Abstract
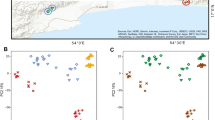
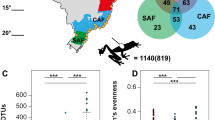
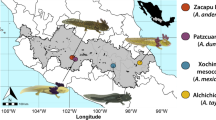
Host microbial communities are increasingly seen as an important component of host health. In amphibians, the first land vertebrates that are threatened by a fungal skin disease globally, our understanding of the factors influencing the microbiome of amphibian skin remains incomplete because recent studies have focused almost exclusively on bacteria, and little information exists on fungal communities associated with wild amphibian species. In this study, we describe the effects of host phylogeny, climate, geographic distance, and infection with a fungal pathogen on the composition and structure of bacterial and fungal communities in seven tropical salamander species that occur in the Trans-Mexican Volcanic Belt of Central Mexico. We find that host phylogenetic relatedness is correlated with bacterial community composition while a composite climatic variable of temperature seasonality and precipitation is significantly associated with fungal community composition. We also estimated co-occurrence networks for bacterial and fungal taxa and found differences in the degree of connectivity and the distribution of negative associations between the two networks. Our results suggest that different factors may be responsible for structuring the bacterial and fungal communities of amphibian skin and that the inclusion of fungi in future studies could shed light on important functional interactions within the microbiome.






Similar content being viewed by others
Data Availability
Sequence data for cytb from Sanger sequencing of salamanders are available in Genbank (ON369403–ON369432). Amplicon sequences for 16S and ITS2 are available in the Short Read Archive (BioProject PRJNA830991).
References
Dastogeer KM, Tumpa FH, Sultana A, Akter MA, Chakraborty A (2020) Plant microbiome–an account of the factors that shape community composition and diversity. Curr Plant Biol 23:100161. https://doi.org/10.1016/j.cpb.2020.100161
Apprill A (2017) Marine animal microbiomes: toward understanding host–microbiome interactions in a changing ocean. Front Mar Sci 4:222. https://doi.org/10.3389/fmars.2017.00222
Kueneman JG, Bletz MC, McKenzie VJ, Becker CG, Joseph MB, Abarca JG, Archer H, Arellano AL, Bataille A, Becker M, Belden LK, Crottini A, Geffers R, Haddad CFB, Harris RN, Holden WM, Hughey M, Jarek M, Kearns PJ, Kerby JL, Kielgast J, Kurabayashi A, Longo AV, Loudon A, Medina D, Nunez JJ, Perl RGB, Pinto-Tomas A, Rabemananjara FCE, Rebollar EA, Rodriguez A, Rollins-Smith L, Stevenson R, Tebbe CC, Vargas Asensio G, Waldman B, Walke JB, Whitfield SM, Zamudio KR, Zuniga Chaves I, Woodhams DC, Vences M (2019) Community richness of amphibian skin bacteria correlates with bioclimate at the global scale. Nat Ecol Evol 3:381–389. https://doi.org/10.1038/s41559-019-0798-1
Rojas CA, Ramírez-Barahona S, Holekamp KE, Theis KR (2021) Host phylogeny and host ecology structure the mammalian gut microbiota at different taxonomic scales. Anim Microbiome 3:33. https://doi.org/10.1186/s42523-021-00094-4
Weinstein SB, Martinez-Mota R, Stapleton TE, Klure DM, Greenhalgh R, Orr TJ, Dale C, Kohl KD, Dearing MD (2021) Microbiome stability and structure is governed by host phylogeny over diet and geography in woodrats (Neotoma spp.). Proc Natl Acad Sci USA 118:e2108787118. https://doi.org/10.1073/pnas.2108787118
Bletz MC, Archer H, Harris RN, McKenzie VJ, Rabemananjara FCE, Rakotoarison A, Vences M (2017) Host ecology rather than host phylogeny drives amphibian skin microbial community structure in the biodiversity hotspot of Madagascar. Front Microbiol 8:1530. https://doi.org/10.3389/fmicb.2017.01530
Barnes EM, Kutos S, Naghshineh N, Mesko M, You Q, Lewis JD (2021) Assembly of the amphibian microbiome is influenced by the effects of land-use change on environmental reservoirs. Environ Microbiol 23:4595–4611. https://doi.org/10.1111/1462-2920.15653
Ellison S, Rovito S, Parra-Olea G, Vásquez-Almazán C, Flechas SV, Bi K, Vredenburg VT (2019) The influence of habitat and phylogeny on the skin microbiome of amphibians in Guatemala and Mexico. Microb Ecol 78:257–267. https://doi.org/10.1007/s00248-018-1288-8
Ross AA, Rodrigues Hoffmann A, Neufeld JD (2019) The skin microbiome of vertebrates. Microbiome 7:79. https://doi.org/10.1186/s40168-019-0694-6
Huffnagle GB, Noverr MC (2013) The emerging world of the fungal microbiome. Trends Microbiol 21:334–341. https://doi.org/10.1016/j.tim.2013.04.002
Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T, North G, Visel A, Partida-Martínez LP, Tringe SG (2016) Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol 209:798–811. https://doi.org/10.1111/nph.13697
Kearns PJ, Fischer S, Fernandez-Beaskoetxea S, Gabor CR, Bosch J, Bowen JL, Tlusty MF, Woodhams DC (2017) Fight fungi with fungi: antifungal properties of the amphibian mycobiome. Front Microbiol 8:2494. https://doi.org/10.3389/fmicb.2017.02494
Longcore JE, Pessier AP, Nichols DK (1999) Batrachochytrium dendrobatidis gen et sp nov, a chytrid pathogenic to amphibians. Mycologia 91:219–227. https://doi.org/10.2307/3761366
Berger L, Speare R, Daszak P, Green DE, Cunningham AA, Goggin CL, Slocombe R, Ragan MA, Hyatt AD, McDonald KR, Hines HB, Lips KR, Marantelli G, Parkes H (1998) Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci U S A 95:9031–9036. https://doi.org/10.1073/pnas.95.15.9031
Lips KR, Brem F, Brenes R, Reeve JD, Alford RA, Voyles J, Carey C, Livo L, Pessier AP, Collins JP (2006) Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proc Natl Acad Sci USA 103:3165–3170. https://doi.org/10.1073/pnas.0506889103
Daszak P, Berger L, Cunningham AA, Hyatt AD, Green DE, Speare R (1999) Emerging infectious diseases and amphibian population declines. Emerg Infect Dis 5:735–748. https://doi.org/10.3201/eid0506.990601
Scheele BC, Pasmans F, Skerratt LF, Berger L, Martel A, Beukema W, Acevedo AA, Burrowes PA, Carvalho T, Catenazzi A, De la Riva I, Fisher MC, Flechas SV, Foster CN, Frias-Alvarez P, Garner TWJ, Gratwicke B, Guayasamin JM, Hirschfeld M, Kolby JE, Kosch TA, La Marca E, Lindenmayer DB, Lips KR, Longo AV, Maneyro R, McDonald CA, Mendelson J 3rd, Palacios-Rodriguez P, Parra-Olea G, Richards-Zawacki CL, Rodel MO, Rovito SM, Soto-Azat C, Toledo LF, Voyles J, Weldon C, Whitfield SM, Wilkinson M, Zamudio KR, Canessa S (2019) Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity. Science 363:1459–1463. https://doi.org/10.1126/science.aav0379
Wake DB, Vredenburg VT (2008) Colloquium paper: are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc Natl Acad Sci U S A 105(Suppl 1):11466–11473. https://doi.org/10.1073/pnas.0801921105
Barnosky AD, Matzke N, Tomiya S, Wogan GO, Swartz B, Quental TB, Marshall C, McGuire JL, Lindsey EL, Maguire KC, Mersey B, Ferrer EA (2011) Has the Earth’s sixth mass extinction already arrived? Nature 471:51–57. https://doi.org/10.1038/nature09678
Harris RN, Brucker RM, Walke JB, Becker MH, Schwantes CR, Flaherty DC, Lam BA, Woodhams DC, Briggs CJ, Vredenburg VT, Minbiole KP (2009) Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J 3:818–824. https://doi.org/10.1038/ismej.2009.27
Vredenburg VT, Briggs CJ, Harris RN (2011) Host-pathogen dynamics of amphibian chytridiomycosis: the role of the skin microbiome in health and disease. In: Institute of Medicine. Fungal diseases: an emerging challenge to human, animal, and plant health. The National Academies Press, Washington, D.C., pp 342–355
Bernardo-Cravo AP, Schmeller DS, Chatzinotas A, Vredenburg VT, Loyau A (2020) Environmental factors and host microbiomes shape host-pathogen dynamics. Trends Parasitol 36:616–633. https://doi.org/10.1016/j.pt.2020.04.010
Zumbado-Ulate H, Bolaños F, Gutiérrez-Espeleta G, Puschendorf R (2014) Extremely low prevalence of Batrachochytrium dendrobatidis in frog populations from Neotropical dry forest of Costa Rica supports the existence of a climatic refuge from disease. EcoHealth 11:593–602. https://doi.org/10.1007/s10393-014-0967-2
Reeder NM, Pessier AP, Vredenburg VT (2012) A reservoir species for the emerging amphibian pathogen Batrachochytrium dendrobatidis thrives in a landscape decimated by disease. PLoS ONE 7:e33567. https://doi.org/10.1371/journal.pone.0033567
Gervasi S, Gondhalekar C, Olson DH, Blaustein AR (2013) Host identity matters in the amphibian-Batrachochytrium dendrobatidis system: fine-scale patterns of variation in responses to a multi-host pathogen. PLoS ONE 8:e54490. https://doi.org/10.1371/journal.pone.0054490
Prado-Irwin SR, Bird AK, Zink AG, Vredenburg VT (2017) Intraspecific variation in the skin-associated microbiome of a terrestrial salamander. Microb Ecol 74:745–756. https://doi.org/10.1007/s00248-017-0986-y
Bird AK, Prado-Irwin SR, Vredenburg VT, Zink AG (2018) Skin microbiomes of California terrestrial salamanders are influenced by habitat more than host phylogeny. Front Microbiol 9:442. https://doi.org/10.3389/fmicb.2018.00442
Jani AJ, Briggs CJ (2014) The pathogen Batrachochytrium dendrobatidis disturbs the frog skin microbiome during a natural epidemic and experimental infection. Proc Natl Acad Sci U S A 111:E5049–E5058. https://doi.org/10.1073/pnas.1412752111
Rebollar EA, Hughey MC, Medina D, Harris RN, Ibanez R, Belden LK (2016) Skin bacterial diversity of Panamanian frogs is associated with host susceptibility and presence of Batrachochytrium dendrobatidis. ISME J 10:1682–1695. https://doi.org/10.1038/ismej.2015.234
Ellison S, Knapp RA, Sparagon W, Swei A, Vredenburg VT (2019) Reduced skin bacterial diversity correlates with increased pathogen infection intensity in an endangered amphibian host. Mol Ecol 28:127–140. https://doi.org/10.1111/mec.14964
Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ (2012) Emerging fungal threats to animal, plant, and ecosystem health. Nature 484:186–194. https://doi.org/10.1038/nature10947
Kueneman JG, Woodhams DC, Van Treuren W, Archer HM, Knight R, McKenzie VJ (2016) Inhibitory bacteria reduce fungi on early life stages of endangered Colorado boreal toads (Anaxyrus boreas). ISME J 10:934–944. https://doi.org/10.1038/ismej.2015.168
Medina D, Hughey MC, Walke JB, Becker MH, Pontarelli K, Sun S, Badgley B, Belden LK (2019) Amphibian skin fungal communities vary across host species and do not correlate with infection by a pathogenic fungus. Environ Microbiol 21:2905–2920. https://doi.org/10.1111/1462-2920.14682
Wake DB (2012) Taxonomy of salamanders of the family Plethodontidae (Amphibia: Caudata). Zootaxa 3484:75–82. https://doi.org/10.11646/zootaxa.3484.1.5
Fitzpatrick BM, Allison AL (2014) Similarity and differentiation between bacteria associated with skin of salamanders (Plethodon jordani) and free-living assemblages. FEMS Microbiol Ecol 88:482–494. https://doi.org/10.1111/1574-6941.12314
Loudon AH, Venkataraman A, Van Treuren W, Woodhams DC, Parfrey LW, McKenzie VJ, Knight R, Schmidt TM, Harris RN (2016) Vertebrate hosts as islands: dynamics of selection, immigration, loss, persistence, and potential function of bacteria on salamander skin. Front Microbiol 7:333. https://doi.org/10.3389/fmicb.2016.00333
AmphibiaWeb (2022) Information on Amphibian Biology and Conservation. www.amphibiaweb.org. Accessed 25 Apr 2022
Rovito SM, Parra-Olea G, Vásquez-Almazán CR, Papenfuss TJ, Wake DB (2009) Dramatic declines in neotropical salamander populations are an important part of the global amphibian crisis. Proc Natl Acad Sci U S A 106:3231–3236. https://doi.org/10.1073/pnas.0813051106
Cheng TL, Rovito SM, Wake DB, Vredenburg VT (2011) Coincident mass extirpation of neotropical amphibians with the emergence of the infectious fungal pathogen Batrachochytrium dendrobatidis. Proc Natl Acad Sci U S A 108:9502–9507. https://doi.org/10.1073/pnas.1105538108
Olivares-Miranda M, Vredenburg VT, García-Sánchez JC, Byrne AQ, Rosenblum EB, Rovito SM (2020) Fungal infection, decline and persistence in the only obligate troglodytic Neotropical salamander. PeerJ 8:e9763. https://doi.org/10.7717/peerj.9763
Hyatt AD, Boyle DG, Olsen V, Boyle DB, Berger L, Obendorf D, Dalton A, Kriger K, Heros M, Hines H, Phillott R, Campbell R, Marantelli G, Gleason F, Coiling A (2007) Diagnostic assays and sampling protocols for the detection of Batrachochytrium dendrobatidis. Dis Aquat Organ 73:175–192. https://doi.org/10.3354/dao073175
Puschendorf R, Bolaños F (2006) Detection of Batrachochytrium dendrobatidis in Eleutherodactylus fitzingeri: effects of skin sample location and histologic stain. J Wildl Dis 42:301–306. https://doi.org/10.7589/0090-3558-42.2.301
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108(Suppl 1):4516–4522. https://doi.org/10.1073/pnas.1000080107
Ihrmark K, Bodeker IT, Cruz-Martinez K, Friberg H, Kubartova A, Schenck J, Strid Y, Stenlid J, Brandstrom-Durling M, Clemmensen KE, Lindahl BD (2012) New primers to amplify the fungal ITS2 region—evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol 82:666–677. https://doi.org/10.1111/j.1574-6941.2012.01437.x
Vo AT, Jedlicka JA (2014) Protocols for metagenomic DNA extraction and Illumina amplicon library preparation for faecal and swab samples. Mol Ecol Resour 14:1183–1197. https://doi.org/10.1111/1755-0998.12269
Boyle DG, Boyle DB, Olsen V, Morgan JA, Hyatt AD (2004) Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis Aquat Org 60:141–148. https://doi.org/10.3354/dao060141
Zhang J, Kobert K, Flouri T, Stamatakis A (2014) PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. https://doi.org/10.1093/bioinformatics/btt593
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodriguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vazquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Nilsson RH, Larsson KH, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glockner FO, Tedersoo L, Saar I, Koljalg U, Abarenkov K (2019) The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47:D259–D264. https://doi.org/10.1093/nar/gky1022
Brown SP, Veach AM, Rigdon-Huss AR, Grond K, Lickteig SK, Lothamer K, Oliver AK, Jumpponen A (2015) Scraping the bottom of the barrel: are rare high throughput sequences artefacts? Fungal Ecol 13:221–225. https://doi.org/10.1016/j.funeco.2014.08.006
Oliver AK, Brown SP, Callaham MAJ, Jumpponen A (2015) Polymerase matters: non-proofreading enzymes inflate fungal community richness estimates by up to 15 %. Fungal Ecol 15:86–89. https://doi.org/10.1016/j.funeco.2015.03.003
Price MN, Dehal PS, Arkin AP (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. https://doi.org/10.1371/journal.pone.0009490
Bray JR, Curtis JT (1957) An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349. https://doi.org/10.2307/1942268
Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. https://doi.org/10.1128/AEM.01996-06
Halwachs B, Madhusudhan N, Krause R, Nilsson RH, Moissl-Eichinger C, Hogenauer C, Thallinger GG, Gorkiewicz G (2017) Critical issues in mycobiota analysis. Front Microbiol 8:180. https://doi.org/10.3389/fmicb.2017.00180
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. https://doi.org/10.1186/gb-2011-12-6-r60
QGIS Development Team (2021) QGIS Geographic Information System. Open Source Geospatial Foundation Project 2.4. http://qgis.osgeo.org
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25:1965–1978. https://doi.org/10.1002/joc.1276
Moritz C, Schneider CJ, Wake DB (1992) Evolutionary relationships within the Ensatina eschscholtzii complex confirm the ring species interpretation. Syst Biol 41:273–291. https://doi.org/10.1093/sysbio/41.3.273
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Can Res 27:209–220
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol) 57:289–300. https://doi.org/10.1111/j.2517-6161.1995.tb02031.x
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2013) Community ecology package. R Package Version 2
Kielak AM, Scheublin TR, Mendes LW, van Veen JA, Kuramae EE (2016) Bacterial community succession in pine-wood decomposition. Front Microbiol 7:231. https://doi.org/10.3389/fmicb.2016.00231
Zamkovaya T, Foster JS, de Crecy-Lagard V, Conesa A (2021) A network approach to elucidate and prioritize microbial dark matter in microbial communities. ISME J 15:228–244. https://doi.org/10.1038/s41396-020-00777-x
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. http://www.genome.org/cgi/doi/10.1101/gr.1239303
Faust K, Raes J (2016) CoNet app: inference of biological association networks using Cytoscape. F1000Res 5:1519. https://doi.org/10.12688/f1000research.9050.2
Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, Weigel D, Kemen EM (2016) Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol 14:e1002352. https://doi.org/10.1371/journal.pbio.1002352
Muletz Wolz CR, Yarwood SA, Campbell Grant EH, Fleischer RC, Lips KR (2018) Effects of host species and environment on the skin microbiome of plethodontid salamanders. J Anim Ecol 87:341–353. https://doi.org/10.1111/1365-2656.12726
Kueneman JG, Parfrey LW, Woodhams DC, Archer HM, Knight R, McKenzie VJ (2014) The amphibian skin-associated microbiome across species, space, and life history stages. Mol Ecol 23:1238–1250. https://doi.org/10.1111/mec.12510
Wilhelm L, Besemer K, Fasching C, Urich T, Singer GA, Quince C, Battin TJ (2014) Rare but active taxa contribute to community dynamics of benthic biofilms in glacier-fed streams. Environ Microbiol 16:2514–2524. https://doi.org/10.1111/1462-2920.12392
Pester M, Bittner N, Deevong P, Wagner M, Loy A (2010) A ‘rare biosphere’ microorganism contributes to sulfate reduction in a peatland. ISME J 4:1591–1602. https://doi.org/10.1038/ismej.2010.75
Hol WH, Garbeva P, Hordijk C, Hundscheid PJ, Gunnewiek PJ, Van Agtmaal M, Kuramae EE, De Boer W (2015) Non-random species loss in bacterial communities reduces antifungal volatile production. Ecology 96:2042–2048. https://doi.org/10.1890/14-2359.1
Jousset A, Bienhold C, Chatzinotas A, Gallien L, Gobet A, Kurm V, Kusel K, Rillig MC, Rivett DW, Salles JF, van der Heijden MG, Youssef NH, Zhang X, Wei Z, Hol WH (2017) Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J 11:853–862. https://doi.org/10.1038/ismej.2016.174
Walke JB, Becker MH, Hughey MC, Swartwout MC, Jensen RV, Belden LK (2017) Dominance-function relationships in the amphibian skin microbiome. Environ Microbiol 19:3387–3397. https://doi.org/10.1111/1462-2920.13850
Walke JB, Becker MH, Loftus SC, House LL, Cormier G, Jensen RV, Belden LK (2014) Amphibian skin may select for rare environmental microbes. Isme J 8:2207–2217. https://doi.org/10.1038/ismej.2014.77
Corsaro D, Valassina M, Venditti D (2003) Increasing diversity within Chlamydiae. Crit Rev Microbiol 29:37–78. https://doi.org/10.1080/713610404
Friedman J, Alm EJ (2012) Inferring correlation networks from genomic survey data. PLoS Comput Biol 8:e1002687. https://doi.org/10.1371/journal.pcbi.1002687
Faust K (2021) Open challenges for microbial network construction and analysis. ISME J 15:3111–3118. https://doi.org/10.1038/s41396-021-01027-4
Matchado MS, Lauber M, Reitmeier S, Kacprowski T, Baumbach J, Haller D, List M (2021) Network analysis methods for studying microbial communities: a mini review. Comput Struct Biotechnol J 19:2687–2698. https://doi.org/10.1016/j.csbj.2021.05.001
Flechas SV, Blasco-Zúñiga A, Merino-Viteri A, Ramírez-Castañeda V, Rivera M, Amézquita A (2017) The effect of captivity on the skin microbial symbionts in three Atelopus species from the lowlands of Colombia and Ecuador. PeerJ 5:e3594. https://doi.org/10.7717/peerj.3594
Paiva G, Abreu P, Proença DN, Santos S, Nobre MF, Morais PV (2014) Mucilaginibacter pineti sp. nov., isolated from Pinus pinaster wood from a mixed grove of pines trees. Int J Syst Evol Microbiol 64:2223–2228. https://doi.org/10.1099/ijs.0.057737-0
Lambiase A (2014) The family Sphingobacteriaceae. The Prokaryotes 4:455
Figueiredo G, Gomes M, Covas C, Mendo S, Caetano T (2021) The unexplored wealth of microbial secondary metabolites: the Sphingobacteriaceae case study. Microb Ecol 1–12. https://doi.org/10.1007/s00248-021-01762-3
Röttjers L, Vandeputte D, Raes J, Faust K (2021) Null-model-based network comparison reveals core associations. ISME Commun 1:1–8. https://doi.org/10.1038/s43705-021-00036-w
Banerjee S, Schlaeppi K, van der Heijden MGA (2018) Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol 16:567–576. https://doi.org/10.1038/s41579-018-0024-1
Rebollar EA, Martínez-Ugalde E, Orta AH (2020) The amphibian skin microbiome and its protective role against chytridiomycosis. Herpetologica 76:167–177. https://doi.org/10.1655/0018-0831-76.2.167
García-Rodríguez A, Basanta MD, García-Castillo MG, Zumbado-Ulate H, Neam K, Rovito S, Searle CL, Parra-Olea G (2021) Anticipating the potential impacts of Batrachochytrium salamandrivorans on Neotropical salamander diversity. Biotropica 54:157–161. https://doi.org/10.1111/btp.13042
Acknowledgements
This research was funded by Conacyt Ciencia Básica grant 221614 and Problemas Nacionales grant 2015-01-721 to Sean Rovito and Belmont Forum project: People, Pollution and Pathogens (P3) NSF 1633948 to Vance Vredenburg. Julio García-Sánchez was funded by a Conacyt graduate fellowship. Omar Becerra and Mirna García-Castillo helped with fieldwork.
Funding
This research was funded by Conacyt Ciencia Básica grant 221614 and Problemas Nacionales grant 2015–01-721 to Sean Rovito and Belmont Forum project: People, Pollution and Pathogens (P3) NSF 1633948 to Vance Vredenburg. Julio García-Sánchez was funded by a Conacyt graduate fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Statement
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted. This research was conducted under animal use protocol SIACUAL 0148–15 to Sean Rovito. The article does not contain any studies with human participants performed by any of the authors.
Conflict of Interest
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
García-Sánchez, J.C., Arredondo-Centeno, J., Segovia-Ramírez, M.G. et al. Factors Influencing Bacterial and Fungal Skin Communities of Montane Salamanders of Central Mexico. Microb Ecol 86, 670–686 (2023). https://doi.org/10.1007/s00248-022-02049-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-022-02049-x