Abstract
Rapid diagnosis of infectious diseases and up-to-the-minute commencement of relevant treatments are important factors that not only promote positive changes in the clinical scenario but also the health of the mass at large. Surpassing the time-consuming conventional, straightforward in vitro methods for diagnosing infectious diseases, biosensors have shown their tremendous potential in the recent era. Current developments concerning biosensing technologies bring point-of-care diagnostics to the forefront. This proves to be advantageous over conventional practices that demand centralized laboratory facilities, experienced personnel, and colossal machinery. Currently, the infectious pandemic caused by the spreading of the novel coronavirus has created an unprecedented adverse effect on both the global economy and health security. The current situation of growing cases of infection despite several measures and the unavailability of testing kits to diagnose every suspected case point toward the need of urgent upgradation of the conventional diagnostic approaches to advanced, robust, and cost-effective diagnosis. Increasing demand in viral vigilance and directive regulatory steps toward the disease transmission also reveals the need for rapid as well as sensitive devices for viral diagnosis. From the last several decades, biosensors for their noteworthy sensitivity and specificity have been considered as a promising and potent tool for precise and quantifiable detection of viruses. Current developments in genetic engineering inclusive the genetic manipulation and material engineering have introduced several approaches to enhance sensitivity, selectivity, and the overall recognition efficiency of biosensors. This chapter presents an overview of the biosensing methodologies, especially focusing on various labeled and label-free techniques that have been used in the past and are being reported in the recent era for diagnosis.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Since the beginning of the twentieth century, outbreaks due to viruses specifically leading to respiratory diseases have caused a major setback. The spread of such viruses at a huge epidemiological scale poses as a serious threat toward the seven billion health and wealth (Donnelly et al. 2019; Knobler et al. 2004; Memish et al. 2020; Ozili and Arun 2020; Wu et al. 2020). Tracing down the outbreak history, the severe acute respiratory syndrome coronavirus (SARS-CoV) was first reported in the year 2002. Ten years later in 2012, the middle-east respiratory syndrome coronavirus (MERS-CoV) again caused a major threat (Zumla et al. 2015). Following which, the more recent outbreak of the SARS-Coronavirus 2 (SARS-CoV-2) has affirmed the fact that despite mankind’s substantial efforts to improve diagnosis, treatment, and prevention strategies toward communicable diseases for the last ten decades, novel contagions remain an inevitable challenge toward global health issues (Fauci and Morens 2012; França et al. 2013). Over the period, the foremost challenge to contain such emerging contagions includes the evolution of novel infectious promoters with the rapid spreading of diseases over the human population.
Similar to several pathogenic diagnosis procedures, methods concerning SARS, MERS, and SARS-CoV-2 detection count on various laboratory-based evaluations like electron and cryo-electron microscopy (Gui et al. 2017), in vitro growth and quantification (Coleman and Frieman 2015; Hui et al. 2004; Matsuyama et al. 2020), immunological assays (Kogaki et al. 2005; Lau et al. 2004; Lee et al. 2017), and amplification of nucleic acid (Corman et al. 2020; Shen et al. 2020; Cotten et al. 2013; Liu et al. 2020) accompanied with radiological analysis (Gogna et al. 2014; Hamimi 2016; Hosseiny et al. 2020; Jardon et al. 2019; Lin et al. 2005; Nasir et al. 2020). The basic dependency upon these in vitro diagnostic procedures is also documented with several shortcomings. For instance, microscopic analysis and radio-imaging deal with inadequate sensitivity, and culturing viral strains in vitro is rather challenging (Yu et al. 2020). Moreover, enzyme-based immunological assays and approaches for nucleic-acid amplification involving polymerase chain reaction (PCR) are often time-consuming and necessitate exhaustive sample preparation steps while falling short to produce multiple detections at a time (Ben-Assa et al. 2020; Chow 2004; Kurstjens et al. 2020). PCR also requires sophistication in sample preparation, which on rare instances might pose false-negative results for tests (Li et al. 2020a; Pan et al. 2020; Xiao et al. 2020).
To date, for diagnosing several coronaviruses, the typical protocol has been followed which involves the collection of biological samples (nasopharyngeal swab, sputum, blood and in some cases tissue swabs) and delivering to a high-equipped laboratory facility for further processing which desires involvement of practiced personnel (Chandra 2020; Chow 2004; Kurstjens et al. 2020). Till the test results become available (usually in days), clinicians deliver experimental antimicrobial treatments to the patients, which further complicates the delivery of evidence-based attention in such cases.
Biosensors break the trend of such cumbersome procedures and can be labeled as one of the best examples of a simple, miniaturized analytical tool that converts the molecular recognition of an analyte of interest into a quantifiable signal through a transducer (Bhatnagar et al. 2018; Chandra 2013; Kumar et al. 2019a, b, c; Mahato et al. 2018a, b, c; Mahato and Chandra 2019). Biosensor reportedly offers inexpensive, rapid, easy-to-detect platform with high sensitivity to effectively identify pathogens for various infectious diseases and, therefore, has proven potent to deliver point-of-care detection (Chua et al. 2011; Kashish et al. 2017; Pejcic et al. 2006; Chandra et al. 2017; Sin et al. 2014; Purohit et al. 2019a, b, c, 2020a, b; Mahato et al. 2020a, b; Kumar et al. 2019a, b, c). Moreover, it also provides a low limit of detection and device portability, consumes less energy, and demands lesser reagents (Prasad et al. 2016; Purohit et al. 2019a, b, c, 2020a, b; Mahato et al. 2019, 2020a, b; Kumar et al. 2019a, b, c). Structural upgradation and enhancement using micro\nanotechnologies have significantly improved the biosensor capability for executing complex assays (molecular, genetic, etc.) for the detection of various infectious diseases (Dai and Choi 2013; Mahato et al. 2018a, b, c; Polizzi 2019; Vaddiraju et al. 2010). Biosensor-based immunoassays enhance the detection sensitivity toward pathogen-specific antigens, while multiplex recognition of host-immune response improves the inclusive specificity compared to the conventional diagnostic procedures (Chandra 2020). Additionally, system assimilation also allows varying evaluation strategies to assimilate both pathogen-specific targets along with biomarker’s illustrative host-immune interaction responses at varying phases of infection (Mohan et al. 2011; Chandra 2016).
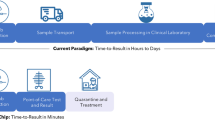
In this chapter, we have focused specifically on the up-to-date biosensor technologies for the detection of coronavirus diseases concentrating on various signal transducer along with their potent medical paraphrase. We have discussed the detection strategies in two distinctive sections in label-free and labeled biosensors (Fig. 1). Briefly, label-free sensors directly analyze the occurrence of an analyte via biochemical reactions (Hunt and Armani 2010; Rapp et al. 2010; Chandra 2020), whereas, for labeled sensors, the analyte is sandwiched in-between the biorecognition molecule and the explicitly labeled detector agent for signal output (Ju et al. 2011). So far, only a few reports have been published concerning biosensor-based detection of human coronaviruses. Here, we have tried to highlight those biosensors in a nutshell.
2 Label-Free Biosensor
A label-free biosensor analyzes intrinsic physicochemical property, such as charge, size, molecular weight, interfacial capacitance, resistance, refractive index, or electrical impedance of the target analyte to detect its presence in the test sample (Cooper 2009; Daniels and Pourmand 2007; Choudhary et al. 2016). Label-free biosensors warrant for a single recognition element. Assay simplification by virtue of this method is hence immense, specifically cutting reagent cost and saving assay time. Also, small molecular targets, owing to their size and reasons of obscurity within the binding region of the capturing element, are often not suitable for a labeled assay. These too can be seamlessly used with this method. Additionally, these label-free systems can carry out quantitative measurements of molecular in real-time. Considering such an advantage, continuous data recording is available. The longevity of target analytes is also increased by label-free biosensing, owing to the use of analytes in their original form devoid of any modifications. The use of label-free biosensing techniques and their applications in several infectious diseases has been discussed below, by categorizing the type of signal generated by the transducer element.
2.1 Electrical Transducer
Electrical transducer-based detection methodologies comprehensively support the broad-spectrum approaches for subtle, miniaturized, and portable biomarker recognition procedures. This is done as it can be easily integrated within standard electronic microfabrication setups and will have the swift emerging capability in microfluidics. It can also perform the simplistic yet diversified heterogeneous detection of several biomolecules even in a limited volume of samples making it utterly advantageous with minimal production cost within a bench top or handheld system (Luo and Davis 2013; Baranwal and Chandra 2018). Ishikawa et al. in a study have shown that antibody mimic proteins, a type of affinity binding agents developed by in vitro selection methods, can be employed as biorecognition elements in nanobiosensors (Ishikawa et al. 2009). Here they have developed a sensor constructed on ln2O3 nanowires and improvised it using an antibody mimic protein (AMP), a class of affinity binding agents produced by in vitro selection techniques. They have used fibronectin-based protein as an example of such AMPs. The sensor demonstrates to have a selective affinity toward the SARS biomarker nuclear (N) protein (Fig. 2) even in subnanomolar concentration. The sensitivity of the sensing device is also comparable to the contemporary immunosensing detection techniques, favorably reduces the time consumption, and excludes any involvement of labeling agents. This study also demonstrated the potential of nanobiosensors. It can be used as an accurate, convenient, and rapid tool to measure the variable entities associated to complex biological systems, such as antigen-antibody, ligand-protein, oligonucleotide interaction, etc.
Fibronectin is immobilized on the surface of an In2O3 nanowire as a capturing agent. The highlighted sections (red) are fibronectin with the engineered peptide sequence. It recognizes and binds the nucleocapsid (N) protein of the SARS-CoV. (Image reused with permission from Ishikawa et al. 2009)
Again in an experiment, Layqah et al. have created and demonstrated a new one-of-a-kind competitive electrochemical immunosensor on a carbon array electrode (DEP). Further, the DEP was modified using gold nanoparticle (AuNP) which enhances the electron transfer efficiency and the electrode surface area which in turn increases the overall sensitivity of the sensor (Layqah and Eissa 2019). For a constant amount of antibody concentration added to the sample, the viral strain and the predetermined MERS-CoV protein compete with each other. The antigen-antibody-affinity interaction generates the signal necessitates for detection. Briefly, the response from the sensor was assessed by evaluating alteration in the highest amperage/peak current of the square wave voltammetry signal while gradually increasing the MERS-CoV antigen concentrations. The recorded results revealed a lower limit of detection value and a higher tendency of selectivity toward the other flu proteins with adequate stability. Moreover, the used DEP array electrodes are disposable, environment friendly, as well as cost-effective.
2.2 Plasmonic Transducer
Plasmonic biosensing provides swift, label-free probing of biological analytes in real-time. This method of biosensing can aid in the detection of very small-sized molecules at ultralow concentrations hence acting as excellent devices for point-of-care analysis (Mejía-Salazar and Oliveira 2018). These biosensors can be divided into two classes: one that uses thin metal films and the other that uses disjunct inorganic plasmon resonant nanostructures. There are different sensing techniques for each class, and additionally, a combination of both classes of the sensors can also be employed in certain platforms. The extensively used plasmonic biosensor uses “surface plasmon resonance” (SPR), which is a film-based sensor and purposed to characterize the interactions between biomolecules.
Qiu et al. recently have reported constructing a highly sensitive, canonical dual-functioning plasmonic biosensor that exhibits swift responsive diagnostic ability for SARS-CoV-2 detection (Qiu et al. 2020). The concept of the dual-functional plasmonic biosensor is to integrate the plasmonic photothermal (PPT) effect with localized surface plasmon resonance (LSPR) sensing platform on a two-dimensional gold nano-island (AuNI) chip. Here, two different wavelengths are projected at two different angles of incidence to stimulate the PPT effect and LSPR, which viciously improved the sensitivity, stability, and reliability of the device. The LSPR sensing unit is modulated to obtain real-time and label-free detection of the viral sequences of SARS-CoV-2, such as RdRp-COVID, ORF1ab-COVID, and E genes. Furthermore, the augmentation on the AuNI chips using the in situ PPT increases the specificity of the device to detect nucleic acid sequences by enhancing the hybridization kinetics. It is anticipated that associating the in situ PPT augmentation technique can precisely distinguish the sequence similarity, for RdRp genes from SARS-CoV and SARS-CoV-2. The LOD value of the biosensor was recorded as low to the concentration of 0.22 pM toward the selected SARS-CoV-2 sequences. Considering the pandemic circumstantial of COVID-19, the purposed dual-functional LSPR biosensor has claimed to deliver a consistent platform with ease of implementation. This new cutting-edge technology will certainly introduce a new trail for diagnosis besides the currently available conforming medical tests and the prolonged PCR analysis.
2.3 Field-Effect Transistor (FET)
The field-effect transistor (FET)-based biosensing devices have revealed quite a lot of advantageous properties compared to the other presently viable diagnostic methods. It has the capability of making exceptionally sensitive as well as rapid quantifications even for a little quantity of analytes (Janissen et al. 2017; Liu et al. 2019). These biosensors are determined as potentially worthwhile for medical diagnosis, point-of-care testing, and on-site detection applications. Seo et al. in a recent study have constructed a graphene-based FET biosensor functionalized with the SARS-CoV-2 spike antibody to detect the SARS-CoV-2 virus (Seo et al. 2020). Graphene-based biosensors can sense the variations of the surrounding on their surface while sustaining an optimal sensing environment intended for ultrasensitive and low-noise recognition. For the purpose, the SARS-CoV-2 spike antibody has been immobilized on the construct using a probe linker, 1-pyrenebutyric acid N-hydroxysuccinimide ester (PANHS). PANHS is an aromatic hydrocarbon and an indeed effective interface coupling agent. The sensor was subjected for in vitro studies on cultured SARS-CoV-2 strain and on associate clinical samples, i.e., nasopharyngeal swabs (Fig. 3). It claims to detect the specific SARS-CoV-2 antigen protein effectively recording the LOD of 1 fg/mL. Further, they have also reported testing the specificity of the sensor by distinguishing the SARS-CoV-2 antigen protein from the MERS-CoV efficiently.
Abstract illustration of a plasmonic biosensor evaluated for SARS-CoV-2 detection. (Image reused with permission from Seo et al. 2020)
3 Labeled Biosensors
Label-based biosensing essays are one of the most popular and prospering methods of biosensing nowadays. In label-based biosensors, the analyte molecule gets sandwiched between an immobilized capturing agent on a solid surface (i.e., micro/nanoparticles, electrodes, sensor-chips, etc.) and detecting agents which are typically tagged with signaling molecules, like luminescence molecules, nanoparticles, enzymes, or fluorophores (Baranwal et al. 2016; Chandra et al. 2010). Generally, the receptor (capture and detector) molecules have distinguished binding sites, which as a result enhances device specificity and reduces background noise. A typical example of ELISA-based biosensing assay uses sandwich immunoassay for the diagnosis of various infectious diseases. It uses an antibody as a captured molecule and another enzymatically modified fluorescence tagged antibody to catalyze the translation reaction of the chromogenic substrate to a visually distinctive colored product. The formed color products are then compared based on their optical densities and the concentration of analytes.
Label-based biosensors explore mechanical, optical, and electrical transducers attached to a signal-tagged molecule. The example of such interactions between sensor and tag includes optical, magneto-resistive, and electrochemical biosensors. Further, detection of fluorescence, luminescent tags, and colorimetric evaluation is done by optical sensors. Electrochemical biosensors detect redox reactions with the help of enzyme tags, whereas magnetically tag-aided detections are done by magneto-resistive biosensors. These assemblies quantitatively and semi-quantitatively detect analytes by relating the signal produced to the amount of analyte captured.
3.1 Optical Biosensor
Optical biosensors are applied to a wide array of performance in the field of detection. These not only include the detection of biological systems to promote ground-breaking advances in diagnostics but also improvements in drug discoveries and environmental supervision. Including the additional advantages of higher sensitivity, reliability, robustness, and integrity, optical biosensors also support to avoid the complication of pretreatment and probable influence on the nature of target molecules. Antibody-based immunosensors are the most viable optical sensors that are employed for the detection of pathogens (Byrne et al. 2009). Polyclonal, monoclonal, and recombinant antibodies have been often selected for immunodiagnostics and biomarker detection. This technique is known as an immunochromatographic test (ICT) (Kogaki et al. 2005). ICT works generally based on a sandwich format by means of double antigens or double antibodies. For the rapid detection of SARS-CoV, Tyson Bioresearch, Inc., Taipei, Taiwan, developed an immunogold-based ICT device by incorporating the recombinant N protein antigen of SARS-CoV in the test (Wu et al. 2004). The construction of the ICT device is such that a nitrocellulose strip is present, wherein a detection zone is allocated at the top. This is where an anti-mouse IgG and SARS-CoV N protein is present in an immobilized state onto the control and test line. In the middle, the strip consists of mouse IgG and SARS-CoV N protein teamed with gold nanoparticles serving as the detector/locator. Two wells for the sample and the buffer are present at the bottom. For the assay, a neat serum sample and testing buffer are added to the sample and the buffer well, respectively. An antibody-antigen-gold complex is formed if the sample comprises specific antibodies to SARS-CoV. After lateral flow along the membrane, the colored complex of antibodies-antigen-gold gets accumulated on the test line, and a red color becomes apparent to the naked eye. Two parallel red lines are seen on a positive result; the control line implies that the device is working fine, and the test line indicates the presence of the SARS-CoV antibody in the serum sample. In case of a contrary result, the red line will only be seen on the control region. The test is invalid if red color is found only at the test line or no lines are visible at all. Following a very similar mechanism, Sino Biological, Beijing, China, had released the first ICT kits for the detection of SARS-CoV-2 N and S proteins (Web reference 1).
Huang et al. previously in a study explained the generation of a dual-monoclonal-antibody system tagged with glutathione S-transferase (GST) to apply against SARS-CoV N protein (Huang et al. 2004). Detection of a low concentration GST-N protein (15 ng/mL and 1 ng/mL in PBS and diluted serum, respectively) was accomplished by the use of conventional antigen capture ELISA initially. Later on, an ultrasensitive localized surface plasmon coupled fluorescence (LSPCF) fiberoptic biosensor was developed to further augment the process of detection. After this development, the detection of GST-N in PBS was recorded at the lowest concentration of 0.1 pg/mL. The LOD recorded for 10-fold diluted human serum was 0.1 pg/mL, that is, comparable to that of in raw serum sample (1 pg/mL). The application of LSPCF enhanced the sensitivity of detection by 104-fold by making use of the same monoclonal antibodies. Taking into account the limit of detection and cost-effectiveness, the use of LSPCF always proved to be a preferred method for SARS-CoV N protein detection from serum sample. Fluorescence can be enhanced and excited by LSPs with greater efficiency close to the GNP surface. As over 40 fluorophores excite simultaneously that are presented on each fluorescence probe collectively amplifies the fluorescence signal. In the coming era, LSPCF will also show immense possibilities through utilizing the promising chip-based evaluation to measure serum protein levels both qualitatively and quantitatively. On contrasting with conventional setups, the LSPCF fiber-optic biosensors can detect fluorescence signals near the reaction region and hence accentuate the collection of fluorescence (Chang et al. 2009; Hsieh et al. 2007). Owing to this elevated quantitative capability and sensitivity, this technique can be employed for easy and prompt identification of diseases.
Recently, based on lateral flow assay, Li et al. developed a point-of-care immunoassay kit to detect IgM and IgG antibodies simultaneously in human blood within 15 min (Fig. 4) (Z. Li et al. 2020b). They had reported a clinical trial involving eight hospitals and Chinese CDC agencies corroborating the kit’s medical efficiency. The results were promising and show the rapid detection of antibodies with greater sensitivity and specificity. Owing to excellent results, it is now being authorized to be used in hospitals, clinics, and laboratories and, thus, become a compelling medium in the fight against SARS-CoV-2.
Graphical representation of rapid IgM-IgG-combined antibody test for SARS-CoV-2. A typical detection device (a); abstract representation of varying testing results (b). G stands for immunoglobulin G (IgG); M stands for immunoglobulin M (IgM); C is the control line. (Image reused with permission from Li et al. 2020a, b)
3.2 Genosensor
In the recent era, the generations of nano-biosensors and their different usage protocol have shown a curve of evolution nevertheless. Such an example of a sensor is a genosensor. A genosensor is a typical gene-based sensor that exploits immobilized genetic probes as the recognition element to evaluate specific binding processes that involves the formation of hybrids, i.e., DNA–DNA or DNA–RNA hybrids, and the interactions between ligand molecules or proteins and DNA at the sensing surface. Further, genosensors can be classified into the following types. A receptor-based genosensor, consisting of a bioreceptor probe, that is typically small oligonucleotide sequences. In the case of an aptamer-based genosensor, the aptamer is made up of a sequence of synthetic oligonucleotides that is rendered immobile at the transducer surface, and in due course of time for their affinity, these oligonucleotide sequences acknowledge the nucleic acids analyte by creating complementary duplexes. Additionally, nanocomposites/nanoparticles can be tagged and coupled with genosensors to collectively improve the process of immobilization and the overall sensitivity of the oligonucleotide sequence on the surface of the transducer. In a study, Abad-Valle et al. had reported designing a simple, sensitive, and cost-effective miniaturized homemade device. It was based on a structural framework made of thin gold film for electrochemical detection in fewer volumes (Abad-Valle et al. 2005). They employed the construct for further development of genosensors especially in the detection of the specific sequence of a SARS-CoV virus. The DNA probe was immobilized on the gold surface, and the process was carried out through a thiol group linked at the 3′ end with an aliphatic spacer. The parameters that affected the immobilization were studied using a double-labeled (biotinylated and thiolated) DNA strand. Enzymatic detection was carried out by alkaline phosphatase-labeled streptavidin. Also, blocking with 1-hexanethiol produced well-defined signals. Subsequently, the solvent was evaporated to achieve optimum results, favoring immobilization while avoiding hybridization. A low limit of detection (6 pM) concerning the previously reported analogous schemes in literature was achieved on enzymatic hydrolysis of 3-indoxyl phosphate. The specificity of the assessment was verified using a 3-base mismatch DNA strand, where a strong inequity was reported while retaining 1 h of hybridization period and 50% of formamide in the buffer. The group has also specified to study in detail the processes to discriminate the 2-base and 1-base mismatching strands as a prospect.
3.3 Luciferase-Based Sensors
Kilianski et al. have reported developing a luciferase-based biosensor to detect MERS-CoV. They employed the biosensor construct for expressing the two MERS-CoV-specific biomarkers, the papain-like protease (PLpro), and the 3-chymotrypsin-like protease (3CLpro) while monitoring their activity in vitro simultaneously (Kilianski et al. 2013). It has been demonstrated that the biosensor recognizes the expressed PLpro in MERS-CoV while processing the recognized CoV-PLpro cleavage site, RLKGG. However, because of the divergent amino acid sequence in the binding site of the drug, the already in-use CoV-PLpro inhibitors were not able to block MERS-CoV PLpro activity. Again, they utilize the luciferase-based biosensors together with the recognized 3CLpro cleavage site VRLQS to understand the activity of 3CLpro, by expressing the protease affixed with nonstructural protein 4 (nsp4) and the amino-terminal portion of nsp6. They have also determined that similar to SARS-CoV and murine CoV, a small-molecule inhibitor inhibits the replication process in the case of MERS-CoV 3CLpro and inhibits the activity. As a whole, the developed biosensor assays involving the proteases permit rapid identification as well as the evaluation of the protease inhibitors and viral protease activity, respectively. It is anticipated that to assess protease activity, the expression of MERS-CoV PLpro and 3CLpro is effective. Luciferase-based biosensor supports such expression of PL and CLpro that in turn enable swift identification of the replication inhibiting small molecule that is specific to MERS-CoV. Accordingly, it may also prove its compelling aspects over other coronaviruses.
4 Conclusion
Biosensing methodologies have shown enormous advancements for viral detection in terms of rapid analysis with low LOD, wide linear detection range, high sensitivity, and specificity. The need to develop point-of-care diagnostic devices for rapid, sensitive, and cost-effective screening even for multiple samples at a time explains the necessity of biosensors. It also rationalizes the increase in demand for compelling epidemiological surveillance along with high-throughput screening tests. So far biosensing techniques despite having such beneficial properties remain inceptive for commercialized diagnosis of infectious diseases. Biosensors when compared with other diagnostics come with an added advantage of being one of the most compact and handheld devices. This makes biosensors probably the best among the race of diagnostic tools. Infectious diseases in urban and suburban areas spread like wildfire in a very short duration of time; hence regulation of such outbreaks becomes a reason for concern. Hence, such rapid miniaturized tests may overcome such delinquency with effective enactment toward initial disease control and would also waive off delayed diagnosis as seen in conventional procedures. Furthermore, novel and technologically advanced nano-sized materials, composites, and polymers with significant biocompatibility also are recognized to provide improved specificity and stability to affinity reagents. It is for a fact that the current advances in technology would accelerate the development of biosensors into such a tool of profound magnificence that it will be a milestone in diagnostics and infection surveillance in medical setups and laboratories.
References
Abad-Valle P, Fernández-Abedul MT, Costa-García A (2005) Genosensor on gold films with enzymatic electrochemical detection of a SARS virus sequence. In: Biosensors and bioelectronics. https://doi.org/10.1016/j.bios.2004.10.019
Baranwal A, Chandra P (2018) Clinical implications and electrochemical biosensing of monoamine neurotransmitters in body fluids, in vitro, in vivo, and ex vivo models. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2018.09.002
Baranwal A, Mahato K, Srivastava A, Maurya PK, Chandra P (2016) Phytofabricated metallic nanoparticles and their clinical applications. RSC Adv. https://doi.org/10.1039/c6ra23411a
Ben-Assa N, Naddaf R, Gefen T, Capucha T, Hajjo H, Mandelbaum N, Elbaum L, Rogov P, King DA, Kaplan S, Rotem A, Chowers M, Szwarcwort-Cohen M, Paul M, Geva-Zatorsky N (2020) SARS-CoV-2 on-the-spot virus detection directly from patients. medRxiv. https://doi.org/10.1101/2020.04.22.20072389
Bhatnagar I, Mahato K, Ealla KKR, Asthana A, Chandra P (2018) Chitosan stabilized gold nanoparticle mediated self-assembled gliP nanobiosensor for diagnosis of invasive Aspergillosis. Int J Biol Macromol. https://doi.org/10.1016/j.ijbiomac.2017.12.084
Byrne B, Stack E, Gilmartin N, O’Kennedy R (2009) Antibody-based sensors: principles, problems and potential for detection of pathogens and associated toxins. Sensors (Switzerland). https://doi.org/10.3390/s90604407
Chandra P (2013) Miniaturized multiplex electrochemical biosensor in clinical bioanalysis. J Bioanal Biomed. https://doi.org/10.4172/1948-593X.1000e122
Chandra P (2016) Nanobiosensors for personalized and onsite biomedical diagnosis. Nanobiosens Pers Onsite Biomed Diagn. https://doi.org/10.1049/PBHE001E
Chandra P (2020) Miniaturized label-free smartphone assisted electrochemical sensing approach for personalized COVID-19 diagnosis. Sens Int. https://doi.org/10.1016/j.sintl.2020.100019
Chandra P, Das D, Abdelwahab AA (2010) Gold nanoparticles in molecular diagnostics and therapeutics. Dig J Nanomater Biostruct 5:363–367
Chandra P, Tan YN, Singh SP (2017) Next generation point-of-care biomedical sensors technologies for cancer diagnosis. Next Gener Point-of-care Biomed Sens Technol Cancer Diagn. https://doi.org/10.1007/978-981-10-4726-8
Chang YF, Chen RC, Lee YJ, Chao SC, Su LC, Li YC, Chou C (2009) Localized surface plasmon coupled fluorescence fiber-optic biosensor for alpha-fetoprotein detection in human serum. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2008.08.019
Choudhary M, Yadav P, Singh A, Kaur S, Ramirez-Vick J, Chandra P, Arora K, Singh SP (2016) CD 59 targeted ultrasensitive electrochemical immunosensor for fast and noninvasive diagnosis of oral cancer. Electroanalysis 28:2565–2574
Chow CB (2004) Post-SARS infection control in the hospital and clinic. Paediatr Respir Rev. https://doi.org/10.1016/j.prrv.2004.07.006
Chua AL, Yean CY, Ravichandran M, Lim BH, Lalitha P (2011) A rapid DNA biosensor for the molecular diagnosis of infectious disease. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2011.02.040
Coleman CM, Frieman MB (2015) Growth and quantification of MERS-CoV infection. Curr Protoc Microbiol. https://doi.org/10.1002/9780471729259.mc15e02s37
Cooper MA (2009) Label-free biosensors: techniques and applications. https://doi.org/10.1017/CBO9780511626531
Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DGJC, Haagmans BL, Van Der Veer B, Van Den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eur Secur. https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045
Cotten M, Watson SJ, Kellam P, Al-Rabeeah AA, Makhdoom HQ, Assiri A, Aal-Tawfiq J, Alhakeem RF, Madani H, AlRabiah FA, Al Hajjar S, Al-Nassir WN, Albarrak A, Flemban H, Balkhy HH, Alsubaie S, Palser AL, Gall A, Bashford-Rogers R, Rambaut A, Zumla AI, Memish ZA (2013) Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet. https://doi.org/10.1016/S0140-6736(13)61887-5
Dai C, Choi S (2013) Technology and applications of microbial biosensor. Open J Appl Biosens. https://doi.org/10.4236/ojab.2013.23011
Daniels JS, Pourmand N (2007) Label-free impedance biosensors: opportunities and challenges. Electroanalysis. https://doi.org/10.1002/elan.200603855
Donnelly CA, Malik MR, Elkholy A, Cauchemez S, Van Kerkhove MD (2019) Worldwide reduction in MERS cases and deaths since 2016. Emerg Infect Dis. https://doi.org/10.3201/eid2509.190143
Fauci AS, Morens DM (2012) The perpetual challenge of infectious diseases. N Engl J Med. https://doi.org/10.1056/NEJMra1108296
França RFO, Da Silva CC, De Paula SO (2013) Recent advances in molecular medicine techniques for the diagnosis, prevention, and control of infectious diseases. Eur J Clin Microbiol Infect Dis. https://doi.org/10.1007/s10096-013-1813-0
Gogna A, Tay KH, Tan BS (2014) Severe acute respiratory syndrome: 11 years later – a radiology perspective. Am J Roentgenol. https://doi.org/10.2214/AJR.14.13062
Gui M, Liu X, Guo D, Zhang Z, Yin CC, Chen Y, Xiang Y (2017) Electron microscopy studies of the coronavirus ribonucleoprotein complex. Protein Cell. https://doi.org/10.1007/s13238-016-0352-8
Hamimi A (2016) MERS-CoV: Middle East respiratory syndrome corona virus: can radiology be of help? Initial single center experience. Egypt J Radiol Nucl Med. https://doi.org/10.1016/j.ejrnm.2015.11.004
Hosseiny M, Kooraki S, Gholamrezanezhad A, Reddy S, Myers L (2020) Radiology perspective of coronavirus disease 2019 (COVID-19): lessons from severe acute respiratory syndrome and Middle East respiratory syndrome. AJR Am J Roentgenol. https://doi.org/10.2214/AJR.20.22969
Hsieh BY, Chang YF, Ng MY, Liu WC, Lin CH, Wu HT, Chou C (2007) Localized surface plasmon coupled fluorescence fiber-optic biosensor with gold nanoparticles. Anal Chem. https://doi.org/10.1021/ac0624389
Huang Q, Yu L, Petros AM, Gunasekera A, Liu Z, Xu N, Hajduk P, Mack J, Fesik SW, Olejniczak ET (2004) Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. https://doi.org/10.1021/bi036155b
Hui DSC, Chan MCH, Wu AK, Ng PC (2004) Severe acute respiratory syndrome (SARS): epidemiology and clinical features. Postgrad Med J 80:373 LP–373381. https://doi.org/10.1136/pgmj.2004.020263
Hunt HK, Armani AM (2010) Label-free biological and chemical sensors. Nanoscale. https://doi.org/10.1039/c0nr00201a
Ishikawa FN, Chang H, Curreli M, Liao H, Olson ЌCA, Chen P, Zhang R, Roberts RW, Sun R, Cote ЌRJ, Thompson ME, Zhou C (2009) Label-free, electrical detection of the SARS virus N-protein with nanowire capture probes. ACS Nano
Janissen R, Sahoo PK, Santos CA, Da Silva AM, Von Zuben AAG, Souto DEP, Costa ADT, Celedon P, Zanchin NIT, Almeida DB, Oliveira DS, Kubota LT, Cesar CL, Souza APD, Cotta MA (2017) InP nanowire biosensor with tailored Biofunctionalization: ultrasensitive and highly selective disease biomarker detection. Nano Lett. https://doi.org/10.1021/acs.nanolett.7b01803
Jardon M, Mohammad SF, Jude CM, Pahwa A (2019) Imaging of emerging infectious diseases. Curr Radiol Rep. https://doi.org/10.1007/s40134-019-0338-4
Ju H, Zhang X, Wang J (2011) Signal amplification for Nanobiosensing. https://doi.org/10.1007/978-1-4419-9622-0_2
Kashish BS, Jyoti A, Mahato K, Chandra P, Prakash R (2017) Highly sensitive in vitro biosensor for Enterotoxigenic Escherichia coli detection based on ssDNA anchored on PtNPs-chitosan Nanocomposite. Electroanalysis. https://doi.org/10.1002/elan.201600169
Kilianski A, Mielech AM, Deng X, Baker SC (2013) Assessing activity and inhibition of Middle East respiratory syndrome coronavirus papain-like and 3C-like proteases using luciferase-based biosensors. J Virol. https://doi.org/10.1128/jvi.02105-13
Knobler S, Mahmoud A, Lemon S, Mack A, Sivitz L, Oberholtzer K (2004) Learning from SARS: preparing for the next disease outbreak. The National Academies Press, Washington
Kogaki H, Uchida Y, Fujii N, Kurano Y, Miyake K, Kido Y, Kariwa H, Takashima I, Tamashiro H, Ling AE, Okada M (2005) Novel rapid immunochromatographic test based on an enzyme immunoassay for detecting nucleocapsid antigen in SARS-associated coronavirus. J Clin Lab Anal. https://doi.org/10.1002/jcla.20070
Kumar A, Purohit B, Maurya PK, Pandey LM, Chandra P (2019a) Engineered nanomaterial assisted signal-amplification strategies for enhancing analytical performance of electrochemical biosensors. Electroanalysis. https://doi.org/10.1002/elan.201900216
Kumar A, Purohit B, Mahato K, Mandal R, Srivastava A, Chandra P (2019b) Gold-iron bimetallic nanoparticles impregnated reduced Graphene oxide based Nanosensor for label-free detection of biomarker related to non-alcoholic fatty liver disease. Electroanalysis. https://doi.org/10.1002/elan.201900337
Kumar A, Roy S, Srivastava A, Naikwade MM, Purohit B, Mahato K, Naidu VGM, Chandra P (2019c) Chapter 10 – Nanotherapeutics: a novel and powerful approach in modern healthcare system. Nanotechnol Mod Anim Biotechnol. https://doi.org/10.1016/B978-0-12-818823-1.00010-7
Kurstjens S, van der Horst A, Herpers R, Geerits MWL, Kluiters-de Hingh YCM, Göttgens E-L, Blaauw MJT, Thelen MHM, Elisen MGLM, Kusters R (2020) Rapid identification of SARS-CoV-2-infected patients at the emergency department using routine testing. https://doi.org/10.1101/2020.04.20.20067512
Lau SKP, Woo PCY, Wong BHL, Tsoi HW, Woo GKS, Poon RWS, Chan KH, Wei WI, Malik Peiris JS, Yuen KY (2004) Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in SARS patients by enzyme-linked immunosorbent assay. J Clin Microbiol. https://doi.org/10.1128/JCM.42.7.2884-2889.2004
Layqah LA, Eissa S (2019) An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim Acta. https://doi.org/10.1007/s00604-019-3345-5
Lee E-Y, Ko HL, Jung S-Y, Nam J-H (2017) Development of a rapid diagnostic kit for antigen and antibody of Middle East respiratory syndrome coronavirus. J Immunol
Li D, Wang D, Dong J, Wang N, Huang H, Xu H, Xia C (2020a) False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based ct diagnosis and insights from two cases. Korean J Radiol. https://doi.org/10.3348/kjr.2020.0146
Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W, Zhang Y, Wang J, Huang B, Lin Y, Yang J, Cai W, Wang X, Cheng J, Chen Z, Sun K, Pan W, Zhan Z, Chen L, Ye F (2020b) Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. https://doi.org/10.1002/jmv.25727
Lin YC, Dong SL, Yeh YH, Wu YS, Lan GY, Liu CM, Chu TC (2005) Emergency management and infection control in a radiology department during an outbreak of severe acute respiratory syndrome. Br J Radiol. https://doi.org/10.1259/bjr/17161223
Liu J, Chen X, Wang Q, Xiao M, Zhong D, Sun W, Zhang G, Zhang Z (2019) Ultrasensitive monolayer MoS 2 field-effect transistor based DNA sensors for screening of down syndrome. Nano Lett. https://doi.org/10.1021/acs.nanolett.8b03818
Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, Carter LJ, Smoot J, Gregg AC, Daniels AD, Jervey S, Albaiu D (2020) Research and Development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci. https://doi.org/10.1021/acscentsci.0c00272
Luo X, Davis JJ (2013) Electrical biosensors and the label free detection of protein disease biomarkers. Chem Soc Rev. https://doi.org/10.1039/c3cs60077g
Mahato K, Chandra P (2019) Paper-based miniaturized immunosensor for naked eye ALP detection based on digital image colorimetry integrated with smartphone. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2018.12.006
Mahato K, Baranwal A, Srivastava A, Maurya PK, Chandra P (2018a) Smart materials for biosensing applications. In: Pawar PM, Ronge BP, Balasubramaniam R, Seshabhattar S (eds) Techno-societal 2016. Springer International Publishing, Cham, pp 421–431
Mahato K, Kumar S, Srivastava A, Maurya PK, Singh R, Chandra P (2018b) Electrochemical immunosensors: fundamentals and applications in clinical diagnostics. In: Handbook of immunoassay technologies. https://doi.org/10.1016/B978-0-12-811762-0.00014-1
Mahato K, Maurya PK, Chandra P (2018c) Fundamentals and commercial aspects of nanobiosensors in point-of-care clinical diagnostics. 3 Biotech. https://doi.org/10.1007/s13205-018-1148-8
Mahato K, Purohit B, Bhardwaj K, Jaiswal A, Chandra P (2019) Novel electrochemical biosensor for serotonin detection based on gold nanorattles decorated reduced graphene oxide in biological fluids and in vitro model. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2019.111502
Mahato K, Purohit B, Kumar A, Chandra P (2020a) Paper-based biosensors for clinical and biomedical applications: emerging engineering concepts and challenges. In: Comprehensive analytical chemistry. https://doi.org/10.1016/bs.coac.2020.02.001
Mahato K, Purohit B, Kumar A, Chandra P (2020b) Clinically comparable impedimetric immunosensor for serum alkaline phosphatase detection based on electrochemically engineered au-nano-Dendroids and graphene oxide nanocomposite. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2019.111815
Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, Nagata N, Sekizuka T, Katoh H, Kato F, Sakata M, Tahara M, Kutsuna S, Ohmagari N, Kuroda M, Suzuki T, Kageyama T, Takeda M (2020) Enhanced isolation of SARS-CoV-2 by TMPRSS2- expressing cells. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.2002589117
Mejía-Salazar JR, Oliveira ON (2018) Plasmonic biosensing. Chem Rev. https://doi.org/10.1021/acs.chemrev.8b00359
Memish ZA, Perlman S, Van Kerkhove MD, Zumla A (2020) Middle East respiratory syndrome. Lancet. https://doi.org/10.1016/S0140-6736(19)33221-0
Mohan R, Mach KE, Bercovici M, Pan Y, Dhulipala L, Wong PK, Liao JC (2011) Clinical validation of integrated nucleic acid and protein detection on an electrochemical biosensor array for urinary tract infection diagnosis. PLoS One. https://doi.org/10.1371/journal.pone.0026846
Nasir MU, Roberts J, Muller NL, Macri F, Mohammed MF, Akhlaghpoor S, Parker W, Eftekhari A, Rezaei S, Mayo J, Nicolaou S (2020) The role of emergency radiology in COVID-19: from preparedness to diagnosis. Can Assoc Radiol J. https://doi.org/10.1177/0846537120916419
Ozili PK, Arun T (2020) Spillover of COVID-19: impact on the global economy. SSRN Electron J. https://doi.org/10.2139/ssrn.3562570
Pan Y, Long L, Zhang D, Yan T, Cui S, Yang P, Wang Q, Ren S (2020) Potential false-negative nucleic acid testing results for severe acute respiratory syndrome coronavirus 2 from thermal inactivation of samples with low viral loads. Clin Chem. https://doi.org/10.1093/clinchem/hvaa091
Pejcic B, De Marco R, Parkinson G (2006) The role of biosensors in the detection of emerging infectious diseases. Analyst. https://doi.org/10.1039/b603402k
Polizzi KM (2019) Biosensors. In: Comprehensive biotechnology. https://doi.org/10.1016/B978-0-444-64046-8.00060-4
Prasad A, Mahato K, Maurya PK, Chandra P (2016) Biomaterials for biosensing applications. J Anal Bioanal Tech. https://doi.org/10.4172/2155-9872.1000e124
Purohit B, Kumar A, Mahato K, Chandra P (2019a) Novel sensing assembly comprising engineered gold dendrites and MWCNT-AuNPs Nanohybrid for acetaminophen detection in human urine. Electroanalysis. https://doi.org/10.1002/elan.201900551
Purohit B, Kumar A, Mahato K, Roy S, Chandra P (2019b) Cancer cytosensing approaches in miniaturized settings based on advanced nanomaterials and biosensors. In: Nanotechnology in modern animal biotechnology: concepts and applications. https://doi.org/10.1016/B978-0-12-818823-1.00009-0
Purohit B, Mahato K, Kumar A, Chandra P (2019c) Sputtering enhanced peroxidase like activity of a dendritic nanochip for amperometric determination of hydrogen peroxide in blood samples. Microchim Acta. https://doi.org/10.1007/s00604-019-3773-2
Purohit B, Kumar A, Mahato K, Chandra P (2020a) Smartphone-assisted personalized diagnostic devices and wearable sensors. Curr Opin Biomed Eng. https://doi.org/10.1016/j.cobme.2019.08.015
Purohit B, Kumar A, Mahato K, Chandra P (2020b) Electrodeposition of metallic nanostructures for biosensing applications in health care. J Sci Res. https://doi.org/10.37398/jsr.2020.640109
Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J (2020) Dual-functional Plasmonic Photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. https://doi.org/10.1021/acsnano.0c02439
Rapp BE, Gruhl FJ, Länge K (2010) Biosensors with label-free detection designed for diagnostic applications. Anal Bioanal Chem. https://doi.org/10.1007/s00216-010-3906-2
Seo G, Lee G, Kim MJ, Baek S-H, Choi M, Ku KB, Lee C-S, Jun S, Park D, Kim HG, Kim S-J, Lee J-O, Kim BT, Park EC, Kim SI (2020) Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. https://doi.org/10.1021/acsnano.0c02823
Shen M, Zhou Y, Ye J, Abdullah AL-maskri AA, Kang Y, Zeng S, Cai S (2020) Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal. https://doi.org/10.1016/j.jpha.2020.02.010
Sin ML, Mach KE, Wong PK, Liao JC (2014) Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev Mol Diagn. https://doi.org/10.1586/14737159.2014.888313
Vaddiraju S, Tomazos I, Burgess DJ, Jain FC, Papadimitrakopoulos F (2010) Emerging synergy between nanotechnology and implantable biosensors: a review. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2009.12.001
Web Refereces 1. https://www.sinobiological.com/research/virus/2019-ncov-antigen
Wu HS, Chiu SC, Tseng TC, Lin SF, Lin JH, Hsu YF, Wang MC, Lin TL, Yang WZ, Ferng TL, Huang KH, Hsu LC, Lee LL, Yang JY, Chen HY, Su SP, Yang SY, Lin TH, Su IJ (2004) Serologic and molecular biologic methods for SARS-associated coronavirus infection. Taiwan Emerg Infect Dis. https://doi.org/10.3201/eid1002.030731
Wu D, Wu T, Liu Q, Yang Z (2020) The SARS-CoV-2 outbreak: what we know. Int J Infect Dis. https://doi.org/10.1016/j.ijid.2020.03.004
Xiao AT, Tong YX, Zhang S (2020) False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J Med Virol. https://doi.org/10.1002/jmv.25855
Yu J, Ding N, Chen H, Liu XJ, Pu ZH, Xu HJ, Lei Y, Zhang HW (2020) Loopholes in current infection control and prevention practices against COVID-19 in radiology department and improvement suggestions. Can Assoc Radiol J. https://doi.org/10.1177/0846537120916852
Zumla A, Hui DS, Perlman S (2015) Middle East respiratory syndrome. Lancet. https://doi.org/10.1016/S0140-6736(15)60454-8
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 The Editor(s) (if applicable) and The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Mahapatra, S., Baranwal, A., Purohit, B., Roy, S., Mahto, S.K., Chandra, P. (2020). Advanced Biosensing Methodologies for Ultrasensitive Detection of Human Coronaviruses. In: Chandra, P., Roy, S. (eds) Diagnostic Strategies for COVID-19 and other Coronaviruses. Medical Virology: From Pathogenesis to Disease Control. Springer, Singapore. https://doi.org/10.1007/978-981-15-6006-4_2
Download citation
DOI: https://doi.org/10.1007/978-981-15-6006-4_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-6005-7
Online ISBN: 978-981-15-6006-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)