Abstract
Circular DNAs are frequent genomic molecules, especially among the simplest life beings, whereas circular RNAs have been regarded as weird nucleic acids in biology. Now we know that eukaryotes are able to express circRNAs, mostly derived from backsplicing mechanisms, and playing different biological roles such as regulation of RNA splicing and transcription, among others. However, a second natural and highly efficient pathway for the expression in vivo of circRNAs has been recently reported, which allows the accumulation of abundant small (100–1000 nt) non-coding RNA circles through the participation of small self-cleaving RNAs or ribozymes called hammerhead ribozymes. These genome-encoded circRNAs with ribozymes seem to be a new family of small and nonautonomous retrotransposable elements of plants and animals (so-called retrozymes), which will offer functional clues to the biology and evolution of circular RNA molecules as well as new biotechnological tools in this emerging field.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Genomic circular DNAs are frequent macromolecules among simple organisms, from small prokaryotic plasmids to the larger genomes of many bacteriophages or viruses, bacteria, archaea and plastids. On the other hand, circular RNAs have been regarded as very rare nucleic acids in biology till very recently. Now we know that numerous life beings express stable circRNAs [1], and among them, it is noteworthy the recent discovery of a myriad of splicing-derived circRNAs in eukaryotes [2,3,4] with diverse functions in regulation of splicing [5] and transcription [6], small RNAs biology [7], RNA-mediated inheritance and epigenetics [8] and some others, as described in this book. However, it has been recently reported that eukaryotes have a second natural pathway that allows the expression in vivo of abundant circular RNAs [9, 10]. This alternative mechanism does not require any classical spliceosome reaction but the involvement of small self-cleaving RNAs or ribozymes called hammerhead ribozymes (HHRs) [11].
The finding of catalytic RNAs or ribozymes more than 30 years ago [12, 13] propelled the revolution in the RNA field and started with the uninterrupted discovery of the many different roles and capabilities of this macromolecule in biology. Moreover, the ground-breaking discovery of ribozymes strongly supported the hypothesis of the prebiotic RNA world [14], where RNAs carried out both informative (RNA genomes) and catalytic (ribozymes) roles. Somehow, it is thought that these primal RNA molecules would have evolved to present organisms based in DNA and proteins as the genetic material and catalytic machines, respectively [15,16,17]. Proofs supporting this hypothesis are the existence of RNA genomes among the simplest organisms (such as RNA viruses and viroids), as well as catalytic and regulatory ribo-functions among all living beings, where RNA itself is the final molecule in charge of the activity. A remarkable example of a catalytic RNA would be the central machine of life, the ribosome [18], which is the universal ribozyme that catalyses the peptide bond formation during protein synthesis in all known living entities. This fact allows to connect DNA and proteins through a catalytic RNA, which offers a solution to the chicken or the egg (or more precisely, the DNA or the protein) causality dilemma. Other key ribozymes and regulatory RNAs considered as ancient relics of the prebiotic RNA world would be the autocatalytic introns [12] and small ribozymes [19], the RNase P [13], the spliceosome [20], the riboswitches [21], most non-coding RNAs (such as those small RNA guides found in the CRISPR [22] and the RNAi [23] pathways) or even the circRNAs described in this book, which altogether confirm the extraordinary potential of any RNA molecule present in a living organism.
2 The Family of Small Self-Cleaving RNAs and the Singular Case of the Hammerhead Ribozyme
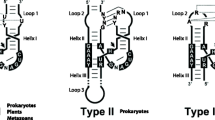
Among the simplest ribozymes so far described, it can be highlighted the enigmatic group of small (50–200 nt) self-cleaving RNAs, which all catalyse a sequence-specific intramolecular reaction of transesterification. This reaction starts by a nucleophilic attack of the 2′ oxygen to the adjacent 3′ phosphate, resulting in cleavage of the phosphodiester bond to form two RNA products with a 5′-hydroxyl and a 2′,3′-cyclic phosphate ends each (Fig. 5.1a). The family of small self-cleaving ribozymes is composed so far by nine different classes: hammerhead (HHR) [24, 25], hairpin (HPR) [26], human hepatitis-δ (HDV) [27], Varkud satellite (VS) [28], GlmS [29], twister [30], twister sister, hatchet and pistol [31] ribozymes. The HHR was the first discovered and one of the best known members of the family of small self-cleaving ribozymes. It is composed of a conserved catalytic core of 15 nucleotides surrounded by three double helixes (I to III), which adopt a γ-shaped fold where helix I interacts with helix II through tertiary interactions required for efficient in vivo activity (Fig. 5.1b) [32,33,34]. There are three possible circularly permuted topologies for the HHR, named type I, type II or type III, depending on the open-ended helix (Fig. 5.1c). The HHR were first found encoded in the small circRNA genomes of a group of infectious subviral agents of plants, such as viral RNA satellites and viroids [24, 25], where it catalyses a self-cleavage transesterification reaction required for the rolling-circle replication of these pathogens. Surprisingly, few other examples of HHR motifs were also found encoded in the genomes of some unrelated eukaryotes such as newts, trematodes or even some mammals, among others [35,36,37,38,39]. In 2010, different labs reported the widespread occurrence of HHR motifs in prokaryotic and eukaryotic genomes [40,41,42,43], including our own genome [44], which confirmed that the HHR was a ubiquitous catalytic RNA motif in all life kingdoms [45, 46]. Interestingly, the occurrence of genomic HHR motifs along the tree of life seems to follow a kind of structural or functional compartmentalization. This way, anyone of the three topologies of the HHR motif (types I, II and III, Fig. 5.1c) can be frequently detected in the genomes of prokaryotes and bacteriophages. However, metazoan genomes mostly show type-I HHR motifs, whereas plant genomes, as well as their subviral agents, almost exclusively show the presence of type-III HHR motifs.
RNA self-cleavage by the hammerhead ribozyme (a) Mechanism of internal transesterification in the RNA. The cleavage reaction starts with an attack of the 2′ hydroxyl to the 3′ phosphate, followed by a bipyramidal transition state. The cleavage products are a 2′,3′-cyclic phosphate at the 5′ RNA product and a 5′-hydroxyl at the 3′ RNA product. (b) Classic two (left)- and three (right)-dimensional diagrams of the hammerhead ribozyme motif. Black boxes indicate the highly conserved nucleotides (in white letters) at the catalytic core. (c) Representation of the three possible hammerhead ribozyme topologies (types I, II and III). Dotted and continuous lines refer to non-canonical and Watson-Crick base pairs, respectively. The three topologies have been reported in the genomes of bacteriophages and prokaryotes. Type-I hammerheads are mostly found in metazoan genomes, whereas typical type-III motifs are found in the plants and their infectious circRNAs (viroidal RNAs). N stands for any nucleotide, whereas R stands for purines (A or G), Y for pyrimidines (U or C) and H for either A, U or C
Other small self-cleaving RNA motifs have been also found widespread in DNA genomes, such as HDV [47] or twister ribozymes [30], which confirms that small catalytic RNAs would be more frequent than previously thought. Although the precise biological roles of all these genomic self-cleaving ribozymes are still under study, a direct connection with retrotransposons and other mobile genetic elements has been reported for most of them [10, 48,49,50,51].
3 Hammerhead Ribozymes in Plant Genomes Promote circRNA Expression: The Retrozymes
Two examples of type-III HHR motifs were originally reported in the genome of A. thaliana [35]. Numerous copies of this ribozyme were also detected in the genomes of diverse flowering plants [40]. In many instances, these HHRs have been found as tandem repeats of several motifs (usually two or three) separated by a few hundred base pairs. These observations have been recently extended in our lab, and we have reported the occurrence of hundreds of type-III HHRs in more than 40 plant species [10]. Comparative genomics revealed that sequences flanked by tandem HHR motifs sized from 600 to 1000 bp with almost no identity. However, these genomic repetitive elements show a similar topology: they are delimited by 4 bp target site duplications (TSDs), whereas HHRs are embedded in direct long terminal repeats (LTRs) of ~350 bp. LTRs delimit a central region (~300–700 bp), which begins with the primer binding site (PBS, corresponding to the tRNAMet) and finishes with the polypurine tract (PPT) sequences characteristic of LTR retrotransposons [52] (Fig. 5.2a). Altogether, these elements were classified as a new family of nonautonomous retrotransposons with hammerhead ribozymes (so-called retrozymes) similar to other nonautonomous retroelements of plants like TRIMs [53] and SMARTs [54]. Most likely, autonomous retrotransposons of the Ty3-Gypsy family would mobilize the small retrozymes based on the sequence similarities (PBS and 5′ and 3′ LTR ends) between both types of retroelements.
Genomic plant retrozymes (a) Schematic representation (top) of a full genomic retrozyme element of plants. Target side duplications (TSDs) delimiting the retrozyme are shown in grey boxes. Long terminal repeats (LTRs) are shown in black boxes. The positions of the primer binding site (PBS), the polypurine tract (PPT), the hammerhead ribozymes (HHR) and the typical sizes encompassed by the ribozymes are indicated. The resulting self-cleaved retrozyme RNA after transcription (middle) and circularization (bottom) is indicated. (b) An example of a northern blot analysis of RNA extracts (~30 μg each) from physic nut (Jatropha curcas) leaves, young seedlings and seeds. Samples were run on a 5% denaturing PAGE and were detected using a digoxigenin-labelled J. curcas retrozyme fragment as a probe, which revealed the presence of both circular and linear RNA forms in each plant tissue as indicated at the right. Ethidium bromide staining of the 5S rRNA is shown at the bottom as a loading control
Northern blot analysis and RT-PCR experiments of diverse somatic and reproductive tissues from several plant species, such as physic nut, strawberry, eucalyptus or citrus plants, revealed the presence of high levels of circular and linear RNAs (up to 1 ng per μg of total RNA) of the precise size encompassed by the HHR motifs (Fig. 5.2b), which strongly indicates an RNA self-processing activity by the ribozymes during in vivo transcription followed by RNA circularization. Although sequence identity between retrozymes from non-related plant species is very low, secondary structure predictions for these circRNAs show similar architecture and high stability (Fig. 5.3a). These structured circRNAs with type-III hammerhead ribozymes highly resemble those infectious circRNAs of plants, such as viral satellite RNAs and viroids (Fig. 5.3b and c), which indicates a clear evolutionary relationship between all of them [9].
Minimum free energy secondary structure predictions for (a) a retrozyme circRNA of Jatropha curcas (Entry KX273075.1), (b) the Nepovirus satellite RNA sTRSV (Entry M14879.1) and (c) the viroid CChMVd (Entry AJ878085.1). HHR sequences are shown in purple (positive polarity) and green (negative polarity). The corresponding structure of the HHRs motifs are shown under each circRNA structure, and dotted lines indicate predicted tertiary interactions between HHR loops based on previous models [60, 61]. Self-cleavage sites are indicated with arrowheads. Kissing-loop interactions described for CChMVd [62] are shown. Numbering for each circRNA starts at the self-cleavage site
4 Tandem Copies of Hammerhead Ribozymes in Metazoan Genomes
Previous bioinformatic searches in metazoan genomes have also revealed the widespread occurrence of the HHR motif in animals [10, 40, 45]. As observed in plants, these ribozymes are usually found in close tandem copies, suggesting that genomic retrozymes in animals may express similar circRNA molecules with HHRs, which would also accumulate in metazoan transcriptomes. However, several differences can be highlighted between plant and animal retrozymes. On the one hand, none of the characteristic sequences of plant retrozymes, such as LTRs, PBS or PPT, are present in their animal counterparts. Moreover, whereas plant retrozymes only show a few HHR motifs (usually, just two copies per retrozyme) of the type III, ribozymes in metazoan retrozymes occur as many copies (dozens to even hundreds) of type-I HHR motifs (Fig. 5.4). These type-I HHRs not only show a characteristic set of tertiary interactions but a very short or even no helix III at all, which indicates that these ribozymes may require in many instances the adoption of dimeric HHR conformations to self-cleave efficiently [55, 24]. The minimal type-I HHRs of metazoan retrozymes highly resemble those described in the pseudo-LTRs of the autonomous Penelope-like retroelements (PLEs) of metazoans and other eukaryotes [48], which somehow links these two families of retrotransposons. On the other hand, animal retrozymes are composed of smaller minimal repeats in tandem (150–300 bp), indicating that the expected animal circRNAs are also smaller than those described in plants. These repeats, however, are frequently, but not always, flanked by TSDs as well, although these are slightly larger (8–12 bp) than those found in plant retrozymes (4 bp) [9, 10].
Metazoan retrozymes
Schematic representation (top) of a typical genomic retrozyme element present in metazoan genomes.
Target side duplications (TSDs) delimiting the retrozyme are shown in grey boxes. Tandem repeats of around 300 bp are indicated with arrows. Typical type-I HHRs are shown. Minimum free energy secondary structure prediction of three examples of circRNAs derived from metazoan retrozymes (rotifers, corals and arthropods) are shown (bottom). HHR sequences in the circRNAs are shown in purple letters, and the self-cleavage sites are indicated with arrowheads. Numbering starts after the HHR self-cleavage site
Recent analysis done in our lab with diverse retrozyme-containing metazoans has confirmed that, as suspected, these organisms accumulate abundant circRNAs in most of the analysed tissues, in a similar way as described for plants (De la Peña and Cervera, to be published). Altogether, the resulting landscape offered by genomic HHRs (either type I or III) in eukaryotes indicates that close tandem copies of this ribozyme allows the expression of small circRNAs (100–1000 nt). Although the presence of other small self-cleaving ribozymes in eukaryotic genomes have been described, such as HDV and twister ribozymes [30, 47], the characteristic occurrence in close tandem repeats seems to be exclusively restricted to the case of the hammerhead ribozyme.
5 A Proposed Mechanism for the Expression and Spreading of Eukaryotic circRNAs with Hammerhead Ribozymes
Retrozymes are a new and atypical group of nonautonomous eukaryotic retroelements with self-cleaving hammerhead ribozymes. In plants, genomic retrozymes resemble other small nonautonomous LTR retrotransposons such as TRIMs [53] and SMARTs [54]. As nonautonomous retrotransposons, retrozymes do not show protein-coding regions but self-cleaving HHR motifs in their LTRs, which, most likely, are responsible of the accumulation in vivo of circular and linear RNAs of the precise size encompassed by the HHRs. Regarding the life cycle of retrozymes, the most plausible model would start with the transcription of the genomic retrozyme. Similar retroelements, such as TRIMs or autonomous LTR retrotransposons, are known to be generally transcribed by RNA Pol II, although examples of RNA Pol III-transcribed retrotransposons have been also reported [56]. Plant and metazoan retrozymes do not seem to contain any recognizable promoter, and in consequence, a feasible hypothesis could be that retrozymes may undergo Pol-driven (either I, II or III) read-through transcription depending on tissues and/or their genomic location. In any case, nascent RNA transcripts would follow co-transcriptional self-processing by tandem self-cleaving HHRs, producing linear RNAs with 5′-OH and 2′,3′-cyclic-phosphate ends. Whereas the step of self-cleavage is expected to occur with high efficiency for plant retrozymes carrying type-III HHRs, in the case of metazoan retrozymes, self-cleavage frequently requires the adoption of a dimeric conformation of minimal type-I HHRs, which is expected to be slightly less efficient than the monomeric version [55]. As summarized in Fig. 5.5, covalent circularization of the resulting self-cleaved RNAs through either the HHR itself or a host RNA ligase factor [57] would finish in stable circRNAs. As the most plausible model, these circRNAs are the final template for retrotranscription, whereas linear retrozyme RNAs would be intermediaries and/or by-products of the circRNAs. In the case of plants, circRNAs derived from LTR-like retrozymes could be primed by any cellular tRNAMet through their PBS motifs. Then, retrotranscriptases encoded by Ty3-Gypsy LTR retrotransposons would produce cDNAs of different lengths, thanks to the circular nature of the RNA template. In the case of metazoan retrozymes, retrotranscriptases encoded by non-LTR retrotransposon (such as PLEs or LINEs) would be responsible of carrying out this latter step of cDNA synthesis. Finally, the resulting cDNAs would be integrated in new genomic locations through the machinery of the autonomous retrotransposons (Fig. 5.5). A last question to be addressed is related to the very high levels of circRNAs with HHRs detected in most organisms analysed. Genomic retrozymes are frequently found as many copies (from dozens to thousands of repeats) within a given genome, which suggests that even low transcription activity would result in abundant levels of circRNAs. However, most of the obtained data indicates that only a few retrozyme copies would be transcriptionally active [10], which suggests that the higher stability of these structured circRNAs with ribozymes compared with linear RNAs would be the reason of their high levels of accumulation in vivo. Moreover, the presence of a high sequence heterogeneity observed for a population of circRNAs in a given organism, together with the presence of retrozyme RNAs of the negative polarity, also suggests the intriguing possibility of replication of the circRNAs through endogenous polymerases.
Model for the life cycle of retrozymes
A full genomic retrozyme containing at least two HHRs in tandem is transcribed (top), and the resulting RNA would self-process through the HHRs to give a linear RNA with 5′-OH and 2′,3′-cyclic phosphate ends. The linear RNA would be circularized through an RNA ligase activity, and the resulting circRNA(+) could be recognized for either endogenous RNA polymerases (replication cycles), other cell factors (new biological roles), or retrotranscriptases encoded by autonomous retrotransposons. In the latter case, the resulting cDNAs from retrotranscription of a circular RNA template would have different lengths depending on the processivity of the retrotranscriptase. In a final step, the machinery of the retrotransposon would integrate the retrozyme DNAs at new genomic loci
6 Functional and Biotechnological Applications of circRNAs with Ribozymes
Retrotransposons, and mobile genetic elements in general, constitute a major fraction of nuclear genomes of most eukaryotes. Historically, these genomic sequences have been regarded as junk DNA, but now we know that retroelements are major drivers of genome evolution with a role in shaping the genomes that they inhabit. In this regard, genomic retrozymes and their associated circRNAs would have similar evolutionary impact as any other retroelement. However, the atypically high accumulation levels of RNA circles encoded by genomic retrozymes in the transcriptomes of most eukaryotic tissues, either somatic or reproductive, suggest that other biological roles can be possible. In this regard, several genic circRNAs have been found to play a role as microRNA sponges [7], and a comparable role for retrozyme circRNAs would be feasible. Moreover, the highly structured circRNAs derived from genomic retrozymes are suitable to be recognized and processed by the RNAi machinery of the cell, and, consequently, these abundant circRNAs with ribozymes would be potential templates for the production of miRNA/siRNAs with specific regulatory roles in the biology of the organisms where they are expressed. At the same time, we already know many examples of co-option or domestication of the transposable elements by their hosts as adaptations to diverse problems [58]. Usually, these domestications are performed with transposon-derived proteins, but also small ribozymes such as 3′ UTR HHRs [39] and intronic HHRs [44] or HDV ribozymes [59] seem to be examples of retroelement domestication. Consequently, circRNAs with HHRs in some organisms could have been specifically co-opted to play precise functions, a possibility that should be studied in the future.
Regarding the biotechnological applications of tandem small ribozymes in the expression of circRNAs, it has to be pointed out that the mechanism of backsplicing described for the synthesis of most genic circRNAs seems to be a complex pathway, which still requires deeper study in order to fully understand and use for practical applications. In this regard, our current knowledge about small ribozymes allow us to design much easier approaches for the expression of circRNAs, which, moreover, could reach higher accumulation levels as observed for most eukaryotic retrozymes so far analysed. Moreover, in vitro synthesis of circRNAs through self-cleaving ribozymes may also offer a straightforward approach for the production and study of specific genic circRNAs from eukaryotes.
7 Conclusions and Future Prospects
Genomic retrozymes are a new family of eukaryotic retrotransposons, which spread through circRNAs with hammerhead ribozymes among plant and animal genomes [10]. In plants, retrozymes seem to be mostly restricted to eudicots, although the presence of putative retrozymes with tandem HHR copies were also detected in some monocots, primitive land plants (like the spikemoss Selaginella moellendorffii) and algae (such as Chlamydomonas reinhardtii) [10, 40]. However, most of these HHRs in the genomes of primitive plants are type-I motifs, which are more related to those found in metazoan than in angiosperm genomes. This observation would indicate that retrozymes in flowering plants may have a different origin, either due to a de novo origin in angiosperms by chance or through horizontal transfer from other organisms containing type-III HHRs such as bacteria [40,41,42]. In any case, all these data suggest that genome-encoded circRNAs with HHRs would be more frequent molecules in eukaryotic transcriptomes than previously thought. Moreover, circRNAs with type-III HHRs in plants allow to propose an evolutionary path for the origin of the small infectious circRNAs with HHRs of plants (viroids and viral satellite RNAs), which may come by chance from the abundant reservoirs of circRNAs present in plant transcriptomes [9]. In contrast, metazoan retrozymes with type-I HHRs seem to indicate that this HHR topology would be more efficient for circRNA expression in animals. Future in vivo experiments will help us to better understand this new tool in the knowledge of the biology and biotechnology of eukaryotic circRNAs.
Abbreviations
- circRNA:
-
circular RNA
- HHR:
-
hammerhead ribozyme
- LTR:
-
long terminal repeat
- PBS:
-
primer binding site
- PPT:
-
polypurine tract
- RT:
-
retrotranscriptase
- TSD:
-
target site duplication
References
Lasda E, Parker R (2014) Circular RNAs: diversity of form and function. RNA 20(12):1829–1842
Salzman J, Gawad C, Wang PL et al (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One 7(2):e30733
Jeck WR, Sorrentino JA, Wang K et al (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19(2):141–157
Wang PL, Bao Y, Yee MC et al (2014) Circular RNA is expressed across the eukaryotic tree of life. PloS One 9(6):e90859
Ashwal-Fluss R, Meyer M, Pamudurti NR et al (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56(1):55–66
Memczak S, Jens M, Elefsinioti A et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–338
Hansen TB, Jensen TI, Clausen BH et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388
Talhouarne GJ, Gall JG (2014) Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA 20(9):1476–1487
De la Peña M, Cervera A (2017) Circular RNAs with hammerhead ribozymes encoded in eukaryotic genomes: the enemy at home. RNA Biol 14(8):985–991
Cervera A, Urbina D, de la Peña M (2016) Retrozymes are a unique family of non-autonomous retrotransposons with hammerhead ribozymes that propagate in plants through circular RNAs. Genome Biol 17(1):135
De la Peña M, Garcia-Robles I, Cervera A (2017) The hammerhead ribozyme: a long history for a short RNA. Molecules 22(1):78–89
Kruger K, Grabowski PJ, Zaug AJ et al (1982) Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31(1):147–157
Guerrier-Takada C, Gardiner K, Marsh T et al (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35(3 Pt 2):849–857
Gilbert W (1986) The RNA world. Nature 319:618
Crick FH (1968) The origin of the genetic code. J Mol Biol 38(3):367–379
Orgel LE (1968) Evolution of the genetic apparatus. J Mol Biol 38(3):381–393
Woese CR (1968) The fundamental nature of the genetic code: prebiotic interactions between polynucleotides and polyamino acids or their derivatives. Proc Natl Acad Sci U S A 59(1):110–117
Steitz TA, Moore PB (2003) RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem Sci 28(8):411–418
Ferre-D’Amare AR, Scott WG (2010) Small self-cleaving ribozymes. Cold Spring Harb Perspect Biol 2(10):a003574
Valadkhan S, Manley JL (2001) Splicing-related catalysis by protein-free snRNAs. Nature 413(6857):701–707
Winkler W, Nahvi A, Breaker RR (2002) Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419(6910):952–956
Mojica FJ, Diez-Villasenor C, Garcia-Martinez J et al (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60(2):174–182
Fire A, Xu S, Montgomery MK et al (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391(6669):806–811
Prody GA, Bakos JT, Buzayan JM et al (1986) Autolytic processing of dimeric plant virus satellite RNA. Science 231(4745):1577–1580
Hutchins CJ, Rathjen PD, Forster AC et al (1986) Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res 14(9):3627–3640
Buzayan JM, Gerlach WL, Bruening G (1986) Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature 323:349–353
Kuo MY, Sharmeen L, Dinter-Gottlieb G et al (1988) Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J Virol 62(12):4439–4444
Saville BJ, Collins RA (1990) A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell 61(4):685–696
Winkler WC, Nahvi A, Roth A et al (2004) Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428(6980):281–286
Roth A, Weinberg Z, Chen AG et al (2014) A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat Chem Biol 10(1):56–60
Weinberg Z, Kim PB, Chen TH et al (2015) New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol 11(8):606–610
De la Peña M, Gago S, Flores R (2003) Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J 22(20):5561–5570
Khvorova A, Lescoute A, Westhof E et al (2003) Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol 10(9):708–712
Martick M, Scott WG (2006) Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126(2):309–320
Przybilski R, Graf S, Lescoute A et al (2005) Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell 17(7):1877–1885
Ferbeyre G, Smith JM, Cedergren R (1998) Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol 18(7):3880–3888
Rojas AA, Vazquez-Tello A, Ferbeyre G et al (2000) Hammerhead-mediated processing of satellite pDo500 family transcripts from Dolichopoda cave crickets. Nucleic Acids Res 28(20):4037–4043
Epstein LM, Gall JG (1987) Self-cleaving transcripts of satellite DNA from the newt. Cell 48(3):535–543
Martick M, Horan LH, Noller HF et al (2008) A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature 454(7206):899–902
De la Peña M, Garcia-Robles I (2010) Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA 16(10):1943–1950
Jimenez RM, Delwart E, Luptak A (2011) Structure-based search reveals hammerhead ribozymes in the human microbiome. J Biol Chem 286(10):7737–7743
Perreault J, Weinberg Z, Roth A et al (2011) Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput Biol 7(5):e1002031
Seehafer C, Kalweit A, Steger G et al (2011) From alpaca to zebrafish: hammerhead ribozymes wherever you look. RNA 17(1):21–26
De la Peña M, Garcia-Robles I (2010) Intronic hammerhead ribozymes are ultraconserved in the human genome. EMBO Rep 11(9):711–716
Hammann C, Luptak A, Perreault J et al (2012) The ubiquitous hammerhead ribozyme. RNA 18(5):871–885
Garcia-Robles I, Sanchez-Navarro J, De la Peña M (2012) Intronic hammerhead ribozymes in mRNA biogenesis. Biol Chem 393(11):1317–1326
Webb CH, Riccitelli NJ, Ruminski DJ et al (2009) Widespread occurrence of self-cleaving ribozymes. Science 326(5955):953
Cervera A, De la Peña M (2014) Eukaryotic penelope-like retroelements encode hammerhead ribozyme motifs. Mol Biol Evol 31(11):2941–2947
Eickbush DG, Eickbush TH (2010) R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA cotranscript. Mol Cell Biol 30(13):3142–3150
Ruminski DJ, Webb CH, Riccitelli NJ et al (2011) Processing and translation initiation of non-long terminal repeat retrotransposons by hepatitis delta virus (HDV)-like self-cleaving ribozymes. J Biol Chem 286(48):41286–41295
Kennell JC, Saville BJ, Mohr S et al (1995) The VS catalytic RNA replicates by reverse transcription as a satellite of a retroplasmid. Genes Dev 9(3):294–303
Gorinsek B, Gubensek F, Kordis D (2004) Evolutionary genomics of chromoviruses in eukaryotes. Mol Biol Evol 21(5):781–798
Witte CP, Le QH, Bureau T et al (2001) Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc Natl Acad Sci U S A 98(24):13778–13783
Gao D, Chen J, Chen M et al (2012) A highly conserved, small LTR retrotransposon that preferentially targets genes in grass genomes. PloS one 7(2):e32010
Forster AC, Davies C, Sheldon CC et al (1988) Self-cleaving viroid and newt RNAs may only be active as dimers. Nature 334(6179):265–267
Kalendar R, Tanskanen J, Chang W et al (2008) Cassandra retrotransposons carry independently transcribed 5S RNA. Proc Natl Acad Sci U S A 105(15):5833–5838
Flores R, Grubb D, Elleuch A et al (2011) Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: variations on a theme. RNA Biol 8(2):200–206
Jangam D, Feschotte C, Betran E (2017) Transposable element domestication as an adaptation to evolutionary conflicts. Trends Genet 33(11):817–831
Salehi-Ashtiani K, Luptak A, Litovchick A et al (2006) A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 313(5794):1788–1792
Chi YI, Martick M, Lares M et al (2008) Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol 6(9):e234
Dufour D, de la Peña M, Gago S et al (2009) Structure-function analysis of the ribozymes of chrysanthemum chlorotic mottle viroid: a loop-loop interaction motif conserved in most natural hammerheads. Nucleic Acids Res 37(2):368–381
Gago S, De la Peña M, Flores R (2005) A kissing-loop interaction in a hammerhead viroid RNA critical for its in vitro folding and in vivo viability. RNA 11(7):1073–1083
Funding
Funding for this work was provided by the Ministerio de Economía y Competitividad of Spain and FEDER funds (BFU2014-56094-P and BFU2017-87370-P).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
de la Peña, M. (2018). Circular RNAs Biogenesis in Eukaryotes Through Self-Cleaving Hammerhead Ribozymes. In: Xiao, J. (eds) Circular RNAs. Advances in Experimental Medicine and Biology, vol 1087. Springer, Singapore. https://doi.org/10.1007/978-981-13-1426-1_5
Download citation
DOI: https://doi.org/10.1007/978-981-13-1426-1_5
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-1425-4
Online ISBN: 978-981-13-1426-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)