Abstract
Autophagy is an important metabolic pathway of cells. Cells degrade harmful intracellular components with the aid of autophagy to maintain a healthy state. In recent decades, the study of non-coding RNA in the regulation of autophagy has been a hot area. Mounting evidence indicates that many ncRNAs are involved in the dynamic process of autophagy, and further studies were undertaken to dissect the detailed cellular and molecular mechanisms underlying this process. In this chapter, we mainly summarized the regulation of different non-coding RNAs in autophagy as well as the detailed mechanisms. Based on these findings, we also discussed the roles of non-coding RNAs in the diagnosis, treatment, and prognosis of diseases with an emphasis on their use as potential biomarkers and therapeutic targets for different diseases.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 MicroRNAs and Autophagy
1.1 MicroRNAs (miRNAs)
MicroRNAs (miRNAs) are endogenous short noncoding RNAs (ncRNAs) of 20–25 nucleotides that mediate gene expression. They are highly conserved among species and are not only expressed in almost all eukaryotes but also in several viruses, suggesting their powerful biological functions.
1.1.1 The Biogenesis of miRNAs
In the nucleus, the miRNA genes are transcribed into polyadenylated primary miRNAs (pri-miRNAs) by RNA polymerase. Pri-miRNAs can be up to several thousand nt long and contain 22–25 nt long sequences of mature miRNA. A nuclear protein complex participates in the processing of pri-miRNA to single hairpins termed precursor miRNA (pre-miRNA). The complex comprises the RNase III enzyme Drosha, DGCR8 and several factors such as p68 and p72. Subsequently, pre-miRNA enters the cytoplasm through a direct interaction with Exp5. In the cytoplasm, the dicer enzyme cuts the pre-miRNA to a mature length, and the molecule remains double-stranded. Finally, one of the two strands will be translated into AGO family proteins and then assembled into the RNA-induced silencing complex (RISC) to participate in gene silencing.
1.1.2 The Function of miRNAs
Although microRNAs were identified in 1993, their role in gene regulation was recognized approximately 10 years later. Studies have shown that assembly into an RNA-induced silencing complex (RISC), which mainly includes AGO and GW182, is required for miRNA function. AGO proteins are highly conserved, and small RBPs (RNA-binding proteins) are composed of four domains: the N-terminal domain, the PAZ domain, the MID domain and the PIWI domain. The MID and PIWI domains can bind to the 5′ end of miRNA, while the PAZ domain can bind to the 3′ end. The mammalian genome encodes four AGO proteins (Ago1–4), but only Ago2 can degrade messenger RNA (mRNA) that is completely complementary to the full miRNA sequence. GW183, as a binding protein of AGO, can bind to AGO through the GW domain at the N-terminus, whereas the silencing domain of the C-terminus of GW183 can be used as a platform to recruit various auxiliary proteins, such as PABP and CCR4-NOT, and then repress translation from the mRNA. The central role of miRNAs in this process is to bind to the complementary sequences at the 3′-UTR of the mRNA through the seed region, which contains 2–8 nucleotides at the 5′-terminal, and then identify the corresponding RNA to regulate gene expression at the posttranscriptional level through RISC. A single miRNA can target multiple genes to regulate their expression, and multiple miRNAs can also regulate the same gene to precisely regulate the expression of a single gene. miRNAs have been confirmed to be involved in many biological processes, such as cell development, proliferation, differentiation, and apoptosis, and they play an extremely important role in the regulation of autophagy.
1.2 MiRNA-Mediated Regulation of Autophagy
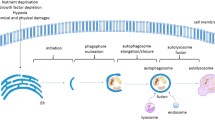
Studies have shown that miRNAs are involved in various stages of autophagy including autophagic induction, vesicle nucleation, vesicle elongation, vesicle retrieval and fusion, and they are also involved in the regulation of upstream signaling pathways that can affect autophagy induction (Fig. 10.1).
1.2.1 Autophagic Induction
In higher mammals, autophagy is mainly activated by ULK (unc-51-like kinase) complexes, which contain ULK 1/2, ATG13, FIP200 and ATG101. Among them, ULK1/2 is the core protein that initiates autophagy. When cellular metabolism is disrupted by starvation and oxidative stress, mTOR activity is inhibited, AMPK kinase is activated, and ULK1/2 is rapidly activated to phosphorylate downstream substrates such as ATG13, Beclin1, and VPS34. ATG13 can further promote the activation of ULK1/2 and mediate the phosphorylation of FIP200. As a scaffold protein, FIP200 is involved in the assembly of ATG proteins. ATG101, as a hydrophilic protein, can stabilize ATG13 and protect it from proteasomal degradation. Therefore, all components of the ULK complex have indispensable functions. miRNA can affect the formation of the ULK complex by regulating the expression of various components of ULK complex proteins.
In the C2C12 myoblast cell line, miR-20a and miR-106b, which belong to the miR-17 family, may participate in regulating leucine deprivation-induced autophagy by inhibiting ULK1 expression (Wu et al. 2012). Other miRNAs belonging to the miR-17 family, such as miR-20b, miR-106a, miR-93, and miR-17-5p, were also confirmed to inhibit the expression of ULK1 and inhibit autophagy. This demonstrated the powerful role of the miR-17 family in regulating the induction phase of autophagy. In addition, miR-489, miR-142-5p, and miR-25 have been reported to affect autophagy by targeting ULK1. However, miR-26a and miR-26b can target and regulate ULK1 and ULK2, respectively. In addition, ULK2 is regulated by miR-885-3p, suggesting that miR-885-3p might contribute to the regulation of squamous cell carcinoma cell autophagy and/or apoptosis upon cisplatin exposure. Furthermore, ATG13 was reported to be regulated by miR-133a-3p, and FIP200 was regulated by miR-20a, miR-20b, miR-224-3p, and miR-309-3p. Due to the negative regulation of autophagy by mTOR, the miR-99 family can indirectly promote autophagy by inhibiting the IGF-1R/Akt/mTOR signaling pathway, while miR-100 can directly target the 3′-UTR of mTOR mRNA to inhibit mTOR and thus activate autophagy. All of the miRNAs described above affect autophagy at the stage of autophagic induction.
1.2.2 Vesicle Nucleation
During vesicle nucleation, the double membrane-bound vesicles of autophagosomes are formed by collecting proteins and liposomes. Vesicle nucleation is induced by the PI3K complex, which is mainly composed of PI3KC3 (hVPS34), Beclin-1, p150, and ATG14L. In addition, several proteins, such as Ambra1, Bif-1, UVRAG and Rubicon, can bind and regulate the PI3K complex. Many miRNAs are involved in the regulation of vesicle nucleation, and previous studies mainly focused on miRNA regulation of Beclin-1 expression. Thus far, multiple miRNAs, including members of the miR-30 family, miR-124-3p, miR-216b, miR-17-5p, and miR-376b, were found to affect Beclin-1 expression and autophagy by targeting the 3′-UTR of Beclin-1 mRNA. miR-23a can target Ambra1 and inhibit the autophagy of fibroblasts during UV-induced photoaging (Zhang et al. 2016). In addition to the direct regulation of Beclin-1, miRNAs can indirectly regulate Beclin-1 to affect autophagy. As a protein with a BH3 domain, Beclin-1 can bind bcl-2/bcl-xl, and PUMA, which also has a BH3 domain, can release Beclin-1 through the competitive binding of bcl-2/bcl-xl to induce autophagy. Studies have shown that miR-143 can inhibit the methylphenidate-induced autophagy of microglia by targeting PUMA, which illustrates the multiple regulatory effects of miRNAs on autophagy. Ambra1 was also targeted by miR-7, leading to the miR-7-mediated promotion of lung cancer cell proliferation by inhibiting autophagy. UVRAG was found to be regulated by the miR-125 family, including miR-183, miR-216b, miR-351, miR-374a, miR-630, and miR-1185. All of these studies demonstrated the powerful regulatory role of miRNAs in the vesicle nucleation stage.
1.2.3 Vesicle Elongation
During vesicle elongation, two ubiquitylation-like conjugation pathways are involved. The first pathway is based on ATG12, which can be activated by the ubiquitin-activating enzyme ATG7 and bind with ATG5 under the influence of the ubiquitin ligase ATG10, finally forming the ATG12-ATG5-ATG16L complex with the addition of ATG16L. This pathway mainly participates in the dilation of the autophagosome. The second pathway involves the conjugation of LC3 to a lipid molecule. LC3 should be cleaved at its carboxy terminus by ATG4 to generate the cytosolic free LC3-I form in which the glycine residue is exposed. After activation by ATG7 and the action of ATG10, LC3-I binds to phosphatidylethanolamine and is modified to LC3-II on the surface of the autophagosome membrane.
Due to the large number of proteins participating in this stage, many of the corresponding miRNAs involved in the regulation of these proteins have been identified. For example, ATG12 is regulated by a variety of miRNAs, such as the miR-30 family members miR-23a, miR-23b, miR-214, miR-505-3p, etc. The miR-30 family is also involved in the regulation of ATG5 by targeting the 3′-UTR of ATG5 mRNA. However, this does not mean that the members of the miR-30 family can regulate autophagy. A recent investigation indicated that ATG5 also has non-autophagic functions. After LPS stimulation in human brain microvascular endothelial cells, miR-30d was downregulated, which subsequently upregulated the expression of ATG5. Instead of activating the autophagic pathway, these biological events promoted the transformation of endothelial cells into mesenchymal cells (Yang et al. 2018). ATG5 was also regulated by miR-9a-5p, miR-142-3p, miR-181a, miR-224-3p, miR-638, etc. Inside the cell, miR-142-3p can also simultaneously regulate the expression of ATG16L1 and exert important effects on autophagy during vesicle elongation. The miR-17 family members miR-106a and miR-106b have two binding sites on the mRNA 3′-UTR of the ATG16L1 gene; however, only miR-106b inhibits starvation-induced autophagy by inhibiting the expression of ATG16L1, whereas the miR-106a regulation of autophagy does not appear to be through ATG16L1 and instead may be via other mechanisms. The miR-17 family can also participate in the regulation of ATG7 expression and inhibit autophagy including miR-137, miR-210, and miR-520b. The 3′-UTR of ATG10 mRNA contains sites targeted by miR-4458, miR-4667-5p and miR-4668-5p, which indicates that they can simultaneously regulate ATG10 expression, thereby inhibiting the autophagy-mediated elimination of Burkholderia pseudomallei in human lung epithelial cells. ATG4D is regulated by miR-101 and plays a negative regulatory role in vesicle elongation by regulating the binding of RAB5AATG12 and ATG5. The above results showed the important role of miRNAs in vesicle elongation.
1.2.4 Vesicle Retrieval
Vesicle retrieval is the stage of recycling some components such as surface receptors in autophagosomes. The mechanism, however, is not yet clear. ATG9, a transmembrane protein, is necessary for this stage. ATG9 continuously shuttles between the autophagosome precursor structure and other structures or organelles (such as the endoplasmic reticulum, Golgi apparatus, and mitochondria). This process also requires ATG2 and ATG18. Unfortunately, research on miRNA regulation at this stage is relatively limited. Until recently, only miR-34a was known to directly target the ATG9A mRNA 3′-UTR and inhibit ATG9A expression and autophagy. These results demonstrated that a miR-34a mutation can prolong the life of Caenorhabditis elegans by enhancing autophagy (Jurong et al. 2013). Recent studies have also suggested that microRNA-130a inhibits autophagy by activating ATG2B and DICER1 and inducing the killing of chronic lymphocytic leukemia cells. These studies suggest the importance of miRNAs in the vesicle retrieval stage.
1.2.5 Lysosomal Fusion
During lysosomal fusion, the engulfed cargo is degraded after the fusion of autophagosomes and lysosomes. In mammals, RAB7 is mainly involved in this process, and LAMP1 and LAMP2 also play important roles. Although RAB27A and LAMP3 have not been thoroughly studied, some research suggests that they are related to the vesicle fusion of autophagosomes. A recent study found that miR-138-5p can indirectly regulate RAB7 by targeting SIRT1, thereby inhibiting autophagy in the vesicle fusion phase in pancreatic cancer. miR-487b-5p can promote lung cancer cell proliferation by targeting LAMP2 to inhibit autophagy. In the process of ischemic stroke, miR-207 and miR-352 can affect autophagy by directly targeting LAMP2. LAMP2 is also regulated by miR-21, allowing miR-21 to inhibit molecular chaperone-induced autophagy in a Parkinson’s disease model. miR-224, miR-373-5p, and miR-379 can target LAMP2 and regulate its expression. Furthermore, miR-205 can inhibit autophagy by regulating RAB27A and LAMP3, thereby affecting the sensitivity of prostate cancer cells to cisplatin.
In addition, studies have shown that certain miRNAs can target multiple proteins at various stages of autophagy. For example, miR-33a-5p and miR-33a-3p have been reported to directly target key autophagy effectors, such as ATG5, ATG12, LC3B and LAMP1, and to inhibit AMPK-dependent autophagic activation and lysosomal gene transcription by targeting FOXO3 and TFEB, which in turn affect autophagy in many ways (Ouimet et al. 2016). miRNAs may also affect autophagy through indirect regulation. miR-378 was reported to activate Akt by targeting PDK1, which indirectly activated mTORC1, inhibited FoxO1 and FoxO3, and ultimately promoted autophagy in skeletal muscle, which provides a potential therapeutic target to treat myopathies.
2 Long Noncoding RNAs and Autophagy
2.1 Long Non-coding RNAs (lncRNAs)
lncRNAs are nonprotein-coding RNA transcripts that are longer than 200 nucleotides and have a similar structure to messenger RNA (mRNA) with a 5′-cap and poly (A) tail. In recent years, with the development of second-generation sequencing technology, lncRNA and its functions have gradually attracted the interest of researchers. Growing evidence shows that lncRNAs are broadly involved in the regulation of human disease such as cancer, autoimmunity, cardiovascular disease, and neural disease. However, the current research on lncRNA is still the tip of the iceberg, and there are still many unknowns awaiting further excavation.
2.1.1 The Biological Characteristics of lncRNAs
lncRNAs exhibit highly spatially and temporally restricted expression patterns in the process of tissue differentiation and development. lncRNAs can be transcribed from various genomic regions such as enhancers, promoters, introns, antisense coding, or intergenic regions of genes. Specifically, lncRNAs show a relatively lower expression level but much more tissue-specific pattern than protein-coding genes, suggesting the specific regulatory role of lncRNAs and potential specific targets for disease treatment. Based on the relative position of the lncRNA coding sequence and protein-coding gene, lncRNAs can be divided into five groups: intergenic, intronic, bidirectional, sense and antisense lncRNAs.
lncRNA expression is controlled by both transcriptional and epigenetic factors. Activating histone marks correlate well with the expression of lncRNA genes, which is similar to protein-coding genes. However, lncRNA genes were found to contain more methylation around the TSS (transcriptional start site) than protein-coding genes regardless of their expression status, suggesting that the epigenetic regulation of lncRNAs at the level of DNA methylation is markedly dissimilar from that of protein-coding genes. To date, multiple transcription factors have been found to transactivate the expression of lncRNAs, including Nanog, Sox2, Oct4, TP53, and ZNF143. Transcribed lncRNAs are further subject to posttranscriptional processing, such as 5′ capping, polyadenylation, alternative splicing, and RNA editing.
Although lncRNAs are present in both the nucleus and the cytoplasm, the fact that many lncRNAs are enriched in the nucleus highlights a potential role for lncRNAs as epigenetic regulators within the nucleus. The biophysical analysis showed that lncRNAs can be folded into a functional secondary structure that binds to DNA/RNA or protein and participates in physiological and pathological processes. Multiple lines of evidence demonstrate that lncRNAs exert their functions at the level of epigenetics via interactions with RNA-binding proteins (RBPs) at specific DNA regions, such as promoters or enhancers. Nevertheless, the role of lncRNAs present in the cytoplasm cannot be ignored. ceRNA (competing endogenous RNA) is the common mechanism for lncRNAs present in the cytoplasm to be involved in physiological activities. When thrombus occurs, WTAPP1 lncRNA inhibits the regulation of miRNA on the downstream target gene MMP-1 by adsorbing miR-3 and miR-120-5p, resulting in the activation of the PI3K/AKT/mTOR pathway and the regulation of endothelial progenitor cell autophagy.
In addition, some lncRNAs have highly conserved sequences and structures among different species and may be involved in the conservation of biological functions between different species. The fact that nonconserved RNAs are present in specific species and tissues highlights their involvement in the species-specific regulation of biological activities.
2.1.2 The Biological Functions of lncRNAs
In contrast to the intensively studied miRNAs, lncRNAs are larger and thus have a complex secondary structure. The complicated structure endows lncRNAs with the ability to bind protein, RNA, and DNA and thus with several regulatory capacities. In addition to these transcriptional and epigenetic regulatory activities, lncRNAs have been found to be important players in posttranscriptional regulation, such as functioning as mRNA editors, mRNA splicing regulators, and small ncRNA reservoirs. Breakthroughs over the past few years have revealed numerous examples of lncRNAs that regulate the progression from DNA to RNA to protein. Some recent excellent reviews have summarized the mechanisms by which lncRNA regulates chromatin structure and transcriptional control.
lncRNAs can regulate gene expression at the epigenetic level, including dose compensation, chromatin modification, and genomic imprinting. The most typical role of lncRNAs involved in dose compensation is participation in the balance of the expression of X-linked genes. The Xist lncRNA is transcribed from the XIST gene on an X chromosome. When an X chromosome is inactivated, the expression of the originally suppressed lncRNA Xist is upregulated (Yan et al. 2018). Chromatin modification refers to the process of adding and removing chemical groups to components of chromatin, such as DNA and histones. As a molecular scaffold, the lncRNA HOTAIR can bind to the histone modification complex to promote histone H3K4 demethylation and regulate the expression of target genes. Genomic imprinting, also known as genetic imprinting, is the biological process of labeling the information of its parental origin on a gene or genome by certain modifications. Gene expression levels and whether a gene is silent depend on whether the gene came from the male or female parent and the chromosome on which it is located The lncRNA H19 is one of the earliest discovered lncRNAs and is also an imprinted gene located on human chromosome 11p15, which is important for allele-specific expression on the imprinted gene cluster.
lncRNAs can regulate gene expression at the transcriptional level. Transcriptional regulation is an important part of eukaryotic gene expression and the most important method to regulate gene expression. lncRNAs are also regulated in a variety of ways at the transcription level such as by competing for transcription factors or recruiting protein complexes to influence gene expression. The lncRNA MUF is highly expressed in liver cancer tissues and activates the Wnt/β-catenin signaling pathway by binding to annexin A2 (ANXA2), leading to epithelial-mesenchymal transformation. However, MUF lncRNA can also act as an endogenous sponge of miR-34a to remove the miR-34a-mediated inhibition of the downstream target gene Snail1 and promote epithelial-mesenchymal transformation (Yan et al. 2017).
lncRNAs can regulate gene expression at the posttranscriptional level and mainly have three functions: splicing, translation, and degradation of mRNA. After transcription, mRNA needs further processing to become mature. Precursor mRNA (pre-mRNA) is the original transcription product including exon and intron sequences and noncoding sequences of a gene. Therefore, RNA exon splicing and intron removal are required for posttranscriptional processing. The lncRNA MALAT1 can regulate the distribution and activity of serine/arginine (SR) splicing factors leading to variable splicing of pre-mRNA. Further analysis found that the lncRNA MALAT1 regulated alternative splicing via SR protein phosphorylation, including the SRSF1, SRSF2, and SRSF3 proteins. Eukaryotic initiation factor 4A (eIF4A) is an RNA helicase. Bc1 lncRNA activates its ATPase to mediate the translation of eIF4A by blocking the double-stranded cleavage activity of eIF4A. lncRNA can be used as a miRNA sponge to block the binding of miRNAs to mRNA 3′-UTRs and affect the degradation of mRNA. Moreover, lncRNAs regulate mRNA stability through the nonsense-mediated mRNA degradation (NMD) pathway.
2.2 Regulation of Autophagy by lncRNAs
2.2.1 lncRNAs Related to Autophagic Initiation
As described in Chaps. 3 and 4 of this book, AMPK and mTORC1 promote autophagic initiation to maintain cellular homeostasis under conditions of hypoxia, ischemia, and malnutrition. Recent studies have found that many lncRNAs regulate autophagy by directly or indirectly affecting the AMPK and mTORC1 molecules. The downregulation of H9 lncRNA in diabetic rats relieves the transcriptional repression of DIRAS3, resulting in the inactivation of the PI3K/AKT/mTOR pathway and autophagic activation. Additionally, the downregulation of H9 lncRNA can increase Beclin 1 and ATG7 expression, which indicates the role of H9 lncRNA in mediating autophagy. In contrast, the exogenous overexpression of H19 lncRNA induces autophagic cell death in cerebral ischemia and reperfusion (I/R) injury. H19 lncRNA induces autophagy by inhibiting DUSP5 expression, a mitogen-activated protein kinase phosphatase. The downregulation of DUSP5 leads to the phosphorylation of ERK1/2, which induces autophagic initiation. The opposite effects of H19 lncRNA on autophagy in diabetic cardiomyopathy and cerebral I/R injury indicate that conditional gene interference targeting H19 lncRNA may be an efficient therapeutic approach to different pathological processes (Fig. 10.2).
lncRNAs are good mediators of AMPK. AMPK promotes its activation by binding to NBR2 lncRNA. Intriguingly, the expression of NBR2 lncRNA can also be induced by the increased activation of AMPK under energy stress. Healthy tissue depends on the combination of AMPK and lncRNA NBR2 to resist energy stress and tumors. NBR2 lncRNA expression is reduced in human cancers, and loss of its function is correlated with poor prognosis for cancer patients. lncRNAs commonly bind proteins to mediate the activity of proteins and participate in physiological functions. However, identifying the mechanisms of this effect is still difficult: does it occur through changing the conformation of the protein or by altering its affinity for other regulatory factors?
In addition to H9 lncRNA and NBR2 lncRNA, many lncRNAs are involved in autophagic initiation such as AD5-AlncRNA, PTENP1 lncRNA, and MEG3 lncRNA. Among them, AD5-A lncRNA is particularly interesting because it is an artificial lncRNA synthesized in vitro. miR-21, miR-216, and miR-27 are key molecules leading to sorafenib resistance, which is a new type of multitargeted oral drug for clinical tumor treatment. The overexpression of Ad5-AlncRNA can sequester these miRNAs from binding to the 3′-UTR of the target mRNA, repressing AKT/mTOR activity, promoting autophagic activation and subsequently reversing sorafenib resistance. Therefore, synthetic lncRNA is similar to the endogenous lncRNA and can regulate autophagy (Tang et al. 2016).
2.2.2 lncRNAs Related to Phagophore Nucleation
Autophagosomes are the core elements of autophagy. The rough endoplasmic reticulum or Golgi membrane encapsulating broken organelles or abnormally folded proteins in the cytoplasm can turn into phospholipid bilayer vesicles. Phagophore nucleation is the first step for cells to recruit proteins and for lipids to form autophagosome membranes. Mammal phagophore nucleation is mainly regulated by the class III PI3K complex, which mainly comprises Vps34, Vps15, Beclin 1, and ATG14 to generate phosphatidylinositol 3-phosphate (PI3P). PI3P recruits double FYVE-containing protein 1 (DFCP1) and other ATG proteins to promote the formation of the omegasome. Previous studies have confirmed that lncRNAs regulate Beclin 1 and affect vesicle nucleation. The lncRNA ROR reversed gemcitabine and tamoxifen resistance in breast cancer by upregulating Beclin 1 expression. Respiratory exposure to a PM2.5 environment-induced reactive oxygen species (ROS). A high level of ROS caused the upregulation of the lncRNA loc146880, which downregulated Beclin 1 mRNA levels, leading to autophagy and lung cell migration in the lungs. Although the ROR and loc146880 lncRNAs are known to mediate vesicle nucleation by Beclin 1, the relationship between other lncRNAs and Beclin 1 or the Vsp family needs to be further studied.
2.2.3 lncRNAs Related to Autophagosome Elongation
The two unique ubiquitin-like conjugation systems have crucial roles in the elongation and closure of the isolated membrane. Driven by ATG7 (E1-like enzyme) and ATG10 (E2-like enzyme), ATG12 conjugates to ATG5 and then interacts with ATG16 (mammal ortholog is ATG16L) to form the ATG12-ATG5-ATG16 complex. Subsequently, the ATG12-ATG5-ATG16 complex, ATG7 and ATG3 (E2-like enzyme) jointly transform LC3 from its cytosolic soluble isoform (LC3-I) to its membrane-anchored isoform (LC3-II).
Many proteins are involved in the autophagy elongation process, which also involves many lncRNAs. A study revealed that the lncRNA TGFB2 can be upregulated by vascular endothelial cell (VEC) inflammation and functions as a sponge for miR-3960, miR-4488, and miR-4459, thereby increasing the expression levels of their targets such as ATG13, ceramide synthase 1 (CERS1), and La ribonucleoprotein domain family member 1 (LARP1). In addition, overexpression of the lncRNA TGFB2 increases ATG3, ATG7, and P62 expression, probably through upregulating LARP1 by sponging miR-4459, an RNA-binding protein related to transcript stability and mRNA translation. Moreover, when lncRNA TGFB2 prevents the miR-3960-mediated repression of CERS1, the production of C18-ceramide is increased to induce mitophagy by directly interacting with LC3-II-containing autophagolysosomes upon Drp1-dependent mitochondrial fission. Given that autophagy and inflammation have an intricate relationship, intervening in the expression of the TGFB2 lncRNA could be a possible treatment strategy for infectious and autoimmune diseases.
The lncRNA GAS5 has been reported to inhibit autophagy and enhance cisplatin sensitivity in NSCLC cells. In contrast to its deletion in several species of tumors, GAS5 lncRNA is upregulated in osteoarthritis (OA), repressing autophagy and stimulating the apoptosis of OA chondrocytes, which is a key determinant responsible for cartilage degradation and thus OA pathogenesis. The increased expression of GAS5 lncRNA in OA represses autophagy possibly through downregulating Beclin 1, ATG3, ATG5, ATG7, ATG12, and LC3B expression. GAS5 lncRNA-mediated repression of autophagy is beneficial for enhancing drug sensitivity, but it also results in the occurrence of OA. Therefore, different models of interference with GAS5 may be essential for distinct therapeutic purposes.
The lncRNA HNF1A-AS1 can sequester miR-30b from binding to its target ATG5 and thereby provoke autophagy in HCC. In addition, Beclin 1 and ATG12 were defined as targets of miR-30b in a previous study. The TGFB2, GAS5, and HNF1A-AS1 lncRNAs promote autophagic progression by regulating key proteins in the process of autophagosome elongation.
2.2.4 lncRNAs Related to Autolysosome Fusion
The final step of autophagic flux is the fusion of the autophagosome and the lysosome to form an autolysosome, where the autophagic cargo is degraded. The core molecules in this stage include the Rab-SNARE system and the lysosome membrane proteins LAMP1 and LAMP2. In addition, adaptor proteins are necessary to link endocytic and autophagic pathways to the lysosome. Pleckstrin homology domain-containing protein family M member 1 (Plekhm1) is one of these adaptor proteins that contain an LC3-interacting region, which mediates the fusion of endosomes and autophagosomes with lysosomes. The lncRNA Chast can suppress autophagy by downregulating Plekhm1 and possibly ATG5 expression to induce cardiomyocyte hypertrophy. The lncRNA MALAT1 represses autolysosome fusion via the downregulation of LAMP1 and LAMP2, leading to autophagic inhibition (Yang et al. 2017).
2.3 lncRNAs Regulated by Autophagy
Growing evidence suggests that lncRNAs play key roles in autophagy and that it is particularly important to investigate whether lncRNAs can be regulated by autophagy. Previous studies have shown that autophagy can degrade several types of RNAs and associated ribonucleoprotein complexes, which implies that lncRNAs may be degraded in autolysosomes (Frankel et al. 2017). The lncRNA PVT1 is the sole lncRNA reported to be regulated by autophagy thus far. lncRNA PVT1 is upregulated in diabetes, and autophagic repression decreases its transcriptional level. PVT1 lncRNA is probably not degraded by autophagy, as it is downregulated when autophagy is repressed. Thus, further extensive investigations are needed to demonstrate what determinants participate in this process.
3 Circular RNAs and Autophagy
3.1 Circular RNAs
Protein-coding genes and their transcripts are the most studied sequences in eukaryotic cells. However, protein-coding genes and RNAs comprise only a small fraction of genomes and transcriptomes. Indeed, the vast majority of sequences in the human genome do not encode proteins, and non-coding RNAs account for almost 95% of the total RNA transcribed from eukaryotic genomes. circRNAs were first identified in 1976 in an electron microscopy-based study of RNA viruses and have since been found in humans, mice, rats, fungi, and other organisms. Unlike linear RNA molecules, circRNAs are closed circular molecules with a covalently closed-loop structure that lack 5′-3′ polarity or a polyadenylated tail. As a large proportion of the non-coding RNA family, circRNAs have drawn intense interest over the last few years. Non-coding RNAs have been increasingly shown to function in gene regulation and contribute to the development of many human disorders.
3.1.1 circRNA Biogenesis
Although circRNAs are derived from precursor mRNAs (pre-mRNAs), their biogenesis remains elusive. circRNAs differ from other RNAs in their remarkable continuous closed-loop structure, which is covalently linked by free 3′ and 5′ ends. This closed-loop structure is also called a “back-splicing” structure.
circRNAs can mainly be classified into three categories: exonic circular RNAs (ecircRNAs), circular intronic RNAs (ciRNAs), and exon-intron circular RNAs (EIciRNAs). Several models have been proposed to explain the possible formation of ecircRNAs, such as lariat-driven circularization and intron pairing-driven circularization models. In a lariat-driven circularization event, RNA is folded during pre-RNA synthesis, making multiple exons close to each other and “jump” to form a ring-shaped RNA intermediate, and then, circRNA is generated. The “intron pair-driven cyclization” model suggests that reverse complementary ALU sequences exist in introns downstream of exons, and their pairing mediates reverse splicing to form circRNA.
The synthesis of circRNA is regulated by many factors. Many RNA-binding proteins (RBPs) are involved in the biosynthesis of circRNA, such as Muscle blind protein (MBL protein), Quaking protein (QKI protein), and adenosine to inosine acting RNA enzyme 1 (ADAR1), which can inhibit circRNA synthesis (Han et al. 2018a).
3.1.2 circRNA Function
Many of the functions of circRNAs have been elucidated over the past few years. For example, circRNAs can act as gene expression regulators via different regulatory modes, regulate transcription and alternative splicing, interact with RBPs, serve as miRNA sponges and function in translation.
3.1.2.1 circRNAs Serve as miRNA Sponges or Competing Endogenous RNAs
circRNAs are mainly distributed in the cytoplasm, which has increased interest in the role of circRNAs in posttranscriptional regulation. In 2013, it was confirmed for the first time that circRNAs may be used as a sponge or competitive endogenous RNA to regulate the expression of target genes of microRNAs. The competitive endogenous RNA (ceRNA) hypothesis holds that there are RNA-binding sites on microRNAs, while there are also RNA-binding sites on circRNAs. By competing with microRNAs, circRNAs can indirectly regulate the translation of RNA. Earlier, the ceRNA hypothesis mainly involved RNAs, including RNA, transcribed pseudogenes and lncRNA. The ceRNA hypothesis also includes circRNA. For example, ciRS-7/CDR1 circRNA contains more than 70 conservative binding sites of microRNA-7. CiRS-7/CDR1 circRNA can regulate the activity of microRNA-7 by binding with microRNA-7 and affect the expression of target genes of microRNA-7 (Qu et al. 2018).
3.1.2.2 circRNA Interactions with RBPs
Some RNA can bind to RNA-binding proteins or act as storage/isolation proteins, thus participating in protein substructure localization. For example, circMbl, derived from muscleblind (MBL/MBNL1), contains multiple MBL binding sites. When the MBL protein is overexpressed, circMbl can absorb excess protein. CircMbl can regulate the MBL protein level in this way.
3.1.2.3 circRNAs Regulate Transcription or Splicing
circRNAs can influence their parent genes through cis or trans actions. circRNAs can directly participate in gene expression regulation by regulating linear RNA transcription and variable splicing. For example, in the exon hopping model, precursor RNAs can also be alternatively spliced to form mature linear RNA while cyclizing to form ecircRNAs. Thus, the synthesis of ecircRNAs will competitively hinder the synthesis of homologous linear RNA, but it may also increase the expression of the circRNA itself or its corresponding linear RNA. In addition, the cyclization and nonlinear splicing of pre-RNA containing translation initiation sites indicates that the formation of circRNA reduces the formation of RNA and the translation of downstream proteins. This effect is called an “RNA trap” (Meng et al. 2017).
3.1.2.4 Role in Translation
In addition to these functions, circRNA has been found to have translational functions. Because circRNA lacks some common features of coding RNA such as the m7GPPPN cap structure at the 5′ end and the polyadenylation tail structure at the 3′ end, it is considered a non-coding RNA and cannot encode the protein. There are many internal ribosome entry sites (RESs) in circRNAs suggesting that circRNAs may have the ability to encode proteins. Circ-ZNF609 was the first circRNA found to directly translate proteins that participate in muscle development (Legnini et al. 2017).
3.2 circRNAs and Autophagy Regulation
Depending on their unique 3-dimensional covalent structure, circRNAs effectively capture or sequester RNAs or proteins and release them in subcellular locations to mediate autophagy regulation. circRNA may be cleaved by autophagic degradation and regulate autophagy accordingly.
Astrocytes, the most abundant cell type in the central nervous system (CNS), play critical roles in regulating and maintaining CNS homeostasis in normal physiological situations. In pathological conditions, astrocytes become activated and are characterized by abnormal morphology with reactive astrogliosis. Astrocyte activation plays a detrimental role in various neurological pathologies, including stroke, Parkinson’s disease, Alzheimer’s disease, and drug abuse. Astrocyte activation is associated with neuronal damage, as there is a close relationship between methamphetamine-induced degeneration of dopaminergic neurons in the striatum and concomitant reactive astrogliosis. circHIPK2 inhibits the activity of endogenous microRNA-124 by blocking the activity of microRNA-124, which leads to the upregulation of sigma-1 receptor expression and finally affects the activation of astrocytes by co-regulating autophagy and endoplasmic reticulum stress (Huang et al. 2017). In stroke, circHECTD1 acts as an endogenous microRNA-142 sponge, inhibiting the activity of microRNA-142, thereby inhibiting the expression of TCDD-induced ADP-ribose polymerase (TIPARP) and astrocyte autophagy (Han et al. 2018b). In addition, circRNA participates in the regulation of peripheral nerve injury through autophagy. The expression of circRNA-2837 was significantly downregulated in a rat sciatic nerve injury model. The downregulation of circRNA-2837 alleviated sciatic nerve injury by inducing autophagy. Mechanistically, the knockdown of circRNA-2837 may protect neurons against neurological injury by acting as a sponge for members of the miR-34 family (Zhou et al. 2018).
4 Other Non-coding RNAs and Autophagy
Non-coding RNAs (ncRNA) include many types of RNAs that do not encode proteins. Based on their function, length and structure, ncRNAs are divided into microRNAs (miRNAs or miRs; 19–23 bp), long noncoding RNAs (lnRNAs; >200 bp and linear), circular RNAs (circRNAs; >200 bp and circular), transfer RNAs (tRNA; 74–95 bp), ribosomal RNAs (rRNAs; 121–5000 bp), small nuclear RNAs (snRNAs; 100–300 bp), small nucleolar RNAs (snoRNAs; 100–300 bp), guide RNAs (gRNAs; 55–70 bp), piwi-interacting RNAs (piRNAs; 24–30 bp), and small interfering RNAs (siRNAs; 21–25 bp).
Over the past few decades, increasing evidence has indicated that ncRNAs, ranging from miRNAs to lncRNAs and even circRNAs, mediate the transcriptional and posttranscriptional regulation of autophagy-related genes by participating in autophagy regulatory networks. This book provides a detailed introduction to the regulatory roles of miRNAs, lncRNAs, and circRNAs in autophagy. Because the relationship between other ncRNAs and autophagy is less studied, this section will briefly introduce recent research progress.
4.1 Transfer RNAs and Autophagy
4.1.1 Transfer RNA
In 1953, Watson and Crick first announced the three-dimensional structure of the DNA double helix and changed people’s understanding of life. Crick thus proposed a central rule to explain the process of genetic information transmission: genetic information is transmitted exclusively to RNA through DNA and then from RNA to protein in the process of transcription and translation. However, in his hypothesis, a key “conjugator”, which is a key molecule that can specifically connect nucleic acid and protein sequences, was not found. Shortly afterward, this key “conjugator” molecule, named transfer RNA (tRNA), was discovered along with an important family of enzymes, amino acid-tRNA synthase, which specifically catalyzes the coupling of amino acids with their transporting RNA carriers. tRNAs are essential adapter molecules in translation that carry specific amino acids and, by complimentary codon–anticodon base pairing, ensure the incorporation of the correct amino acid sequence in the nascent polypeptide.
4.1.2 The Relationship Between tRNAs and Autophagy
Current studies on the relationship between tRNA and autophagy are limited. Recent studies have shown that the interaction between tRNA and autophagy plays a key role in the stability and translation of RNA, the regulation of translation efficiency and aging.
The cytoplasmic localization, stability, and translation of messenger RNA (messenger RNA) are influenced by its binding proteins. These binding proteins mainly include cold shock domain (CSD) structural proteins, nuclear heterogeneous ribonucleoproteins and serine/arginine-rich proteins. Recent studies have shown that the most abundant RNA-binding proteins in the human body, including the CSD-containing Y-box-binding protein 1 (YBX1), the closely related YBX3 protein and other RNA-binding proteins, such as SRSF1, SRSF2, SRSF3, and hnRNP A1 and H, are specifically and closely related to mitochondrial tRNAs (mt tRNAs). Although the function of mt tRNA in the cytoplasm is not yet clear, the dynamic interaction between mt tRNA and RNA-binding proteins may affect the stability or translation of cytoplasmic RNA. Y-box-binding proteins play a wide range of biological roles in the process of nuclear DNA replication, DNA repair, and transcription, as well as in the process of mRNA processing through their interaction with nucleic acids. YBX1 and YBX3, together with many hnRNP proteins and serine/arginine-rich proteins, can bind to newly synthesized precursor messengers, thereby regulating the splicing and polyadenylation of precursor messengers, promoting the transfer of messengers to the cytoplasm and controlling the translation, stabilization, and localization of cytoplasmic messengers. Recent studies have shown that tRNA and autophagy are involved in these processes. Autophagy induces the release of mt RNA into the cytoplasm and promotes the abovementioned processes. Although the biological function of mt tRNAs in the cytoplasm is still uncertain, the dynamic interaction between mt tRNAs and RNA-binding proteins plays an important role in the stability and translation of RNA (Jady et al. 2018).
Aging is a complex biological process that all organisms must undergo, but its exact molecular mechanism is not yet clear. Because of its short life span, conservative aging pathway, and easy genetic and environmental control, single-cell eukaryotic budding yeast has become the main aging research model. Furthermore, budding yeast is the only system in which the number of times a cell divides, also known as replicative lifespan, can be accurately determined. Protein synthesis is a key factor affecting cell growth, replication, and survival, which is widely regulated by external and internal factors. Translation efficiency is closely related to cell life span. In old cells of budding yeast, the phosphorylation level of eIF2a mediated by stress kinase Gcn2 increased, which reduced the overall translation efficiency of yeast cells, while the expression of the downstream transcription activator Gcn4 did not decrease significantly. Overexpression of tRNA in young yeast cells can activate the stress kinase Gcn2 and prolong cell life through a Gcn4-dependent pathway. In addition, overexpression of Gcn4 can prolong cell life in an autophagy-dependent manner without altering the overall translation efficiency; that is, autophagy-mediated by overexpression of tRNA can prolong cell life (Hu et al. 2018).
4.2 Ribosomal RNAs and Autophagy
4.2.1 Ribosomal RNA
Ribosomal RNA (rRNA) is the most abundant type of RNA, accounting for approximately 82% of the total RNA. It binds to proteins to form ribosomes, whose function is to synthesize amino acids into peptide chains under the guidance of RNA. When rRNA exists alone, it does not perform its function. rRNA binds to various proteins to form ribosomes and serves as an ‘assembly machine’ for protein biosynthesis.
4.2.2 The Relationship Between rRNAs and Autophagy
There are few studies on the relationship between rRNA and autophagy. A few studies have shown that autophagy-dependent rRNA degradation plays an important role in maintaining nucleotide stability and animal development. During autophagy, protein aggregates and organelles are transferred to lysosomes for degradation, and the degradation products are reused by cells to maintain intracellular stability. Many cell components, such as organelles, can be used as substrates for autophagy. Ribosomes have long been observed in autophagy by electron microscopy as markers of cytoplasmic degradation. Except for selective autophagy, mature ribosomes in yeasts can also be selectively phagocytosed and scavenged under long-term nitrogen deficiency. In this process, ribosome subunits are likely to be independent degradation targets that involve ubiquitination and deubiquitination. Ribosomes contain approximately 50% of cellular proteins and 80% of total RNA. In the process of autophagy-mediated degradation, these proteins and RNA molecules are degraded in lysosomes, and their degradation products can be used as the main source of amino acids and nucleotides in nutrient deficiency. Therefore, autophagic degradation of ribosomes is essential for the survival of yeast cells under nutritional deficiency. RNST-2, a ribonucleic acid endonuclease of the T2 family of nematodes, is a key enzyme for the degradation of rRNA in lysosomes. Recent studies have shown that RNST-2 deficiency can induce the aggregation of rRNA and protein in lysosomes, which indicates that RNST-2 mediates the autophagic degradation of rRNA in lysosomes. Lack of RNST-2 can cause developmental defects of embryos and larvae of nematodes and shorten the lifespan of nematodes. In addition, the absence of RNST-2 and pyrimidine nucleotides at the same time would lead to the death of nematode embryos. Supplementation of uridine or cytidine could inhibit the death of embryos (Liu et al. 2018b).
In addition, the process of anabolism and catabolism of intracellular substances is strictly regulated by the energy supply of cells. Energy stress can inhibit the biosynthesis of rRNA and induce autophagy. However, the relationship between rRNA biosynthesis and autophagy remains unclear. Nucleoprotein NAT10 plays a key role in the biosynthesis of rRNA and the transformation between autophagy. Under physiological conditions, NAT10 acetylation activates rRNA biosynthesis and inhibits autophagy. In conditions with an adequate energy supply, NAT10 binds to the autophagy regulator Che-1 K288 and acetylates it, inhibiting the transcriptional activation of the Che-1-mediated downstream genes Redd 1 and Deptor. Under energy stress, NAT10 deacetylates under the action of Sirt 1, inhibiting the biosynthesis of rRNA activated by NAT10. In addition, deacetylation of NAT10 renders its inhibition of Che-1-mediated autophagy ineffective. These results suggest that the acetylation of NAT10 is of great significance for the conversion of metabolic reactions to synthesis and catabolism and provide a new mechanism for the regulation of rRNA biosynthesis and autophagy by nucleoproteins (Liu et al. 2018a).
4.3 Small Nuclear RNAs and Autophagy
4.3.1 Small Nuclear RNA
Small nuclear RNA, also known as intranuclear small RNA, is involved in the processing of RNA in the nucleus of eukaryotic cells. snRNA binds with many proteins to form small ribonucleoproteins, which participate in the splicing of pro-messenger RNA and process it into mRNA.
4.3.2 The Relationship Between snRNAs and Autophagy
In some aging-related diseases, mutations in pathogenic genes not only cause the occurrence of positive symptoms but also change the monoclonal conformation of the proteins or metabolites encoded by them, which leads to the increase and accumulation of potentially toxic proteins with an anti-hydrolysis ability. The lysosomal system is the most important protein hydrolysis system in mammals and is responsible for the treatment of abnormal proteins. In the lysosomal system, abnormal proteins are decomposed by two main pathways: (1) the autophagic lysosome system is mainly responsible for the degradation of proteins with a long half-life and is also the only way to degrade organelles and protein aggregates or inclusions; (2) the transfer of membrane components and extracellular substances to lysosomes under specific targeted signals. Many chemicals can regulate the level of autophagy at different stages of autophagy. Rapamycin can increase autophagy by inhibiting mTOR, and 3-methyladenine (3-MA) can inhibit autophagic formation by blocking PI3K. Genetic analysis of neurodegenerative diseases shows that there is a close relationship between neurodegenerative diseases and lysosomal network function. During the development of AD, a series of pathological changes take place in the network of the neuronal lysosome system, including increased lysosome biosynthesis and inhibition of lysosome ingestion, which ultimately lead to the destruction of the lysosome clearance mechanism. In AD patients, there are many autophagic vacuoles (AVs) and autophagosomes in the brain. This suggests that autophagy is impaired in AD patients because AVs are rare in the brains of healthy people. In addition, AVs mainly accumulate in amyloid precursor protein (APP), and Aβ peptides are produced by APP during autophagy. In normal cells, these Aβ peptides are degraded in lysosomes immediately after they are produced. Presenilin 1 (PS1) is a component of gamma-secretase and an essential part of lysosomal acidification. Recent studies have shown that PS1 can increase the expression of U1 snRNA and cause adverse changes in the expression of APP, the production of Aβ peptides and cell apoptosis. Overexpression of U1 snRNA can significantly activate autophagy, leading to dysfunction of the autophagic lysosomal system and exacerbating AD symptoms (Cheng et al. 2018).
5 Summary and Prospect
In the past decade, the study of ncRNA-mediated autophagic regulation has been a research hotspot in the field of biology. As research progresses, many miRNAs that can regulate autophagy have been discovered, and the regulatory mechanisms of miRNAs on autophagy are becoming increasingly clear. miRNAs regulate the autophagic process mainly at the posttranscriptional level by affecting the expression of autophagy-related genes. However, the miRNAs currently involved in the regulation of autophagy are likely to only be the tip of the iceberg, and more autophagy-related miRNAs will be discovered in the near future. With the increasing development of research, the regulatory mechanism of miRNA on autophagy will likely become clearer, which will provide a new strategy for the clinical treatment of various diseases related to autophagy. Current studies indicate that most lncRNAs regulate autophagy-related protein expression by a ceRNA mechanism. In addition to chromatin and histone remodeling, transcriptional regulation and protein–protein interactions, lncRNAs have more complex functions associated with autophagic regulation to be elucidated. Due to the high spatial and temporal specificity and tissue specificity, lncRNAs may be used as biomarkers for autophagy-related diseases and may be used to develop potential therapeutic measures. There are still few studies on circRNAs and autophagy. However, given the unique biological characteristics of circRNAs, circRNAs also have the potential to become an autophagy-related research tool.
Abbreviations
- 3-MA:
-
3-methyladenine
- ADAR1:
-
Adenosine to inosine acting on RNA enzyme 1
- ANXA2:
-
Annexin A2
- APP:
-
Amyloid precursor protein
- ceRNA:
-
Competitive endogenous RNA
- circRNA:
-
Circular RNA
- ciRNAs:
-
Circular intronic RNAs
- CSD:
-
Cold shock domain
- DFCP1:
-
Double FYVE-containing protein 1
- EcircRNAs:
-
Exonic circRNAs
- EIcircRNAs:
-
Exonic-intronic circRNAs
- eIF4A:
-
Eukaryotic initiation factor 4A
- gRNA:
-
Guide RNA
- lncRNA:
-
Long non-coding RNA
- miRNA:
-
Micro RNA
- mRNA:
-
Message RNA
- mt tRNAs:
-
Mitochondrial tRNAs
- ncRNA:
-
Noncoding RNA
- NMD:
-
Nonsense-mediated mRNA decay
- PI3P:
-
Phosphatidylinositol 3-phosphate
- piRNA:
-
Piwi-interacting
- Plekhm1:
-
Pleckstrin homology domain-containing protein family M member 1
- pre-miRNA:
-
Precursormicro RNA
- pri-mRNA:
-
Primary micro RNA
- PS1:
-
Presenilin 1
- RBP:
-
RNA-binding protein
- RES:
-
Ribsome entrysite
- RISC:
-
RNA-induced silencing complex
- ROS:
-
Reactive oxygen species
- rRNA:
-
Ribosomal RNA
- siRNA:
-
Small interfering
- snoRNA:
-
Small nucleolar RNA
- snRNA:
-
Small nuclear RNA
- SR:
-
Serine/argine
- TIPARP:
-
TCDD inducible poly[ADP-ribose] polymerase
- tRNA:
-
Transfer RNA
- ULK:
-
Unc-51-like kinase
- UTR:
-
Untranslation region
- Vps:
-
Vesicular protein sorting
References
Cheng Z, Du ZQ, Zhai BH et al (2018) U1 small nuclear RNA overexpression implicates autophagic-lysosomal system associated with AD. Neurosci Res 136:48–55
Frankel LB, Lubas M, Lund AH (2017) Emerging connections between RNA and autophagy. Autophagy 13:3–23
Han B, Chao J, Yao HH (2018a) Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther 187:31–44
Han B, Zhang Y, Zhang YH et al (2018b) Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy 14:1164–1184
Hu Z, Xia B, Postnikoff SDL et al (2018) Ssd1 and Gcn2 suppress global translation efficiency in replicatively aged yeast while their activation extends lifespan. Elife 7
Huang RR, Zhang Y, Han B et al (2017) Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy 13:1722–1741
Jady BE, Ketele A, Kiss T (2018) Dynamic association of human mRNP proteins with mitochondrial tRNAs in the cytosol. RNA 24:1706–1720
Jurong Y, Dapeng C, Yani H et al (2013) MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age 35:11–22
Legnini I, Timoteo GD, Rossi F et al (2017) Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66:22–37
Liu XF, Cai SY, Zhang CF et al (2018a) Deacetylation of NAT10 by Sirt1 promotes the transition from rRNA biogenesis to autophagy upon energy stress. Nucleic Acids Res 46:9601–9616
Liu YB, Zou W, Yang PG et al (2018b) Autophagy-dependent ribosomal RNA degradation is essential for maintaining nucleotide homeostasis during C. elegans development. Elife 7
Meng XW, Li X, Zhang PJ et al (2017) Circular RNA: an emerging key player in RNA world. Brief Bioinform 18:547–557
Ouimet M, Koster S, Sakowski E et al (2016) Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat Immunol 17:677–686
Qu SB, Liu ZC, Yang XS et al (2018) The emerging functions and roles of circular RNAs in cancer. Cancer Lett 414:301–309
Tang SY, Tan G, Jiang X et al (2016) An artificial lncRNA targeting multiple miRNAs overcomes sorafenib resistance in hepatocellular carcinoma cells. Oncotarget 7:73257–73269
Wu H, Wang F, Hu S et al (2012) MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell Signal 24:2179–2186
Yan XL, Zhang DD, Wu W et al (2017) Mesenchymal stem cells promote hepatocarcinogenesis via lncRNA-MUF interaction with ANXA2 and miR-34a. Can Res 77:6704–6716
Yan F, Wang X, Zeng Y (2018) 3D genomic regulation of lcnRNA and Xist in X chromosome. In: Seminars in cell & developmental biology, S1084952118301599
Yang LX, Wang HY, Shen Q et al (2017) Long non-coding RNAs involved in autophagy regulation. Cell Death Dis 8
Yang L, Han B, Zhang Y et al (2018) Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy 14:404–418
Zhang JA, Zhou BR, Xu Y et al (2016) MiR-23a-depressed autophagy is a participant in PUVA- and UVB-induced premature senescence. Oncotarget 7:37420–37435
Zhou ZB, Niu YL, Huang GX et al (2018) Silencing of circRNA.2837 plays a protective role in sciatic nerve injury by sponging the miR-34 family via regulating neuronal autophagy. Mol Ther-Nucl Acids 12:718–729
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Science Press and Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Yao, H., Han, B., Zhang, Y., Shen, L., Huang, R. (2019). Non-coding RNAs and Autophagy. In: Qin, ZH. (eds) Autophagy: Biology and Diseases. Advances in Experimental Medicine and Biology, vol 1206. Springer, Singapore. https://doi.org/10.1007/978-981-15-0602-4_10
Download citation
DOI: https://doi.org/10.1007/978-981-15-0602-4_10
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-0601-7
Online ISBN: 978-981-15-0602-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)