 1768:913–922
Lagerström MC, Schiöth HB (2008) Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov 7:339–357
Rosenbaum DM, Rasmussen SGF, Kobilka BK (2009) The structure and function of G-protein-coupled receptors. Nature 459:356–363
Gudermann T, Nürnberg B, Schultz G (1995) Receptors and G proteins as primary components of transmembrane signal transduction. Part 1. G-protein-coupled receptors: structure and function. J Mol Med 73:51–63
Overington JP, Al-Lazikani B, Hopkins AL (2006) How many drug targets are there? Nat Rev Drug Discov 5:993–996
van der Kant R, Vriend G (2014) Alpha-Bulges in G Protein-Coupled Receptors. Int J Mol Sci 15:7841–7864
Isberg V, de Graaf C, Bortolato A, Cherezov V, Katritch V, Marshall FH, Mordalski S, Pin J, Stevens RC, Vriend G, Gloriam DE (2015) Generic GPCR residue numbers-aligning topology maps while minding the gaps. Trends Pharmacol Sci 36(1):22–31
Avlani VA, Gregory KJ, Morton CJ, Parker MW, Sexton PM, Christopoulos A (2007) Critical role for the second extracellular loop in the binding of both orthosteric and allosteric G protein-coupled receptor ligands. J Biol Chem 282:25677–25686
Peeters MC, van Westen GJP, Li Q, IJzerman AP (2011) Importance of the extracellular loops in G protein-coupled receptors for ligand recognition and receptor activation. Trends Pharmacol Sci 32:35–42
Klco JM, Wiegand CB, Narzinski K, Baranski TJ (2005) Essential role for the second extracellular loop in C5a receptor activation. Nat Struct Mol Biol 12:320
Nanevicz T, Wang L, Chen M, Ishii M, Coughlin SR (1996) Thrombin receptor activating mutations: alteration of an extracellular agonist recognition domain causes constitutive signaling. J Biol Chem 271:702–706
Pantel J, Legendre M, Cabrol S, Hilal L, Hajaji Y, Morisset S, Nivot S, Vie-Luton M, Grouselle D, Kerdanet Md, Kadiri A, Epelbaum J, Bouc YL, Amselem S (2006) Loss of constitutive activity of the growth hormone secretagogue receptor in familial short stature. J Clin Invest 116:760–768
Wheatley M, Wootten D, Conner M, Simms J, Kendrick R, Logan R, Poyner D, Barwell J (2012) Lifting the lid on GPCRs: the role of extracellular loops. Br J Pharmacol 165:1688–1703
Chien EYT, Liu W, Zhao Q, Katritch V, Won Han G, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC (2010) Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science 330:1091–1095
Wu B, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC (2010) Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330:1066–1071
Chrencik JE, Roth CB, Terakado M, Kurata H, Omi R, Kihara Y, Warshaviak D, Nakade S, Asmar-Rovira G, Mileni M, Mizuno H, Griffith MT, Rodgers C, Han GW, Velasquez J, Chun J, Stevens RC, Hanson MA (2015) Crystal structure of antagonist bound human lysophosphatidic acid receptor 1. Cell 161:1633–1643
Hanson MA, Roth CB, Jo E, Griffith MT, Scott FL, Reinhart G, Desale H, Clemons B, Cahalan SM, Schuerer SC, Sanna MG, Han GW, Kuhn P, Rosen H, Stevens RC (2012) Crystal structure of a lipid G protein-coupled receptor. Science 335:851–855
Shao Z, Yin J, Chapman K, Grzemska M, Clark L, Wang J, Rosenbaum DM (2016) High-resolution crystal structure of the human CB1 cannabinoid receptor. Nature 540:602
Zhang J, Zhang K, Gao Z, Paoletta S, Zhang D, Han GW, Li T, Ma L, Zhang W, Müller CE, Yang H, Jiang H, Cherezov V, Katritch V, Jacobson KA, Stevens RC, Wu B, Zhao Q (2014) Agonist-bound structure of the human P2Y12 receptor. Nature 509:119
Wang C, Wu H, Katritch V, Han GW, Huang X, Liu W, Siu FY, Roth BL, Cherezov V, Stevens RC (2013) Structure of the human smoothened receptor bound to an antitumour agent. Nature 497:338
Murakami M, Kouyama T (2008) Crystal structure of squid rhodopsin. Nature 453:363
Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H, Kuhn P, Weis WI, Kobilka BK, Stevens RC (2007) High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318:1258–1265
Cheng RKY, Fiez-Vandal C, Schlenker O, Edman K, Aggeler B, Brown DG, Brown GA, Cooke RM, Dumelin CE, Doré AS, Geschwindner S, Grebner C, Hermansson N, Jazayeri A, Johansson P, Leong L, Prihandoko R, Rappas M, Soutter H, Snijder A, Sundström L, Tehan B, Thornton P, Troast D, Wiggin G, Zhukov A, Marshall FH, Dekker N (2017) Structural insight into allosteric modulation of protease-activated receptor 2. Nature 545:112
Zhang C, Srinivasan Y, Arlow DH, Fung JJ, Palmer D, Zheng Y, Green HF, Pandey A, Dror RO, Shaw DE, Weis WI, Coughlin SR, Kobilka BK (2012) High-resolution crystal structure of human protease-activated receptor 1. Nature 492:387
Ma Y, Yue Y, Ma Y, Zhang Q, Zhou Q, Song Y, Shen Y, Li X, Ma X, Li C, Hanson MA, Han GW, Sickmier EA, Swaminath G, Zhao S, Stevens RC, Hu LA, Zhong W, Zhang M, Xu F (2017) Structural basis for apelin control of the human apelin receptor. Structure 25:866.e4
Lu J, Byrne N, Wang J, Bricogne G, Brown FK, Chobanian HR, Colletti SL, Salvo JD, Thomas-Fowlkes B, Guo Y, Hall DL, Hadix J, Hastings NB, Hermes JD, Ho T, Howard AD, Josien H, Kornienko M, Lumb KJ, Miller MW, Patel SB, Pio B, Plummer CW, Sherborne BS, Sheth P, Souza S, Tummala S, Vonrhein C, Webb M, Allen SJ, Johnston JM, Weinglass AB, Sharma S, Soisson SM (2017) Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat Struct Mol Biol 24:570
Glukhova A, Thal DM, Nguyen AT, Vecchio EA, Jörg M, Scammells PJ, May LT, Sexton PM, Christopoulos A (2017) Structure of the adenosine A1 receptor reveals the basis for subtype selectivity. Cell 168:877.e13
Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, IJzerman AP, Cherezov V, Stevens RC (2012) Structural basis for allosteric regulation of GPCRs by sodium ions. Science 337:232–236
Pándy-Szekeres G, Munk C, Tsonkov TM, Mordalski S, Harpsøe K, Hauser AS, Bojarski AJ, Gloriam DE (2018) GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res 46:D446
Michino M, Abola E, Brooks CL, Dixon JS, Moult J, Stevens RC (2009) Community-wide assessment of GPCR structure modeling and docking understanding. Nat Rev Drug Discov 8:455–463
Kufareva I, Rueda M, Katritch V, Stevens RC, Abagyan R (2011) Status of GPCR modeling and docking as reflected by community-wide GPCR Dock 2010 assessment. Structure 19:1108–1126
Chen KM, Sun J, Salvo JS, Baker D, Barth P (2014) High-resolution modeling of transmembrane helical protein structures from distant homologues. PLoS Comput Biol 10:e1003636
Baker D, Sali A (2001) Protein structure prediction and structural genomics. Science 294:93–96
Siu FY, He M, Graaf Cd, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang M, Stevens RC (2013) Structure of the human glucagon class B G-protein-coupled receptor. Nature 499:444
Jazayeri A, Doré AS, Lamb D, Krishnamurthy H, Southall SM, Baig AH, Bortolato A, Koglin M, Robertson NJ, Errey JC, Andrews SP, Teobald I, Brown AJH, Cooke RM, Weir M, Marshall FH (2016) Extra-helical binding site of a glucagon receptor antagonist. Nature 533:274–277
Zhang H, Qiao A, Yang D, Yang L, Dai A, Graaf Cd, Reedtz-Runge S, Dharmarajan V, Zhang H, Han GW, Grant TD, Sierra RG, Weierstall U, Nelson G, Liu W, Wu Y, Ma L, Cai X, Lin G, Wu X, Geng Z, Dong Y, Song G, Griffin PR, Lau J, Cherezov V, Yang H, Hanson MA, Stevens RC, Zhao Q, Jiang H, Wang M, Wu B (2017) Structure of the full-length glucagon class B G-protein-coupled receptor. Nature 546:259
Jazayeri A, Rappas M, Brown AJH, Kean J, Errey JC, Robertson NJ, Fiez-Vandal C, Andrews SP, Congreve M, Bortolato A, Mason JS, Baig AH, Teobald I, Doré AS, Weir M, Cooke RM, Marshall FH (2017) Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature 546:254
Song G, Yang D, Wang Y, de Graaf C, Zhou Q, Jiang S, Liu K, Cai X, Dai A, Lin G, Liu D, Wu F, Wu Y, Zhao S, Ye L, Han GW, Lau J, Wu B, Hanson MA, Liu Z, Wang M, Stevens RC (2017) Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature 546:312–315
Liang Y, Khoshouei M, Glukhova A, Furness SGB, Zhao P, Clydesdale L, Koole C, Truong TT, Thal DM, Lei S, Radjainia M, Danev R, Baumeister W, Wang M, Miller LJ, Christopoulos A, Sexton PM, Wootten D (2018) Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature 555:121–125
Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T (2012) Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature 482:547
Kruse AC, Ring AM, Manglik A, Hu J, Hu K, Eitel K, Hübner H, Pardon E, Valant C, Sexton PM, Christopoulos A, Felder CC, Gmeiner P, Steyaert J, Weis WI, Garcia KC, Wess J, Kobilka BK (2013) Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504:101–106
Thorsen TS, Matt R, Weis WI, Kobilka BK (2014) Modified T4 lysozyme fusion proteins facilitate G protein-coupled receptor crystallogenesis. Structure 22:1657–1664
Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka BK (2012) Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature 482:552
Qin L, Kufareva I, Holden LG, Wang C, Zheng Y, Zhao C, Fenalti G, Wu H, Han GW, Cherezov V, Abagyan R, Stevens RC, Handel TM (2015) Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science 347:1117–1122
Hua T, Vemuri K, Nikas SP, Laprairie RB, Wu Y, Qu L, Pu M, Korde A, Jiang S, Ho J, Han GW, Ding K, Li X, Liu H, Hanson MA, Zhao S, Bohn LM, Makriyannis A, Stevens RC, Liu Z (2017) Crystal structures of agonist-bound human cannabinoid receptor CB1. Nature 547:468–471
Hua T, Vemuri K, Pu M, Qu L, Han GW, Wu Y, Zhao S, Shui W, Li S, Korde A, Laprairie RB, Stahl EL, Ho J, Zvonok N, Zhou H, Kufareva I, Wu B, Zhao Q, Hanson MA, Bohn LM, Makriyannis A, Stevens RC, Liu Z (2016) Crystal structure of the human cannabinoid receptor CB1. Cell 167:762.e14
Zhang K, Zhang J, Gao Z, Zhang D, Zhu L, Han GW, Moss SM, Paoletta S, Kiselev E, Lu W, Fenalti G, Zhang W, Müller CE, Yang H, Jiang H, Cherezov V, Katritch V, Jacobson KA, Stevens RC, Wu B, Zhao Q (2014) Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature 509:115–118
Byrne EFX, Sircar R, Miller PS, Hedger G, Luchetti G, Nachtergaele S, Tully MD, Mydock-McGrane L, Covey DF, Rambo RP, Sansom MSP, Newstead S, Rohatgi R, Siebold C (2016) Structural basis of smoothened regulation by its extracellular domains. Nature 535:517–522
Weierstall U, James D, Wang C, White TA, Wang D, Liu W, Spence JCH, Bruce Doak R, Nelson G, Fromme P, Fromme R, Grotjohann I, Kupitz C, Zatsepin NA, Liu H, Basu S, Wacker D, Han GW, Katritch V, Boutet S, Messerschmidt M, Williams GJ, Koglin JE, Marvin Seibert M, Klinker M, Gati C, Shoeman RL, Barty A, Chapman HN, Kirian RA, Beyerlein KR, Stevens RC, Li D, Shah STA, Howe N, Caffrey M, Cherezov V (2014) Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat Commun 5:3309
Wang C, Wu H, Evron T, Vardy E, Han GW, Huang X, Hufeisen SJ, Mangano TJ, Urban DJ, Katritch V, Cherezov V, Caron MG, Roth BL, Stevens RC (2014) Structural basis for smoothened receptor modulation and chemoresistance to anticancer drugs. Nat Commun 5:4355
Fenalti G, Giguere PM, Katritch V, Huang X, Thompson AA, Cherezov V, Roth BL, Stevens RC (2014) Molecular control of δ-opioid receptor signalling. Nature 506:191
Fenalti G, Zatsepin NA, Betti C, Giguere P, Han GW, Ishchenko A, Liu W, Guillemyn K, Zhang H, James D, Wang D, Weierstall U, Spence JCH, Boutet S, Messerschmidt M, Williams GJ, Gati C, Yefanov OM, White TA, Oberthuer D, Metz M, Yoon CH, Barty A, Chapman HN, Basu S, Coe J, Conrad CE, Fromme R, Fromme P, Tourwé D, Schiller PW, Roth BL, Ballet S, Katritch V, Stevens RC, Cherezov V (2015) Structural basis for bifunctional peptide recognition at human δ-opioid receptor. Nat Struct Mol Biol 22:265–268
Thompson AA, Liu W, Chun E, Katritch V, Wu H, Vardy E, Huang X, Trapella C, Guerrini R, Calo G, Roth BL, Cherezov V, Stevens RC (2012) Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature 485:395
Miller RL, Thompson AA, Trapella C, Guerrini R, Malfacini D, Patel N, Han GW, Cherezov V, Caló G, Katritch V, Stevens RC (2015) The importance of ligand-receptor conformational pairs in stabilization: spotlight on the N/OFQ G protein-coupled receptor. Structure 23:2291–2299
Zhang H, Han GW, Batyuk A, Ishchenko A, White KL, Patel N, Sadybekov A, Zamlynny B, Rudd MT, Hollenstein K, Tolstikova A, White TA, Hunter MS, Weierstall U, Liu W, Babaoglu K, Moore EL, Katz RD, Shipman JM, Garcia-Calvo M, Sharma S, Sheth P, Soisson SM, Stevens RC, Katritch V, Cherezov V (2017) Structural basis for selectivity and diversity in angiotensin II receptors. Nature 544:327
Egloff P, Hillenbrand M, Klenk C, Batyuk A, Heine P, Balada S, Schlinkmann KM, Scott DJ, Schütz M, Plückthun A (2014) Structure of signaling-competent neurotensin receptor 1 obtained by directed evolution in Escherichia coli. Proc Natl Acad Sci USA 111:655
White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG, Grisshammer R (2012) Structure of the agonist-bound neurotensin receptor. Nature 490:508
Krumm BE, White JF, Shah P, Grisshammer R (2015) Structural prerequisites for G-protein activation by the neurotensin receptor. Nat Commun 6:7895
Krumm BE, Lee S, Bhattacharya S, Botos I, White CF, Du H, Vaidehi N, Grisshammer R (2016) Structure and dynamics of a constitutively active neurotensin receptor. Sci Rep 6:38564
Warne T, Moukhametzianov R, Baker JG, Nehmé R, Edwards PC, Leslie AGW, Schertler GFX, Tate CG (2011) The structural basis for agonist and partial agonist action on a β(1)-adrenergic receptor. Nature 469:241–244
Moukhametzianov R, Warne T, Edwards PC, Serrano-Vega MJ, Leslie AG, Tate CG, Schertler GF (2011) Two distinct conformations of helix 6 observed in antagonist-bound structures of a beta1-adrenergic receptor. Proc Natl Acad Sci USA 108:8228–8232
Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GF (2008) X. Structure of a β1-adrenergic G-protein-coupled receptor. Nature 454:486
Christopher JA, Brown J, Doré AS, Errey JC, Koglin M, Marshall FH, Myszka DG, Rich RL, Tate CG, Tehan B, Warne T, Congreve M (2013) Biophysical fragment screening of the β1-adrenergic receptor: identification of high affinity arylpiperazine leads using structure-based drug design. J Med Chem 56:3446–3455
Warne T, Edwards PC, Leslie AGW, Tate CG (2012) Crystal structures of a stabilized β1-adrenoceptor bound to the biased agonists bucindolol and carvedilol. Structure 20:841–849
Miller-Gallacher JL, Nehmé R, Warne T, Edwards PC, Schertler GFX, Leslie AGW, Tate CG (2014) The 2.1 Å resolution structure of cyanopindolol-bound β1-adrenoceptor identifies an intramembrane Na+ ion that stabilises the ligand-free receptor. PLoS ONE 9:e92727
Huang J, Chen S, Zhang JJ, Huang X (2013) Crystal structure of oligomeric β1-adrenergic G protein-coupled receptors in ligand-free basal state. Nat Struct Mol Biol 20:419–425
Sato T, Baker J, Warne T, Brown GA, Leslie AGW, Congreve M, Tate CG (2015) Pharmacological analysis and structure determination of 7-methylcyanopindolol-bound β1-adrenergic receptor. Mol Pharmacol 88:1024–1034
Rasmussen SGF, Choi H, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VRP, Sanishvili R, Fischetti RF, Schertler GFX, Weis WI, Kobilka BK (2007) Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature 450:383–387
Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien VP, Velasquez J, Kuhn P, Stevens RC (2008) A specific cholesterol binding site is established by the 2.8 Å structure of the human beta2-adrenergic receptor. Structure 16:897–905
Wacker D, Fenalti G, Brown MA, Katritch V, Abagyan R, Cherezov V, Stevens RC (2010) Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J Am Chem Soc 132:11443–11445
Bokoch MP, Zou Y, Rasmussen SGF, Liu CW, Nygaard R, Rosenbaum DM, Fung JJ, Choi H, Thian FS, Kobilka TS, Puglisi JD, Weis WI, Pardo L, Prosser RS, Mueller L, Kobilka BK (2010) Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature 463:108–112
Rasmussen SGF, Choi H, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK (2011) Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature 469:175–180
Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SGF, Choi H, Devree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK (2011) Structure and function of an irreversible agonist-β(2) adrenoceptor complex. Nature 469:236–240
Rasmussen SGF, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah STA, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK (2011) Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477:549–555
Ring AM, Manglik A, Kruse AC, Enos MD, Weis WI, Garcia KC, Kobilka BK (2013) Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nature 502:575
Zou Y, Weis WI, Kobilka BK (2012) N-terminal T4 lysozyme fusion facilitates crystallization of a G protein coupled receptor. PLoS ONE 7:e46039
Weichert D, Kruse AC, Manglik A, Hiller C, Zhang C, Hübner H, Kobilka BK, Gmeiner P (2014) Covalent agonists for studying G protein-coupled receptor activation. Proc Natl Acad Sci USA 111:10744–10748
Staus DP, Strachan RT, Manglik A, Pani B, Kahsai AW, Kim TH, Wingler LM, Ahn S, Chatterjee A, Masoudi A, Kruse AC, Pardon E, Steyaert J, Weis WI, Prosser RS, Kobilka BK, Costa T, Lefkowitz RJ (2016) Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature 535:448–452
Huang CY, Olieric V, Ma P, Howe N, Vogeley L, Liu X, Warshamanage R, Weinert T, Panepucci E, Kobilka B, Diederichs K, Wang M, Caffrey M (2016) In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures. Acta Crystallogr D 72:93–112
Ma P, Weichert D, Aleksandrov LA, Jensen TJ, Riordan JR, Liu X, Kobilka BK, Caffrey M (2017) The cubicon method for concentrating membrane proteins in the cubic mesophase. Nat Protoc 12:1745–1762
Shihoya W, Nishizawa T, Okuta A, Tani K, Dohmae N, Fujiyoshi Y, Nureki O, Doi T (2016) Activation mechanism of endothelin ETB receptor by endothelin-1. Nature 537:363
Shihoya W, Nishizawa T, Yamashita K, Inoue A, Hirata K, Kadji FMN, Okuta A, Tani K, Aoki J, Fujiyoshi Y, Doi T, Nureki O (2017) X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog. Nat Struct Mol Biol 24:758–764
Okada T, Sugihara M, Bondar A, Elstner M, Entel P, Buss V (2004) The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J Mol Biol 342:571–583L; This paper is dedicated to Dr Yoshimasa Kyogoku
Li J, Edwards PC, Burghammer M, Villa C, Schertler GF (2004) Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol 343:1409–1438
Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y (2002) Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci USA 99:5982–5987
Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M (2000) Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289:739–745
Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE (2001) Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs). Biochemistry 40:7761–7772
Nakamichi H, Okada T (2006) Crystallographic analysis of primary visual photochemistry. Angew Chem Int Ed Engl 45:4270–4273
Nakamichi H, Buss V, Okada T (2007) Photoisomerization mechanism of rhodopsin and 9-cis-rhodopsin revealed by X-ray crystallography. Biophys J 92:106
Standfuss J, Xie G, Edwards PC, Burghammer M, Oprian DD, Schertler GF (2007) Crystal structure of a thermally stable rhodopsin mutant. J Mol Biol 372:1179–1188
Nakamichi H, Okada T (2006) Local peptide movement in the photoreaction intermediate of rhodopsin. Proc Natl Acad Sci USA 103:12729–12734
Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA, Palczewski K (2006) Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci USA 103:16123–16128
Standfuss J, Edwards PC, D’Antona A, Fransen M, Xie G, Oprian DD, Schertler GF (2011) The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature 471:656–660
Makino CL, Riley CK, Looney J, Crouch RK, Okada T (2010) Binding of more than one retinoid to visual opsins. Biophys J 99:2366–2373
Park JH, Scheerer P, Hofmann KP, Choe H, Ernst OP (2008) Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454:183–187
Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe H, Hofmann KP, Ernst OP (2008) Crystal structure of opsin in its G-protein-interacting conformation. Nature 455:497–502
Choe H, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP (2011) Crystal structure of metarhodopsin II. Nature 471:651–655
Stenkamp RE (2008) Alternative models for two crystal structures of bovine rhodopsin. Acta Crystallogr D D64:902–904
Blankenship E, Vahedi-Faridi A, Lodowski DT (2015) The high-resolution structure of activated opsin reveals a conserved solvent network in the transmembrane region essential for activation. Structure 23:2358–2364
Park JH, Morizumi T, Li Y, Hong JE, Pai EF, Hofmann KP, Choe H, Ernst OP (2013) Opsin, a structural model for olfactory receptors? Angew Chem Int Ed Engl 52:11021–11024
Singhal A, Ostermaier MK, Vishnivetskiy SA, Panneels V, Homan KT, Tesmer JJG, Veprintsev D, Deupi X, Gurevich VV, Schertler GFX, Standfuss J (2013) Insights into congenital stationary night blindness based on the structure of G90D rhodopsin. EMBO Rep 14:520–526
Szczepek M, Beyrière F, Hofmann KP, Elgeti M, Kazmin R, Rose A, Bartl FJ, von Stetten D, Heck M, Sommer ME, Hildebrand PW, Scheerer P (2014) Crystal structure of a common GPCR-binding interface for G protein and arrestin. Nat Commun 5:4801
Deupi X, Edwards P, Singhal A, Nickle B, Oprian D, Schertler G, Standfuss J (2012) Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc Natl Acad Sci USA 109:119–124
Gulati S, Jastrzebska B, Banerjee S, Placeres ÁL, Miszta P, Gao S, Gunderson K, Tochtrop GP, Filipek S, Katayama K, Kiser PD, Mogi M, Stewart PL, Palczewski K (2017) Photocyclic behavior of rhodopsin induced by an atypical isomerization mechanism. Proc Natl Acad Sci USA 114:E2615
Singhal A, Guo Y, Matkovic M, Schertler G, Deupi X, Yan EC, Standfuss J (2016) Structural role of the T94I rhodopsin mutation in congenital stationary night blindness. EMBO Rep 17:1431–1440
Lebon G, Warne T, Edwards PC, Bennett K, Langmead CJ, Leslie AGW, Tate CG (2011) Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature 474:521–525
Doré AS, Robertson N, Errey JC, Ng I, Hollenstein K, Tehan B, Hurrell E, Bennett K, Congreve M, Magnani F, Tate CG, Weir M, Marshall FH (2011) Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure 19:1283–1293
Jaakola V, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, IJzerman AP, Stevens RC (2008) The 2.6 Angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 322:1211–1217
Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao Z, Cherezov V, Stevens RC (2011) Structure of an agonist-bound human A2A adenosine receptor. Science 332:322–327
Congreve M, Andrews SP, Doré AS, Hollenstein K, Hurrell E, Langmead CJ, Mason JS, Ng IW, Tehan B, Zhukov A, Weir M, Marshall FH (2012) Discovery of 1,2,4-triazine derivatives as adenosine A(2A) antagonists using structure based drug design. J Med Chem 55:1898–1903
Hino T, Arakawa T, Iwanari H, Yurugi-Kobayashi T, Ikeda-Suno C, Nakada-Nakura Y, Kusano-Arai O, Weyand S, Shimamura T, Nomura N, Cameron AD, Kobayashi T, Hamakubo T, Iwata S, Murata T (2012) G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature 482:237–240
Lebon G, Edwards PC, Leslie AGW, Tate CG (2015) Molecular determinants of CGS21680 binding to the human adenosine A2A receptor. Mol Pharmacol 87:907–915
Segala E, Guo D, Cheng RKY, Bortolato A, Deflorian F, Doré AS, Errey JC, Heitman LH, IJzerman AP, Marshall FH, Cooke RM (2016) Controlling the dissociation of ligands from the adenosine A2A receptor through modulation of salt bridge strength. J Med Chem 59:6470–6479
Carpenter B, Nehmé R, Warne T, Leslie AGW, Tate CG (2016) Structure of the adenosine A(2A) receptor bound to an engineered G protein. Nature 536:104–107
Sun B, Bachhawat P, Chu ML, Wood M, Ceska T, Sands ZA, Mercier J, Lebon F, Kobilka TS, Kobilka BK (2017) Crystal structure of the adenosine A2A receptor bound to an antagonist reveals a potential allosteric pocket. Proc Natl Acad Sci USA 114:2066–2071
Batyuk A, Galli L, Ishchenko A, Han GW, Gati C, Popov PA, Lee M, Stauch B, White TA, Barty A, Aquila A, Hunter MS, Liang M, Boutet S, Pu M, Liu Z, Nelson G, James D, Li C, Zhao Y, Spence JCH, Liu W, Fromme P, Katritch V, Weierstall U, Stevens RC, Cherezov V (2016) Native phasing of X-ray free-electron laser data for a G protein-coupled receptor. Sci Adv 2:e1600292
Dore AS, Bortolato A, Hollenstein K, Cheng R, Marshall HF, Read J (2018) Decoding corticotropin-releasing factor receptor type 1 crystal structures. http://www.eurekaselect.com/149116/article. Accessed 1 May 2018
Thal DM, Sun B, Feng D, Nawaratne V, Leach K, Felder CC, Bures MG, Evans DA, Weis WI, Bachhawat P, Kobilka TS, Sexton PM, Kobilka BK, Christopoulos A (2016) Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature 531:335
Burg JS, Ingram JR, Venkatakrishnan AJ, Jude KM, Dukkipati A, Feinberg EN, Angelini A, Waghray D, Dror RO, Ploegh HL, Garcia KC (2015) Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science 347:1113–1117
Zheng Y, Han GW, Abagyan R, Wu B, Stevens RC, Cherezov V, Kufareva I, Handel TM (2017) Structure of CC chemokine receptor 5 with a potent chemokine antagonist reveals mechanisms of chemokine recognition and molecular mimicry by HIV. Immunity 46:1017.e5
Zhang D, Gao Z, Zhang K, Kiselev E, Crane S, Wang J, Paoletta S, Yi C, Ma L, Zhang W, Han GW, Liu H, Cherezov V, Katritch V, Jiang H, Stevens RC, Jacobson KA, Zhao Q, Wu B (2015) Two disparate ligand-binding sites in the human P2Y1 receptor. Nature 520:317
Wu H, Wang C, Gregory KJ, Han GW, Cho HP, Xia Y, Niswender CM, Katritch V, Meiler J, Cherezov V, Conn PJ, Stevens RC (2014) Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344:58–64
Yin J, Babaoglu K, Brautigam CA, Clark L, Shao Z, Scheuermann TH, Harrell CM, Gotter AL, Roecker AJ, Winrow CJ, Renger JJ, Coleman PJ, Rosenbaum DM (2016) Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors. Nat Struct Mol Biol 23:293
Kim DE, Chivian D, Baker D (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32:526
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242
Gerber PR, Müller K (1995) MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry. J Comput Aided Mol Des 9:251–268
Jakalian A, Jack DB, Bayly CI (2002) Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem 23:1623–1641
Case D, Darden T, Cheatham T, Simmerling C, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz K, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Götz A, Kolossváry I, Wong KF, Paesani F, Kollman PA (2012) AMBER 12. University of California, San Francisco, vol 79, pp 926–935
Canutescu AA, Dunbrack RL (2003) Cyclic coordinate descent: a robotics algorithm for protein loop closure. Protein Sci 12:963–972
Wang C, Bradley P, Baker D (2007) Protein-protein docking with backbone flexibility. J Mol Biol 373:503–519
Mandell DJ, Coutsias EA, Kortemme T (2009) Sub-Angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat Methods 6:551–552
Stein A, Kortemme T (2013) Increased sampling of near-native protein conformations. PLoS ONE 8(5):e63090
Berkholz DS, Driggers CM, Shapovalov MV, Dunbrack RL, Karplus PA (2012) Nonplanar peptide bonds in proteins are common and conserved but not biased toward active sites. Proc Natl Acad Sci USA 109:449–453
Leaver-Fay A, O’Meara MJ, Tyka M, Jacak R, Song Y, Kellogg EH, Thompson J, Davis IW, Pache RA, Lyskov S, Gray JJ, Kortemme T, Richardson JS, Havranek JJ, Snoeyink J, Baker D, Kuhlman B (2013) Scientific benchmarks for guiding macromolecular energy function improvement. Methods Enzymol 523:109–143
O’Meara MJ, Leaver-Fay A, Tyka MD, Stein A, Houlihan K, DiMaio F, Bradley P, Kortemme T, Baker D, Snoeyink J, Kuhlman B (2015) Combined covalent-electrostatic model of hydrogen bonding improves structure prediction with Rosetta. J Chem Theory Comput 11:609–622
Park H, Bradley P, Greisen P, Liu Y, Mulligan VK, Kim DE, Baker D, DiMaio F (2016) Simultaneous optimization of biomolecular energy functions on features from small molecules and macromolecules. J Chem Theory Comput 12:6201–6212
Alford RF, Leaver-Fay A, Jeliazkov JR, O’Meara MJ, DiMaio FP, Park H, Shapovalov MV, Renfrew PD, Mulligan VK, Kappel K, Labonte JW, Pacella MS, Bonneau R, Bradley P, Dunbrack RL, Das R, Baker D, Kuhlman B, Kortemme T, Gray JJ (2017) The Rosetta all-atom energy function for macromolecular modeling and design. J Chem Theory Comput 13:3031–3048
Boomsma W, Hamelryck T (2005) Full cyclic coordinate descent: solving the protein loop closure problem in Cα space. BMC Bioinformatics 6:159
Rotkiewicz P, Skolnick J (2008) Fast procedure for reconstruction of full-atom protein models from reduced representations. J Comput Chem 29:1460–1465
Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE (2004) Friesner, R. A. A hierarchical approach to all-atom protein loop prediction. Proteins 55:351–367
Kufareva I, Katritch V, Stevens RC, Abagyan R (2014) Advances in GPCR modeling evaluated by the GPCR Dock 2013 assessment: meeting new challenges. Structure 22:1120–1139
Arora B, Coudrat T, Wooten D, Christopoulos A, Noronha SB, Sexton PM (2016) Prediction of loops in G protein-coupled receptor homology models: effect of imprecise surroundings and constraints. J Chem Inf Model 56:671–686
Kmiecik S, Jamroz M, Kolinski M (2014) Structure prediction of the second extracellular loop in G protein-coupled receptors. Biophys J 106:2408–2416
Funding
This study was funded by National Institute of Mental Health (Grant No. R15 MH109034).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wink, L.H., Baker, D.L., Cole, J.A. et al. A benchmark study of loop modeling methods applied to G protein-coupled receptors. J Comput Aided Mol Des 33, 573–595 (2019). https://doi.org/10.1007/s10822-019-00196-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00196-x