Abstract
Key message
The genes coding for wheat ATG4 and ATG8 were cloned and their roles in autophagy were verified. Implications of ATG4/ATG8 in wheat responses to stresses were suggested by expression profiling.
Abstract
Autophagy-related proteins ATG4 and ATG8 are crucial for autophagy biogenesis. ATG4 processes ATG8 precursor to expose its C-terminal glycine for phosphatidyl ethanolamine (PE) lipidation. ATG8, in the form of ATG8-PE adduct, functions in the organization dynamics of autophagic membranes. Here, we report the identification of two/nine members of the ATG4/ATG8 family from common wheat (Triticum aestivum L.). Expression of each wheat ATG4/ATG8 could complement the autophagy activity of yeast atg4/atg8 mutant cells. GFP fusion proteins of ATG8s, especially of ATG8s with innate C-terminal-exposed glycines, localized to punctate autophagic membranes. Both of purified ATG4s could cleave ATG8s in vitro, but they had different activities and different preferences for ATG8 substrates. Two times of transcript accumulation, an early one and a late one, of ATG4s/ATG8s were detected in the early phases of the Pm21- and Pm3f-triggered wheat incompatible reactions to the powdery mildew causal fungus Blumeria graminis f. sp. tritici (Bgt), and fluorescence microscopy also revealed a Bgt-induced enhanced wheat autophagy level in the Pm21-triggered incompatible reaction. Only one time of Bgt-induced transcript accumulation of ATG4s/ATG8s, corresponding to but much higher than the late one in incompatible reactions, was detected in a susceptible line isogenic to the Pm21 resistance line. These results suggested positive roles of ATG4/ATG8-associated autophagy process in the early stage and possible negative roles in the late stage of wheat immunity response to Bgt. In addition, expression of wheat ATG4s/ATG8s was also found to be upregulated by abiotic stress factors and distinctively regulated by different phytohormones.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Autophagy is a degradative process adopted by eukaryotic cells to clean up excessive or damaged cellular structures and macromolecules for recycling of nutrients (Liu and Bassham 2012; Li and Vierstra 2012; Klionsky et al. 2012). In autophagy, batch of cytoplasmic materials are engulfed into double membrane-bound autophagosomes and eventually delivered as inner membrane-enclosed autophagic bodies into the vacuole/lysosome for breakdown. So far, 36 autophagy-related (ATG) proteins have been identified in yeast, among which 17 core ATGs are involved in the biogenesis of autophagosomes (Xie and Klionsky 2007; Reggiori and Klionsky 2013). Two ubiquitin-like proteins, ATG8 and ATG12, and their conjugation systems which are essential for autophagy have been studied in detail (Mizushima et al. 1998; Ohsumi 2001; Reggiori and Klionsky 2013). Newly synthesized ATG8 precursor needs cleavage by the cysteine protease ATG4 to expose a C-terminal glycine residue, whereas ATG12 has an innate C-terminal glycine residue. The conjugation systems begin with the ATP-dependent linkage between the ATG8/ATG12 C-terminal glycine and a conservative cystine in the E1-like protein ATG7. Then, ATG8 and ATG12 are, respectively, transferred to the E2-like protein ATG3 and ATG10. Finally, ATG8 is donated to the lipid phosphatidyl ethanolamine (PE) to form ATG8-PE adduct (ATG8 lipidation); ATG12 is connected with ATG5. ATG12–ATG5 directly associates with ATG16 and forms a large complex through ATG16 oligomerization, which may function as an E3-like enzyme to catalyze the last step of ATG8-PE production (Hanada et al. 2007; Romanov et al. 2012). ATG8 lipidation and its autophagic membrane positioning are crucial for autophagic membrane assembly, extension, closure and fusion with vacuole (Nakatogawa et al. 2007; Xie et al. 2008). Besides processing ATG8 to expose its C-terminal glycine, ATG4 can also deconjugate ATG8-PE to release ATG8 from autophagic membranes, which is an important step facilitating autophagosome maturation and converting it into the fusion-capable form (Nair et al. 2012).
The autophagy process is highly conserved and ATG orthologues retain in fungi, plants and mammals (Bassham 2009; Liu and Bassham 2012). As to plants, large scale identifications have suggested a total of 36 ATG genes in Arabidopsis (Doelling et al. 2002; Hanaoka et al. 2002; Yoshimoto et al. 2004; Bassham et al. 2006) and certain numbers in rice (Xia et al. 2011) and maize (Chung et al. 2009). ATG4/ATG8 and other key factors in the two conjugation systems are also essential for plant autophagy (Hanaoka et al. 2002; Ketelaar et al. 2004; Yoshimoto et al. 2004; Yoshimoto 2012). The formation processes of Arabidopsis ATG8-PE and ATG12–ATG5·ATG16s complex have been simulated using recombinant proteins in vitro (Fujioka et al. 2008). Preliminary work on the identification of ATG4/ATG8 families has also been reported in crop plants as rice (Su et al. 2006; Xia et al. 2011), corn (Chung et al. 2009) and soybean (Xia et al. 2012).
Autophagy has been suggested to play important roles in plant growth, development and responses to biotic and abiotic stresses including nutrition deficiency, oxidation, osmosis, drought and pathogen infections (Bassham et al. 2006; Hayward and Dinesh-Kumar 2011). So far, few studies on mechanisms and physiological functions of autophagy have been reported in crop plants, especially few in common wheat (Triticum aestivum L.) which is one of the most important food crops in the world. Here, for the first time, we report the identification of ATG4 and ATG8 family members from common wheat. Evidences from yeast atg mutant complementation, subcellular localization of GFP fusion proteins and in vitro enzyme digestion analysis of ATG8s by ATG4s were provided to demonstrate the essential roles of ATG4s/ATG8s we identified on autophagy biogenesis in wheat. Expression profiles of wheat ATG4s/ATG8s in response to the powdery mildew causal fungus Blumeria graminis f. sp. tritici (Bgt), to various abiotic stresses and to exogenously applied phytohormones were also presented in detail. These results provided crucial clues for deciphering the underlying roles of ATG4/ATG8 and their associated autophagy process in wheat responses to severe environmental conditions.
Materials and methods
Plant materials and fungal isolate
Two pairs of wheat near isogenic lines were used in this study. One pair was 92R137/Yangmai 1587 (carrying the broad-spectrum powdery mildew resistance gene Pm21) and its recurrent parent Yangmai 158, and the other pair was Michigan Amber/Chancellor8 (carrying the isolate-specific powdery mildew resistance gene Pm3f) and its recurrent parent Chancellor. The prevalent Chinese Bgt isolate E09, which is avirulent to Pm21 and Pm3f but virulent to Yangmai 158 and Chancellor, was maintained on seedlings of the susceptible cultivar Sumai 3 in a spore-proof greenhouse.
Growth conditions and stress/chemical treatments
Wheat seeds were sown in 9-cm pots filled with a mixture of peat, vermiculite and perlite and grown under a 20 °C, 16-h light/8-h dark regime in a controlled climate chamber. Two-leaf stage seedlings were subject to different treatments. Bgt inoculation was conducted by heavily shaking off fresh Bgt conidiospores from diseased plants onto experiment seedlings. For treatments of exogenous phytohormones, 2 mM salicylic acid (SA), 1 mM Methyl jasmonate (MeJA), 200 μM Ethylene (ET) generator ethephon or 100 μM abscisic acid (ABA) each in 0.05 % Tween-20 were sprayed onto leaves. Seedlings sprayed with 0.05 % Tween-20 were used as controls. For abiotic treatments, seedlings were transferred to Murashige and Skoog (MS) salt solution for several days and then respectively adding in the MS salt solution of 200 mM NaCl (high salinity stress), 20 % PEG-6000 (drought stress) or by depleting NH4NO3 and replacing KNO3 with KCl (nitrogen deficiency). For low temperature/darkness treatment, seedlings were transferred to a 4 °C refrigerator and kept in darkness. Leaves harvested at defined time points were frozen by liquid nitrogen and then stored at −80 °C.
Gene cloning and sequence analysis
Protein sequences of ATG4/ATG8 family members from Arabidopsis (Hanaoka et al. 2002) and rice (Xia et al. 2011) were used as electronic probes for TBLASTN search against the dbEST division of GenBank. Wheat ESTs putatively encoding ATG4/ATG8 were downloaded from the TBLASTN results and assembled into contigs in Vector NTI Advance 11.5. If needed, contigs were electronically extended by searching for new terminal ESTs to cover full-length ORFs. Primers (Table S1) were designed based on the assembled contigs for subsequent amplification of cDNAs by RT-PCR. Total RNA was prepared using Trizol (Invitrogen) from leaves of 92R137/Yangmai 1587 collected 48 h after inoculation of Bgt. Here, pathogen inoculation was used as an inducer for enhanced transcription of ATG genes of interest. Extracted RNA was treated with DNaseI (RNase-free), and used as templates for first-strand cDNA synthesis by Oligo dT primers and the Quantscript RT Kit (Tiangen). Wheat ATG4/ATG8 cDNAs were amplified with the high-fidelity Pfu DNA polymerase. Amplified fragments were A-tailed and cloned into the T-tailed vector pGEM®-T Easy (Promega). Giving that wheat is a hexaploid with three homoeologous subgenomes (AABBDD) and thus different members of a gene family may share identical sequences at primer binding sites, more than 10 recombinant clones from each PCR product were picked to sequencing. The serial number for each wheat ATG4/ATG8 member was assigned according to their relationship with the members of Arabidopsis ATG4/ATG8 family (Doelling et al. 2002; Hanaoka et al. 2002). Gene sequences were stored in GenBank.
Conservative domains and motifs were predicted in SMART (http://smart.embl-heidelberg.de). Multiple sequence alignment was performed with ClustalX 2.1. Phylogenetic trees were constructed in Mega 5 by the Neighbor-joining method (bootstrap test with 1,000 replicates). Genomic sequences covering upstream promoter regions were obtained by iterative steps of BLAST searching (E ≤ 10−5) and assembling in the wheat draft genome database (http://www.cerealsdb.uk.net) using cDNA sequences as the first input. cDNA and genomic sequences were compared by Splign (NCBI) to analyze the exon–intron structures of genomic ORFs. Cis-acting elements in promoter sequences were predicted in the PlantCare website (http://bioinformatics.psb.ugent.be/webtools/plantcare/) with a matrix score ≥5.
Complementation test of yeast atg mutants
ORFs of wheat ATG4s/ATG8s were amplified as NotI-NotI fragments by primers (Table S1) using the pfu DNA polymerase. PCR fragments were each digested and cloned into the yeast expression vector pFL61 in which constitutive expression of each inserted gene was driven by the yeast phosphoglycerate kinase (PGK) gene promoter. Recombinant vectors were sequenced to prove the correct cDNA insertion direction (initiation codon of cDNA adjacent and downstream to the PGK promoter). Wild-type yeast BY4741 and two mutants, atg4 (BY4741, atg4Δ:: kanMX, MAT a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) and atg8 (BY4741, atg8Δ:: kanMX, MAT a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) were purchased from Open Biosystems (Thermo Scientific). Recombinant vectors were each introduced into yeast atg8 or atg4 cells according to the LiAc/SS-DNA/PEG TRAFO protocol (Gietz and Schiestl 2007). Positive transformants were screened on SC-U medium. Yeast autophagy complementation test was performed according to Fujiki et al. (2007). Briefly, Yeast cells were cultured with shaking at 30 °C in SC medium to mid-log phase (OD 600 = 1). Cells were then collected, washed and incubated for another 5 h in nutrient deprivation medium (0.67 % nitrogen base, without glucose, amino acids and ammonium sulfate) to induce autophagy. 1 mM phenylmethylsulfonyl fluoride (PMSF) was added in the medium to accumulate autophagic bodies in the vacuoles. The appearance of autophagic bodies was observed using a differential interference contrast microscope (DM5000B, Leica).
Subcellular localization of wheat ATG8s
ORFs of wheat ATG8s were amplified using the pfu DNA polymerase by primers with additional restriction sites (Table S1). PCR fragments were each digested and ligated downstream and in-frame with the GFP ORF in the vector pA7-GFP, in which expression of each GFP-ATG8 fusion gene was driven by two tandem-arranged CaMV 35S promoters. Constructed vectors were verified by sequencing. Plasmids were introduced into onion epidermal cells by particle bombardment with PDS1000/He (BioRad) (Scott et al. 1999). GFP fluorescence was visualized by epifluorescence microscopy (DM5000B, Leica) 24 h after bombardment.
In vitro analysis of protease activity of wheat ATG4s on ATG8s
Each of the Not I-Not I PCR fragments of wheat ATG4s/ATG8s amplified in the process of yeast expression vector construction was also cloned into the prokaryotic expression vector pET30a. Recombinant vectors were sequenced to prove the correct cDNA insertion direction (initiation codon of cDNA adjacent and downstream to the T7 promoter). Expression vectors were, respectively, transformed into E. coli BL21 (DE3) by the heat shock method. Purified recombinant proteins were obtained through 1 mM IPTG-induced expression and Ni affinity chromatography under non-denaturing conditions. In vitro enzyme activity analysis was carried out according to Wu et al. (2012). Briefly, 20 μl reaction buffer (150 mM Nacl, 1 mM EDTA, 1 mM DTT, 50 mM Tris–HCl, pH 7.2) containing purified 0.025 mg mL−1 ATG4 and 0.5 mg mL−1 ATG8 was kept at 37 °C for 3 h. Then bromophenol blue sample buffer was added to terminate the reaction. Cleaved products were resolved by 15 % SDS-PAGE with 6 M urea in the gel mix to improve resolution.
Expression analysis by quantitative real-time PCR (qRT-PCR)
Gene-specific qRT-PCR primers (Table S1) were designed, and qRT-PCR was conducted in IQ5 (Biorad) using the RealMasterMix (SYBR Green) kit (Tiangen). Each sample was analyzed in triplicate and the experiment was repeated three times. Relative expression level was calculated using the 2−ΔΔCt method (Livak and Schmittgen 2001) with the amplicon of wheat β-tubulin gene as an internal control.
LysoTracker Red staining and microscopy
Wheat leaf segments were infiltrated with 100 μM E-64d (Sigma) and then stained with 2 μM LysoTracker Red DND-99 (Invitrogen) according to the procedure described previously (Liu Y et al. 2005). Florescence was observed under a confocal microscope (ECLIPSE 90i, Nikon) with 543 nm excitation of a 1 mW helium:neon laser with 565–625 nm band pass.
Results
Identification of wheat ATG8 genes
Plant genomes often contain a plurality of ATG8 paralogs. Likewise, we identified a family of nine ATG8 genes from wheat, eight of which (TaATG8a–8 h, GenBank accession KF294807–KF294814) were identified from the cDNA pool of Bgt-challenged leaves of the powdery mildew resistant wheat line 92R137/Yangmai 1587 and one (TaATG8i) by assembly of ESTs (HX189738, CK196170 and HX250137). Nine TaATG8s were divided into three subfamilies according to their sequence similarity and evolutionary relationship (Fig. 1a, Fig. S1). The six members (TaATG8a–8f) of subfamily I encode only two amino acid sequences (119 A.A.), one by TaATG8a, 8d and 8e and the other by TaATG8b, 8c and 8f, with only one residue difference. TaATG8g, the sole member of subfamily II, encodes an amino acid sequence (119 A.A.) with 93 % similarity to members of subfamily I. Wheat TaATG8 g has identical nucleotide sequence with the reported ATG8 from Triticum dicoccoides (AABB) which is a wheat wild progenitor (Kuzuoglu-Ozturk et al. 2012). There are two members in subfamily III, TaATG8h and 8i, and their nucleotide sequences (351 bp) and encoded amino acid sequences (116 A.A.), respectively, have 96.2 and 97 % identity/similarity. Subfamily III nucleotide/amino acid sequences are 61.5–64.1 %/68 % and 75–76 %/70 % identical/similar to subfamily I and subfamily II, respectively. All of the nine TaATG8s contain an ATG8 ubiquitin domain (pfam: PF02991), within which six essential residues for the N-terminal microtubule binding site, three for the ATG7 binding site and the C-terminal conservative glycine residue for lipidation were predicted (Fig. 1b, Fig. S1).
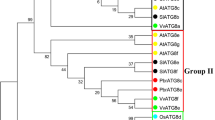
Phylogenetic relationships of eukaryotic ATG4s or ATG8s. Phylogenetic trees were constructed in Mega 5 by the Neighbor-joining method. The reliability of internal branches was assessed by bootstrapping, with 1,000 bootstrap replicates, and values are shown in percentages, with a branching cut-off at 50 %. a Phylogenetic tree of eukaryotic ATG8s. b Alignment of the C-terminal sequences of plant ATG8s. The conservative glycine residue for PE-conjugation is indicated by an arrowhead. Sequences in a and b include the nine wheat ATG8s here identified, AER27507 (TdATG8) from Triticum dicoccoides, XP_003581707 (BdATG8a) and XP_003571383 (BdATG8b) from Brachypodium distachyon, Os07g0512200 (OsATG8a), Os04g0624000 (OsATG8b), Os08g0191600 (OsATG8c) and Os02g0529150 (OsATG8d) from Oryza sativa, ACJ73920 (ZmATG8a), ACJ73921 (ZmATG8b), ACJ73922 (ZmATG8c), ACJ73923 (ZmATG8d), and ACJ73925 (ZmATG8e) from Zea mays, At4g21980 (AtATG8a), At4g04620 (AtATG8b), At1g62040 (AtATG8c), At2g05630 (AtATG8d), At2G45170 (AtATG8e), At4g16520 (AtATG8f), At3g60640 (AtATG8 g), At3g06420 (AtATG8 h) and At3g15580 (AtATG8i) from Arabidopsis thaliana, ACU13796 (GmATG8a), ACU14633 (GmATG8b), ACU17086 (GmATG8c), BAH22449 (GmATG8d), ACU15101 (GmATG8e), ACU19559 (GmATG8f), ACU13862 (GmATG 8 g), ACU16419 (GmATG8 h), BAH22448 (GmATG8i) and ACU19611 (GmATG8 k) from Glycine max, NP_009216 (HsATG8, GATE16/GABARAPL2) from Human and NP_009475 (ScATG8) from Yeast. Names of wheat ATG8s are indicated by asterisks. c Phylogenetic tree of eukaryotic ATG4s. Sequences in the tree include the two wheat ATG4s here identified, Os03g0391000 (OsATG4a) and Os04g0682000 (OsATG4b) from Oryza sativa, ACJ73912 (ZmATG4a) and ACJ73914 (ZmATG4b) from Zea mays, At2g44140 (AtATG4a) and At3g59950 (AtATG4b) from Arabidopsis thaliana, NP_443168 (HsATG4a) from Human and NP_014176 (ScATG4) from Yeast
In the phylogenetic tree, plant ATG8s are clustered into two main clades, Clade I and II (Fig. 1a). Clade I covers the majority of plant ATG8 family members comprising all the members from wheat ATG8 subfamily I and II, 7/9 members from Arabidopsis, 3/4 from rice, 8/10 from soybean and 5/5 from maize, which suggests that the ancestor of Clade I and its descents duplicated more frequently within genomes during evolution. However, high similarities were retained in clade I between paralogs (81–100 %) and between orthologues (80–100 %). Clade II covers 1–2 plant ATG8 family members from each species including TaATG8h–8i of wheat subfamily III. Long branches within clade II suggest that the ancestor of Clade II and its descents experienced rapid evolution processes before and after the differentiation of monocots and dicots. The similarity between Clade I and Clade II is 64–71 %. The ancestor of clade II is related to the production of human ATG8. All plant ATG8s hold the C-terminal conservative glycine residue waiting for PE-conjugation (Fig. 1b, Fig. S1). For ATG8s in clade I, the glycine residue is generally protected by additional 2–6 residues and needs ATG4 cleavage to expose, whereas ATG8s in clade II (except ATG8h and 8i of soybean) have post-translationally exposed C-terminal glycine residues.
BLAST searching against the draft genome sequences of the wheat A-genome progenitor Triticum urartu (Ling et al. 2013) and the D-genome progenitor Aegilops tauschii (Jia et al. 2013) revealed several ATG8 sites on chromosomes 2AL, 2AS, 2D and 5D. Previously, copies of T. dicoccoides ATG8 were mapped on chromosomes 1B, 2A and 2D (Kuzuoglu-Ozturk et al. 2012). The genomic ORF sequences of TaAT8a and 8g have similar four-intron structures with a slight difference in intron size (Fig. 2). The genomic ORF sequence of TaATG8h also contains four introns, but its introns are greatly extended (the second intron) or shortened (the first and third introns) compared to TaATG8a and 8g (Fig. 2).
Identification of wheat ATG4 genes
Two wheat ATG4 genes (TaATG4a–4b, GenBank accession KF294797–KF294798) were identified from the cDNA pool of Bgt-challenged leaves of 92R137/Yangmai 1587. TaATG4a has an ORF of 1,452 bp encoding a 483 A.A. protein, and TaATG4b has an ORF of 1,455 bp encoding a 484 A.A. protein. Three consecutive nucleotides encoding the N-terminus Ala32 of TaATG4b were deleted in TaATG4a (Fig. 2, Fig. S2). The nucleotide and amino acid sequence similarities between TaATG4a and 4b are 96.7 and 97 %, respectively. Both of TaATG4a and 4b have a typical Peptidase_C54 domain (Pfam: PF03416) and a canonical catalytic triad of cysteine protease with three conserved residues: Cys, Asp and His (Fig. S2).
In the phylogenetic tree, ATG4s from different plant species were clustered in separate branches (Fig. 1c), suggesting that ATG4 duplication occurred after the respective speciation of Arabidopsis, wheat, rice and maize. For entire sequences, wheat ATG4s share 83–84, 79–80 and 64–67 % similarities with those of rice, maize and Arabidopsis, respectively. Wheat ATG4s share only 30–31 and 38–39 % similarities with orthologues of yeast and human, much lower than the corresponding ATG8 s similarities of 72–90 % (between wheat and yeast) and 71–78 % (between wheat and human). The Peptidase_C54 domain sequences are conserved among plant ATG4s and some key residues conserved among all eukaryotic ATG4s (Fig. S2). However, the N-terminal sequences before the Peptidase_C54 domain are much diverged between species, and the human ATG4a has a greatly shortened N-terminal sequence. The C-terminal sequences after the Peptidase_C54 domain differ mainly between monocot and dicot ATG4s (Fig. S2).
Results from BLAST searching in the mapped wheat EST database at Graingenes (http://wheat.pw.usda.gov) showed that a homologous EST of TaATG4 mapped on wheat chromosomes 2AL, 2BL and 2DL. The genomic ORF sequences of TaATG4a and 4b have very similar structures with seven introns (Fig. 2).
Wheat ATG4s and ATG8s rescue yeast autophagy
Yeast expression vectors were constructed for wheat ATG4s and ATG8s and introduced, respectively, into yeast atg4 and atg8 mutant cells. After 5-h nutrient starvation induction, wild-type yeast (BY4741) cells accumulated numerous autophagic bodies in vacuoles, which were not (atg8) or rarely (atg4) observed in mutant cells (Fig. 3). Expression of wheat ATG genes partially recovered the autophagy function of yeast atg8 cells (expressing TaATG8a, 8b, 8g or 8h) or atg4 cells (expressing TaATG4a), evidenced by the significantly increased number of autophagic bodies in vacuoles (Fig. 3). Successful complementation verified that wheat TaATG8a, 8b, 8g and 8h are functional homologues of yeast ATG8, and TaATG4a functional homologue of yeast ATG4.
Functional complementation of atg4 or atg8 mutant yeast by wheat ATG4s or ATG8s. Each wheat ATG4/ATG8 gene was cloned in the plasmid pFL61 and, respectively, expressed in atg4 or atg8 yeast cells. Cells grown to mid-log phase were collected, washed, and subsequently incubated for another 5 h in nutrient deprivation medium in the presence of 1 mM PMSF. The appearance of autophagic bodies was observed using a differential interference contrast microscope. Scale bar 5 μm
GFP-TaATG8s localize to autophagic membranes
Localization of ATG8 on the surface of autophagic membranes is essential for autophagy biogenesis. TaATG8a and 8b, 8g and 8h, respectively, representing the wheat ATG8 Subfamily I, Subfamily II and Subfamily III, were selected for subcellular localization analysis. Punctate structures were clearly observed in onion epidermal cells expressing GFP-TaATG8h, which may indicate autophagosomes or their immature structures (Fig. 4). Fluorescence of the other three constructs, GFP-TaATG8a, 8b and 8g, showed few punctate structures and sometimes were even diffusely distributed throughout the cell like the fluorescence in cells expressing GFP alone (Fig. 4). Subcellular localization results suggest that the four wheat ATG8s could be recruited to autophagic membranes and thus involved in autophagy biogenesis.
Wheat ATG4s cleave ATG8s in vitro
ATG8 precursor needs cleavage by the protease ATG4 to expose its N-terminal glycine for PE lipidation. To prove this for wheat ATG4s and ATG8s, purified recombinant proteins of TaATG4a, 4b, 8a, 8b and 8g were obtained through IPTG-induced expression in E. coli and subsequent Ni affinity chromatography. In vitro digestion experiments showed that TaATG8a, 8b, 8g can all be used as cleavage substrates by the two wheat ATG4s (Fig. 5). TaATG4b cleaved the three substrates more efficiently than TaATG4a, and have the highest efficiency towards TaATG8a (Fig. 5).
Wheat ATG8s are cleaved at their C termini by wheat ATG4 proteases. Each wheat ATG4/ATG8 was cloned in the vector pET30a and, respectively, expressed through 1 mM IPTG induction in E. coli BL21 (DE3). Purified recombinant proteins were obtained by Ni affinity chromatography under non-denaturing conditions. Protease cleavage analysis was conducted in a 20-μl reaction buffer containing 0.025 mg mL−1 TaATG4 and 0.5 mg mL−1 TaATG8 at 37 °C for 3 h. Cleaved products were resolved by 15 % SDS-PAGE with 6 M urea in the gel mix to improve resolution. Arrowheads indicate the uncleaved forms of ATG8s (upper bands) and ATG4s-cleaved forms of ATG8s (lower bands), respectively
Expression of wheat ATG4s and ATG8s positively responds to abiotic stresses
Autophagy has been reported to participate in plant responses to abiotic stresses (Bassham et al. 2006). Thus, we exposed the line Yangmai 158 to several abiotic stresses and characterized the expression profiles of TaATG4a, 4b, 8a, 8g and 8h by qRT-PCR. All these five genes showed upregulated expression upon exposure to high salinity, drought, low temperature/darkness (carbon deficiency imposed by impaired photosynthesis) or nitrogen deficiency (Fig. 6a). Drought treatment resulted in the strongest activation of the five genes and had a prolonged effect on TaATG4a and 4b. Low temperature/darkness treatment upregulated the expression of the three TaATG8s more quickly than the other treatments. Among the three TaATG8s, TaATG8a showed the highest transcript accumulation under the treatment of high salinity, and TaATG8h showed the highest transcript accumulation under the treatments of drought and low temperature/darkness, but the lowest transcript accumulation under the treatment of high salinity. As to the two TaATG4s, TaATG4a showed higher transcript accumulation under the treatments of low temperature/darkness and nitrogen deficiency, and TaATG4b showed higher transcript accumulation under the treatment of drought.
Expression patterns of wheat ATG4s and ATG8s. a Expression of wheat ATG4s and ATG8s in responses to abiotic stresses. b Expression of wheat ATG4s and ATG8s in response to Bgt infection. c Expression of wheat ATG4s and ATG8s in responses to phytohormone treatments. Two-leaf stage seedlings were subject to different treatments. For abiotic stress treatments (a), Yangmai 158 seedlings cultured in Murashige and Skoog (MS) salt solution were treated by, respectively, adding in the MS salt solution of 200 mM NaCl (high salinity), 20 % PEG-6000 (drought) or by depleting the NH4NO3 and replacing the KNO3 with KCl (nitrogen deficiency). For low temperature/darkness treatment, seedlings were transferred to 4 °C refrigerator and kept in darkness. Pathogen inoculation (b) was conducted by heavily shaking off fresh Bgt conidiospores from diseased plants onto seedlings of the isogenic lines 92R137/Yangmai 1587 (Pm21 +) and Yangmai 158 (Pm21 -) and the isogenic lines Michigan Amber/Chancellor8 (Pm3f +) and Chancellor (Pm3f −). For treatments of exogenous phytohormones (c), 2 mM SA, 1 mM MeJA, 200 μM ethephon (ET generator) or 100 μm ABA each in 0.05 % Tween-20 were sprayed onto leaves of the isogenic lines 92R137/Yangmai 1587 and Yangmai 158. Samples were collected at the indicated time points after the initiation of treatment. Total RNA was extracted and quantitative real-time PCR (qRT-PCR) analyses were performed with wheat ATG gene-specific primers. The relative expression was normalized by the wheat β-tubulin gene and relative to the control value measured at 0 h. Data represent the average of three independent experiments ± standard deviation (SD, n = 3)
Expression of wheat ATG4s and ATG8s responds to fungus infection
Pathogen infections can induce the expression of plant ATG genes (Hayward and Dinesh-Kumar 2011). To determine if infection by the powdery mildew causal fungus Bgt has such effect on wheat, expression of TaATG4a, 4b, 8a, 8g and 8h in Bgt-challenged wheat leaves was profiled by qRT-PCR in a time period of 0–36 h after inoculation (hai) of Bgt. Two powdery mildew resistance types, Pm21-triggered broad-spectrum resistance and Pm3f-triggered isolate-specific resistance, and two susceptible wheat genotypes were considered in this analysis.
In the Pm21- and Pm3f-triggered incompatible reactions (Fig. 6b), the five wheat ATG genes exhibited two Bgt-induced transcript accumulation (TA) events, an early one and a late one. The early TAs peaked at 6–10 hai, which is around the time when Bgt appressorium germ tubes initially contact and recognize wheat leaf epidermal cells and attempt to invade them. On the contrary to the early TAs in the incompatible reactions, all five genes showed continuous decreased expression during 0–16 hai in the Yangmai 158-Bgt compatible reaction, and four of them showed such decreased expression during 0–6 hai in the Chancellor-Bgt compatible reaction (Fig. 6b). The late TAs in the two incompatible reactions peaked at 16–24 hai or beyond the last investigation time point of 36 hai. Similarly, this time of TA also occurred in the two compatible reactions (Fig. 6b). Around the period of 16–24 hai is the time when invaded structures of Bgt strive in host cells for successful haustorium formation and parasite establishment. The late TAs suggested a second time of ATG4s/ATG8s recruitment occurring after 16 hai in wheat response to Bgt infection. In addition, the late TAs of the analyzed genes were much faster and stronger in the susceptible line Yangmai 158 than in the Yangmai 158-isogenic resistance line 92R137/Yangmai 1587 (Fig. 6b). These Bgt-induced expression profiles imply that ATG4s/ATG8s are universally required in wheat broad-spectrum and isolate-specific resistance responses and in susceptible responses to Bgt.
Wheat autophagy is induced by Bgt infection
Since expression of TaATG4s/TaATG8s positively responds to Bgt infection, we then infiltrated Bgt-inoculated leaves with E-64d and stained them with LysoTracker Red to determine if upregulated expression of ATG genes is in line with elevated autophagy level. E-64d is a membrane-permeable cysteine protease inhibitor and has been successfully used in plant to accumulate autophagic bodies inside autolysosomes including vacuoles and small lytic organelles (Inoue et al. 2006; Bassham 2007). LysoTracker Red is dye which labels acidic organelles such as lysosomes and endosomes. This dye also stains autophagosomes/autolysosomes and dotted structures labeled by this dye have widely been recognized as indicative of plant autophagy activity (Moriyasu et al. 2003; Liu Y et al. 2005; Hofius et al. 2009; Lai et al. 2011; Kwon et al. 2013). Before Bgt inoculation, LysoTracker Red-stained autolysosome-like structures were rarely observed in leaf epidermal cells of the resistant line 92R137/Yangmai 1587 (Fig. 7). After Bgt inoculation, weak (8 hai) or strong (24 hai) level of punctate autolysosome-like structures were detected (Fig. 7), suggesting an enhanced autophagy level required in the Pm21-triggered wheat immune response to Bgt. For the susceptible line Yangmai 158, however, LysoTracker Red-stained punctate structures were rarely detected regardless of Bgt infection (data not shown). Thinking of the enhanced expression of TaATG4s/TaATG8s in Yangmai 158 during 16–24 hai, we cannot exclude that autophagy might become activated in this wheat-Bgt compatible interaction, but failed to be detected by us.
Autophagy is induced in the Pm21-triggered resistance response to Bgt infection. Leaf segments of 92R137/Yangmai 1587 were infiltrated with 100 μM E-64d and then stained with 2 μM LysoTracker Red DND-99. Florescence signals were observed under a confocal microscope (ECLIPSE 90i, Nikon). LysoTracker Red-stained punctate autolysosome-like structures (bright red spots) were not observed in leaf epidermal cells of uninfected 92R137/Yangmai 1587, but weakly at 8 hai and strongly at 24 hai accumulated in leaf epidermal cells of Bgt-challenged 92R137/Yangmai 1587. Results were reproduced in three independent experiments using three or more plants in each experiment. hai, hours after inoculation of Bgt conidiospores. Scale bar 50 µm (color figure online)
Expression of wheat ATG4s and ATG8s responds to exogenously applied phytohormones
Phytohormone signaling pathways are implicated in the involvement of autophagy in plant responses to biotic or abiotic stresses (Liu and Bassham 2012). We hypothesized that the expression of wheat ATG4s and ATG8s can be regulated by phytohormones and that the regulating patterns by same phytohormones have some difference between powdery mildew resistance lines and susceptible lines. Expression analysis (Fig. 6c) revealed that ET and SA treatments inhibited the expression of TaATG4a, 4b, 8a, 8g and 8h in the resistance line 92R137/Yangmai 1587, whereas significantly activated their expression in the susceptible line Yangmai 158. Contrarily, ABA had an activation effect on the resistant line, but an inhibition effect on the susceptible line. MeJA showed inhibitory effects on both of the two lines with a longer duration on the resistance line. For in-depth analysis of the regulation mechanisms of wheat ATG genes in response to pathogen infection and to phytohormone signaling pathways, cis-acting elements were examined on TaATG4b and 8a promoter sequences 2 kb upstream of the predicted transcription start sites. There are three MeJA-response elements (CGTCA motif), one ABA/drought-response element (ABRE) and three defense reaction response elements (one Box-W1, one TC-rich repeats and one CCAAT Box) in the TaATG4b promoter. There are two ABA/drought-response elements (ABRE) in the TaATG8a promoter. In addition, the TaATG4b promoter contains 23 light-response elements, and four tissue-specific expression control elements. The TaATG8a promoter contains 11 light-response elements, four tissue-specific expression control elements, two low temperature response elements and one heat-stress response element.
Discussion
Wheat has functional homologues of yeast ATG4 and ATG8 essential for autophagy
In contrast to yeast, which has a single ATG8, higher eukaryotes usually contain a family of ATG8 members. For examples, there are nine ATG8s in Arabidopsis (Hanaoka et al. 2002), presumably five in maize (Chung et al. 2009), five in rice (Xia et al. 2011) and 11 in soybean (Xia et al. 2012). Here, we identified nine members of wheat ATG8 family. Since common wheat is a hexaploid species (AABBDD), its ATG8 family may contain more members than we identified. Genes for the six members in subfamily I (TaATG8a–8f), for their extreme similarity, may distribute on homoeologous regions of the A, B and D genomes or arise from recent gene duplication events. All wheat ATG8s hold the C-terminal conservative glycine waiting for PE-conjugation. Expanding of the ATG4 family in wheat genome is not so fast like the ATG8 family. There are four ATG4s in mammals (Mariño et al. 2003) and a relatively constant number of two in plant species Arabidopsis (Hanaoka et al. 2002), rice (Xia et al. 2011) and maize (Chung et al. 2009). In this study, we identified two wheat ATG4s, TaATG4a and 4b, both of which contain a conserved protease activity center.
Successful functional complementation of yeast atg4 or atg8 mutants by plant ATG4 or ATG8 have been reported in Arabidopsis (Ketelaar et al. 2004), soybean (Xia et al. 2012) and T. dicoccoides (Kuzuoglu-Ozturk et al. 2012). Here, expression of wheat TaATG8a, 8b, 8g, 8h and 4a also restored autophagy activity in corresponding gene mutant yeast cells. In onion epidermal cells, GFP-TaATG8a, 8b, 8g and 8h showed strong or weak levels of punctate structures, representing autophagosomes and their precursors. Arabidopsis GFP-ATG8h/8i with innate C-terminal-exposed glycine appeared in vacuoles more quickly than other GFP-ATG8s (Yoshimoto et al. 2004). Likewise, GFP-TaATG8h with innate C-terminal-exposed glycine exhibited the most obvious punctate distribution. One reason for this situation may be that ATG8s with innate C-terminal-exposed glycine, without the need of ATG4 processing, quickly go through the conjugation reaction cascade and deposit on autophagic membranes, while other ATG8s need an extra step of cleavage by ATG4 which sustains a very limited level under normal conditions (Yoshimoto et al. 2004). In view of the trademark decoration of ATG8-PE on phagophores, extending and mature autophagic membranes and engulfed autophagic bodies in vacuole, observation of GFP-ATG8 puncta has become a routine means for autophagy activity determination (Mitou et al. 2009; Klionsky et al. 2012). Here, we show that GFP-TaATG8h is a potential indicator for determining the dynamics of autophagy in wheat cells. The C-terminal myc-tagged Arabidopsis ATG8 recombinant proteins can be cleaved in vitro by ATG4 to remove the tag and protective residues for glycine exposure (Yoshimoto et al. 2004). Our in vitro digestion experiments also showed that both of TaATG4a and 4b can cleave TaATG8a, 8b, and 8g. TaATG4b had a higher efficiency on all the three substrates than TaATG4a, and most efficiently cleaved TaATG8a. Members of ATG4 family from worm and Arabidopsis also have different activity and different preference for ATG8 substrates (Wu et al. 2012; Woo et al. 2014). Collectively, we identified wheat ATG4 and ATG8 family members and provide evidences for their involvement in wheat autophagy process.
Expression profiles suggest positive roles of ATG4 and ATG8 in wheat immunity to Bgt
The induced expression profiles suggest that TaATG4s and TaATG8s are implicated in wheat responses to abiotic stresses and to infection by the biotrophic pathogen fungus Bgt. Two TA events of TaATG4s and TaATG8s chronologically occurred in the Pm21- and Pm3f-triggered wheat incompatible reactions to Bgt during 0–36 hai. The late TAs of analyzed genes were also detected in the compatible reactions, but the former early TAs in the incompatible reactions formed a sharp contrast to the downregulation patterns in the compatible reactions. These two Bgt-induced TA events, especially the early one, implied that wheat ATG4s and ATG8s are employed in the early stage of wheat resistance response to Bgt. Pathogen-induced expression of plant ATG genes is consistent with enhanced autophagy levels (Liu Y et al. 2005; Patel and Dinesh-Kumar 2008). Likewise, accumulation of LysoTracker Red-stained autolysosome-like structures was detected in Bgt-challenged leaf epidermal cells of the resistance line 92R137/Yangmai 1587, which links the effects of ATG4 s/ATG8 s on wheat immunity to Bgt with their associated cellular process, autophagy. A positive role of autophagy on plant immunity has been reported in plant-pathogen systems as Nicotiana benthamiana (N)-TMV (incompatible interaction) (Liu Y et al. 2005), Arabidopsis-Pseudomonas syringae (compatible interaction) (Patel and Dinesh-Kumar 2008; Hofius et al. 2009), Arabidopsis-Botrytis cinerea/Alternaria brassicicola (compatible interactions) (Lenz et al. 2011; Lai et al. 2011) and Arabidopsis-Sclerotinia sclerotiorum (incompatible interaction) (Kabbage et al. 2013). Two Bgt-induced TA events have been found for many wheat resistance-related genes in wheat resistance responses, which also match the two pathogen-induced oxygen burst processes (Hückelhoven and Kogel 2003; Liu GS et al. 2005). ROS (reactive oxygen species) and cellular redox state may play key roles on autophagy regulation, and autophagy in turn negatively controls the production of ROS induced by pathogen infection (Yoshimoto et al. 2009; Pérez-Pérez et al. 2012).
Expression of TaATG4s/TaATG8s can be regulated by exogenously applied phytohormones. The ET, SA or ABA-regulated expression patterns were different and even contrast between powdery mildew resistant lines and susceptible lines. These differences may also occur responding to endogenous phytohormone signaling pathways triggered in wheat response to Bgt, and thus contribute to the different Bgt-induced levels of TaATG4s/TaATG8s between resistance and susceptible lines.
Wheat ATG4 and ATG8 may also contribute to the pathogenicity of Bgt
Much higher levels of enhanced expression were detected for TaATG4s/TaATG8s in the Yangmai 158-Bgt compatible reaction at the late stage around 16–36 hai. Since more Bgt conidiospores should have successfully penetrated into host cells of susceptible lines after 16 hai, there is a possibility that the high accumulation levels of TaATG4s/TaATG8s after 16 hai in susceptible lines are accepted and their associated autophagy process hijacked by Bgt to help with its pathogenicity including assimilation of nutrients from host cells. Thus, at least in the genetic background of Yangmai 158, ATG4s/ATG8s and probably their associated autophagy process may play a negative role on wheat immunity to Bgt after 16 hai. Negative roles of autophagy in plant immunity have been reported on plant-(semi-)biotrophic pathogen systems as Arabidopsis-P. syringae and Arabidopsis-powdery mildew fungus Golovinomyces cichoracearum (Lenz et al. 2011; Wang et al. 2011).
Abbreviations
- ATG:
-
Autophagy-related gene
- PE:
-
Phosphatidyl ethanolamine
- EST:
-
Expressed sequence tag
- ORF:
-
Open reading frame
- GFP:
-
Green fluorescent protein
- ET:
-
Ethylene
- SA:
-
Salicylic acid
- MeJA:
-
Methyl jasmonate
- ABA:
-
Abscisic acid
- PEG:
-
Polyethylene glycol
- qRT-PCR:
-
Quantitative real-time PCR
- TA:
-
Transcript accumulation
References
Bassham DC (2007) Plant autophagy-more than a starvation response. Curr Opin Plant Biol 10:587–593
Bassham DC (2009) Function and regulation of macroautophagy in plants. Biochim Biophys Acta 9:1397–1403
Bassham DC, Laporte M, Marty F, Moriyasu Y, Ohsumi Y, Olsen LJ, Yoshimoto K (2006) Autophagy in development and stress responses of plants. Autophagy 2:2–11
Chung T, Suttangkakul A, Vierstra RD (2009) The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol 149:220–234
Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277:33105–33114
Fujiki Y, Yoshimoto K, Ohsumi Y (2007) An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol 143:1132–1139
Fujioka Y, Noda NN, Fujii K, Yoshimoto K, Ohsumi Y, Inagaki F (2008) In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J Biol Chem 283:1921–1928
Gietz RD, Schiestl RH (2007) Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2:35–37
Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T et al (2007) The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282:37298–37302
Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D et al (2002) Leaf senescence and starvation induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129:1181–1193
Hayward AP, Dinesh-Kumar SP (2011) What can plant autophagy do for an innate immune response? Annu Rev Phytopathol 49:557–576
Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O et al (2009) Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137:773–783
Hückelhoven R, Kogel KH (2003) Reactive oxygen intermediates in plant-microbe interactions: who is who in powdery mildew resistance? Planta 216:891–902
Inoue Y, Suzuki T, Hattori M, Yoshimoto K, Ohsumi Y, Moriyasu Y (2006) AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol 47:1641–1652
Jia J, Zhao S, Kong X, Li Y, Zhao G, He W et al (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496:91–95
Kabbage M, Williams B, Dickman MB (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog 9:e1003287
Ketelaar T, Voss C, Dimmock SA, Thumm M, Hussey PJ (2004) Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett 567:302–306
Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K et al (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8:445–544
Kuzuoglu-Ozturk D, Cebeci Yalcinkaya O, Akpinar BA, Mitou G, Korkmaz G, Gozuacik D, Budak H (2012) Autophagy-related gene, TdAtg8, in wild emmer wheat plays a role in drought and osmotic stress response. Planta 236:1081–1092
Kwon SI, Cho HJ, Kim SR, Park OK (2013) The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol 161:1722–1736
Lai Z, Wang F, Zheng Z, Fan B, Chen Z (2011) A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J 66:953–968
Lenz HD, Haller E, Melzer E, Kober K, Wurster K, Stahl M et al (2011) Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J 66:818–830
Li F, Vierstra RD (2012) Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 17:526–537
Ling HQ, Zhao S, Liu D, Wang J, Sun H, Zhang C et al (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496:87–90
Liu Y, Bassham DC (2012) Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63:215–237
Liu GS, Sheng XY, Greenshields DL, Ogieglo A, Kaminskyj S, Selvaraj G, Wei YD (2005) Profiling of wheat class III peroxidase genes derived from powdery mildew-attacked epidermis reveals distinct sequence-associated expression patterns. Mol Plant Microbe Interact 18:730–741
Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell 121:567–577
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25:402–408
Mariño G, Uría JA, Puente XS, Quesada V, Bordallo J, López-Otín C (2003) Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem 278:3671–3678
Mitou G, Budak H, Gozuacik D (2009) Techniques to study autophagy in plants. Int J Plant Genomics 2009:451357
Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD et al (1998) A protein conjugation system essential for autophagy. Nature 395:395–398
Moriyasu Y, Hattori M, Jauh G, Rogers JC (2003) Alpha tonoplast intrinsic protein is specifically associated with vacuole membrane involved in an autophagic process. Plant Cell Physiol 44:795–802
Nair U, Yen WL, Mari M, Cao Y, Xie Z, Baba M et al (2012) A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy 8:780–793
Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130:165–178
Ohsumi Y (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2:211–216
Patel S, Dinesh-Kumar SP (2008) Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4:20–27
Pérez-Pérez ME, Lemaire SD, Crespo JL (2012) Reactive oxygen species and autophagy in plants and algae. Plant Physiol 160:156–164
Reggiori F, Klionsky DJ (2013) Autophagic processes in Yeast: mechanism, machinery and regulation. Genetics 194:341–361
Romanov J, Walczak M, Ibiricu I, Schüchner S, Ogris E, Kraft C, Martens S (2012) Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J 31:4304–4317
Scott A, Wyatt S, Tsou PL, Robertson D, Allen NS (1999) Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26(1125):1128–1132
Su W, Ma H, Liu C, Wu J, Yang J (2006) Identification and characterization of two rice autophagy associated genes, OsAtg8 and OsAtg4. Mol Biol Rep 33:273–278
Wang Y, Nishimura MT, Zhao T, Tang D (2011) ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J 68:74–87
Woo J, Park E, Dinesh-Kumar SP (2014) Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc Natl Acad Sci USA 111:863–868
Wu F, Li Y, Wang F, Noda NN, Zhang H (2012) Differential function of the two Atg4 homologues in the aggrephagy pathway in Caenorhabditis elegans. J Biol Chem 287:29457–29467
Xia K, Liu T, Ouyang J, Wang R, Fan T, Zhang M (2011) Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res 18:363–377
Xia T, Xiao D, Liu D, Chai W, Gong Q, Wang NN (2012) Heterologous expression of ATG8c from Soybean Confers Tolerance to Nitrogen Deficiency and Increases Yield in Arabidopsis. PLoS one 7:e37217
Xie Z, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9:1102–1109
Xie Z, Nair U, Klionsky DJ (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 19:3290–3298
Yoshimoto K (2012) Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol 53:1355–1365
Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8 s, ubiquitin-Like Proteins, and their deconjugation by ATG4 s are essential for plant autophagy. Plant Cell 16:2967–2983
Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R et al (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21:2914–2927
Acknowledgments
This work was supported by the Natural Science Foundation of Tianjin, China (Grant number 12JCZDJC23000); the Fok Ying-Tong Education Foundation for Young Teachers in the Higher Education Institutions of China (Grant number 131026); and the Open Fund of Tianjin Key Laboratory of Animal and Plant Resistance, Tianjin Normal University (Grant number 52LX12).
Conflict of interest
The authors do not have any conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Jeong Sheop Shin.
Electronic supplementary material
Below is the link to the electronic supplementary material.
For supplementary materials, please refer to the online file Supporting Information.docx which contains:
Table S1. List of primers used in this paper.
Figure S1. Multiple alignments of wheat ATG8s and homologues from other species. The alignment was produced in ClustalX 2.1 software. Sequences in the alignment include the nine wheat ATG8s here identified, AER27507 (TdATG8) from Triticum dicoccoides, XP_003581707 (BdATG8a) and XP_003571383 (BdATG8b) from Brachypodium distachyon, Os07g0512200 (OsATG8a), Os04g0624000 (OsATG8b), Os08g0191600 (OsATG8c) and Os02g0529150 (OsATG8d) from Oryza sativa, ACJ73920 (ZmATG8a), ACJ73921 (ZmATG8b), ACJ73922 (ZmATG8c), ACJ73923 (ZmATG8d) and ACJ73925 (ZmATG8e) from Zea mays, At4g21980 (AtATG8a), At4g04620 (AtATG8b), At1g62040 (AtATG8c), At2g05630 (AtATG8d), At2G45170 (AtATG8e), At4g16520 (AtATG8f), At3g60640 (AtATG8 g), At3g06420 (AtATG8h) and At3g15580 (AtATG8i) from Arabidopsis thaliana, ACU13796 (GmATG8a), ACU14633 (GmATG8b), ACU17086 (GmATG8c), BAH22449 (GmATG8d), ACU15101 (GmATG8e), ACU19559 (GmATG8f), ACU13862 (GmATG8g), ACU16419 (GmATG8h), BAH22448 (GmATG8i) and ACU19611 (GmATG8k) from Glycine max, NP_009216 (HsATG8/GATE16/GABARAPL2) from Human and NP_009475 (ScATG8) from Yeast. Names of wheat ATG8s are boxed. One hundred percent similarity, black; 80–100 % similarity, dark gray; 60–80 % similarity, light gray; <60 % similarity, white. Hashes and asterisks, respectively, indicate conserved essential residues at the N-terminal microtubule binding site and at the ATG7 binding site. The C-terminal conservative glycine for PE-conjugation is indicated by an arrowhead.
Figure S2. Multiple alignments of wheat ATG4s and homologues from other species. The alignment was produced in ClustalX 2.1 software. Sequences in the alignment include the two wheat ATG4s here identified, Os03g0391000 (OsATG4a) and Os04g0682000 (OsATG4b) from Oryza sativa, ACJ73912 (ZmATG4a) and ACJ73914 (ZmATG4b) from Zea mays, At2g44140 (AtATG4a) and At3g59950 (AtATG4b) from Arabidopsis thaliana, NP_443168 (HsATG4a) from Human and NP_014176 (ScATG4) from Yeast. One hundred percent similarity, black; 80 to 100 % similarity, dark gray; 60 to 80 % similarity, light gray; <60 % similarity, white. The conserved Peptidase_C54 domain is under thick line;Arrowheads mask the canonical catalytic triad of cysteine protease: Cys, Asp and His.
Rights and permissions
About this article
Cite this article
Pei, D., Zhang, W., Sun, H. et al. Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep 33, 1697–1710 (2014). https://doi.org/10.1007/s00299-014-1648-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-014-1648-x