Abstract
MicroRNA (miRNA or miR) is a small noncoding RNA molecule ~22 nucleotides in size, which is found in plants, animals, and some viruses. miRNAs are thought to primarily down regulate gene expression by binding to 3′ untranslated regions of target transcripts, thereby triggering mRNA cleavage or repression of translation. Recently, evidence has emerged that miRNAs can interact with the promoter and activate gene expression. This mechanism, called RNA activation (RNAa), is a process of transcriptional activation where the direct interaction of miRNA on the promoter triggers the recruitment of transcription factors and RNA-Polymerase-II on the promoter to activate gene transcription. To date, very little is known about the mechanism by which miRNA regulates RNA activation (RNAa) and their role in tumor progression. This is an emerging field in RNA biology. In this chapter, we describe the mechanisms utilized by miRNAs to activate transcription.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Current research in the microRNA (miRNA) field largely focuses on the actions of miRNAs on the 3′ untranslated regions of genes that result in the suppression of target expression either by degradation of mRNA or inhibition of translation [1, 5]. Recently, several groups including ours have demonstrated that small noncoding RNAs can influence gene transcription by direct interaction of small RNAs with the promoter [8, 16, 19]. This mechanism, also called RNA activation (RNAa), is a process of transcriptional activation where microRNAs through their direct interaction with the promoter sequence activate gene transcription. Most of the studies reported on RNAa are on cancer models [17, 18, 25]; however, recent studies have extended these findings to other conditions such as ischemia [28, 31] and erectile dysfunction in diabetes [34]. Together, the current research provides evidence that RNA can precisely target selective genes to activate signaling pathways .
RNA interference caused by siRNA and/or miRNA leads to suppression of transcription or degradation of mRNA and/or inhibition of translation within hours after the transfection of small RNAs [9]. In contrast, transcriptional activation induced by miRNAs requires several days to turn on, but can persist for several days once it is activated [26]. This delay could be due to the epigenetic changes mediated by miRNA as the promoter of target genes, which then facilitates long-term effects on gene expression . Thus RNAa is potentially an important unexplored mechanism employed by cells to activate gene expression for a persistent period of time. This chapter will address the current state of RNAa research and the underlying mechanisms that are responsible for this process.
2 History
In 1998, Craig Mello and Andrew Fire reported the gene-silencing effects of double-stranded small RNAs (dsRNAs) in C. elegans , where they demonstrated that dsRNAs successfully silenced the targeted gene and coined the term RNA interference (RNAi) [10]. This striking discovery in RNA biology was rewarded with a Nobel Prize in Physiology or Medicine in 2006. In contrast to the term RNAi , RNA activation (RNAa) refers to the process of transcriptional activation mediated by small RNAs . The new term RNAa was coined first by Long-Cheng Li and Rajvir Dahiya [17] who designed and synthesized 21-nt dsRNAs targeting selected promoter regions of human genes E-CADHERIN , p21 WAF1/CIP1 (p21 ), and vascular endothelial growth factor (VEGF) . Intriguingly, transfection of those dsRNAs into human prostate cancer cell lines caused long-lasting and sequence-specific induction of all three genes [17]. Shortly thereafter, Janowski et al. identified the induction of progesterone receptor (PR) and major vault protein by promoter-targeting dsRNA [16]. Since then, similar observations have been made by several other groups in different mammalian species including human [17], rat [11], and mouse [12], suggesting that RNAa is a general mechanism of gene regulation conserved across mammalian species. Although RNAa was proposed by Li et al., in 2006, the theory of gene activation by the action of RNAs was initially proposed by Britten and Davidson in their article entitled “Gene regulation for higher cells: This theory postulates the existence of gene regulation by small non-coding RNA” published in Science [6]. In this theory, they proposed that RNA molecules form a sequence-specific complex with the nontranscribed sequences that reside upstream of the sequences transcribed into RNA molecules [6]. To date, very little is known about the precise mechanism of how RNAa occurs in normal cells and the importance of this process in physiology and disease. Because small RNAs can interact with gene promoters, which influence various transcriptional processes, the current hypothesis is that the RNAa mechanism includes changes in the occupancy of transcription factors (TFs) or RNA polymerases on the promoter, and the epigenetic changes associated with this occupancy process. We recently found some evidence in support of this hypothesis . We identified that miR551b, an miRNA located in the 3q26.2 amplicon, a region that is frequently amplified in ovarian cancer patients, interacts with complementary sequences on the promoter of STAT3 transcription factor. This miR551b interaction with STAT3 promoter affects STAT3 transcription by the recruitment of RNA Polymerase II (RNA Pol II) and the Twist1 transcription factor to the STAT3 promoter. To help understand the RNAa mechanism, we will outline next the various steps in the process of miRNA synthesis and biology that are directly relevant to RNAa.
3 Maturation of miRNAs
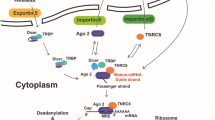
In northern blot analyses for most miRNAs, two species of nucleotides (nt) are often noticed: a larger (~70 nt) and a smaller (~22 nt) RNA. The smaller RNA (~22 nt) is the mature form of miRNAs [1, 24]. It is known that more than two thirds of all human miRNAs are encoded in the intervening regions (introns ) of protein-coding genes as well as in long noncoding transcripts. MiRNAs can also be encoded in exons or introns, or can be located in intergenic region, and within the chromosomal regions encompassing two genes [2, 29]. During the biogenesis, the primary precursor (pri-miRNA) is processed into an approximately 70-nt-long stem-loop structure by nuclear RNase III Drosha present in the microprocessor complex, which is the complex of proteins involved in the processing and maturation of miRNAs. The two RNase domains of Drosha help cleave the 5′ and 3′ ends of the pri-miRNA , which is then exported to the cytoplasm by Exportin-5 and Ran-GTP complexes. Later the final maturation of miRNA occurs with Dicer, another RNase III nuclease that processes the pre-miRNA into a 22-bp dsRNA in the cytoplasm. Subsequently, Dicer-cleaved miRNAs are loaded with RNA-induced silencing complex (RISC) that includes Argonaute (AGO) proteins, transactivating response RNA-binding protein (TRBP) , protein kinase R-activating protein (PACT), and Dicer .
3.1 Transport of Mature miRNA to the Nucleus for RNAa
Since the major process of miRNA maturation occurs in the cytoplasm, it is expected that miRNAs are needed to translocate into the nucleus for RNAa. However, the exact mechanism underlying the transport of miRNA from the cytoplasm to the nucleus is not well understood. Argonaute (AGO) proteins that facilitate the transport of miRNAs from the cytoplasm to the nucleus are widely considered to promote this process. AGO proteins are highly conserved proteins and ubiquitously expressed in all higher eukaryotes. In humans, eight AGO genes are known to exist, of which family members AGO1 , 2, 3, and 4 have been extensively studied [14]. AGO proteins encode four functional domains, namely, N-terminal, PAZ , Mid, and C-terminal PIWI domains [14]. Using all of its functional domains, AGO proteins bind to different classes of small RNAs including miRNAs, siRNAs, and PIWI-interacting RNAs (piRNAs ), which further bind with their specific targets through sequence complementarity base pairing [3, 15]. Among the domains in AGO protein, the PAZ domain binds to the 3′ end of both siRNA and miRNA, which facilitates the binding of small RNAs on target mRNA by base pair interaction [22]. The PIWI domain of AGO proteins contains the catalytic residues which are essential for cleavage [22], and also mediates protein-protein interaction with the RNase III domain of Dicer protein, suggesting that PIWI is a critical domain that is responsible for miRNA maturation function. Within the mid domain, AGO protein encodes for MC motif and is involved in binding cap structures of RNA, thereby suggesting a role in controlling translation of capped mRNA. Altogether, the AGO protein family with multiple domains and multiple protein interactive motifs allows miRNA-guided AGO proteins to interact with several proteins directly or indirectly. For example, AGOs recruit TFs and RNA Pol II to the promoter of the target genes, and this complex recruitment leads to conformational changes that enhance the occupancy of TFs and RNA Pol II to STAT3 oncogene promoter to turn on transcription (Fig. 6.1) [8]. In addition to AGO proteins, a second class of proteins, namely, importin8 (IPO8), a member of the karyopherin family [35], has been identified for miRNA transport from the cytoplasm to the nucleus. Therefore, both IPO8 and AGO proteins have an important role in the transport of mature miRNA from the cytoplasm to the nucleus [35]. AGO proteins have been more extensively studied in the context of miRNA transport. When AGO1 or AGO2 proteins are depleted individually, the respective member of its family (AGO2 for AGO1 depleted and AGO1 for AGO2 depleted) is preferentially retained in the nucleus [20]. Furthermore, Matsui et al. demonstrated that silencing AGO1 reduces nuclear Dicer levels by 90% and nuclear TRBP levels by >50% [20], suggesting that in addition to nuclear import of miRNAs, the AGO protein family may also be involved in the maturation of miRNAs in the nucleus .
4 Target Specificity and Selection for miRNA-Mediated RNAa
Once in the nucleus , the miRNA needs to interact with its target. The principles governing the genomic target site recognition of small RNAs are thought to be sequence dependent in the sense of antisense strand [27]. One of the first studies reported on RNAa, were usedduplex RNAs target promoter sequences designed based on the rules of functional siRNAs, where the rules included low GC content , lack of repeated or inverted sequences, and avoidance of targetingCpG -rich regions [17]. In consequence, authors have identified that, four dsRNAs that met these rules targeting E-cadherin , p21, and VEGF promoters activated the expression of E-cadherin, p21, and VEGF at the mRNA and protein levels [17]. The same group later identified miR -373 complementary target sites in the promoters of E-cadherin and cold shock domain containing protein C2 (CSDC2) , thereby inducing the expression of both E-Cadherin and CSDC2 [25]. Of note, the site-specific mutations on the miR-373 sequences abrogated any increase in the expression of both E-Cadherin and CSDC [25]. In a recent study, Vera Huang and her team tested the effects of miR-774 and miR-1186 which have sequence complementarity to the sequences located in the CyclinB1 promoter [12]. Similarly, miRNA-589 has complementarity to adjacent sites in the COX2 gene promoter where it binds to and activates COX2 transcription. Our recent studies concur with these reports, and indeed, miR551b-3p interacts with the promoter sequences on the STAT3 promoter and activates STAT3 transcription [8].
Although most studies report proximal targeting of the miRNA on the promoter site, for example, overlapping the transcriptional start site (TSS ) [11, 16, 33], there are reports of miRNA targeting 200–1200 bps upstream of the TSS. This upstream targeting region is considered optimal for RNAa. Altogether, the studies in the past 10 years on the transcriptional activation by small RNAs suggest that sequence specificity is a critical factor and miRNA-mediated transcriptional activation largely depends on the sequence complementarity between the miRNAs and the promoter so as to engage the interaction between miRNAs and the promoter for transcriptional activation.
4.1 Mechanism of Transcriptional Activation
The majority of studies report miRNAs’ role in gene inhibition, but only a handful of reports have described their role in the RNAa mechanism. Exactly how RNAa is facilitated is not clear, but studies are beginning to show that the mechanism includes binding of miRNA on the promoter and improve occupancy of transcription factor and RNA Polymerase II (RNA Pol II ). We hypothesize that AGO proteins that are responsible for transporting miRNAs to the nucleus are also critical for facilitating RNAa. This hypothesis is partly based on published studies where AGO1 has been shown to interact with RNA Pol II [13, 20]. Similarly, we have also reported that miRNA-551b-AGO1 complex interacts with RNA Pol II, which in turn facilitates the recruitment of TWIST1 transcription factor to the STAT3 promoter to activate STAT3 transcription [8]. Interestingly, the miR551b-3p-mediated STAT3 activation occurs within 24 h but lasts for more than 10 days. Others have reported similar effects with RNAa occurring within 24 h, and further elevated to the maximum levels by 72 h, which persists for at least one and a half weeks in cells [26]. The persistent nature of this activation is intriguing given that gene-silencing effects by siRNA last for 24 h, and are turned off within 3–4 days [9]. One potential explanation is the alteration of chromatin structures or epigenetic machineries by RNAa. The transcriptional state of genes is dependent on the state of chromatin occupancy, which is modified via histone protein and DNA modifications . Recent CHIP data for AGO1 protein identified AGO1 peaks that mapped within 65 kb of transcription start sites (TSS ) , and overlapped with H3K4me3 marks [13]. Trimethylation of lysine 4 on histone H3 protein subunit (H3K4me3) is an important histone marker in epigenetic studies, and is indicative of the active gene promoters [4]. Similar increases in trimethylated H3K4 were also noticed in PR and CyclinB1 promoters that were facilitated by duplex short RNAs [11, 16]. In addition to the H3K4 methylation , there is a second class of methylation, H3K27 group, that is responsible for transcriptional silencing . In contrast to the methylation of H3K4, trimethyation of H3K27 at the promoter is associated with inactive gene promoters. H3K27 trimethylation is catalyzed by a class of proteins that belongs to the family of Polycomb Group of proteins (PcG). The PcG proteins form two distinct Polycomb-repressive multiprotein complexes: PRC1 and PRC2. Trimethylation of H3K27 is mainly catalyzed by EZH2, which is the catalytic subunit of the PRC2 core complex. It also contains two other PcG proteins, SUZ12 and EED , which are critical proteins required for the process of transcriptional modulation by PRC2 [7, 23]. The precise mechanisms that facilitate miRNA-induced trimethylation of H3K4 or H3K27 on histone proteins located at the promoter sites are still unknown. In light of the current research on histone methylation and analysis of multiple histone marks by small RNAs , activating the active histone marks such as H3K4 is a dynamic process when compared to the loss of methylation of H3K27 . Apart from the histone methylation, there is a paucity of data on the role of DNA methylation during the process of RNAa. The AGO proteins discussed earlier for miRNA transport from the cytoplasm to the nucleus also may play a role in DNA methylation . For example, AGO4 is functionally distinct from AGO1 –AGO3 and incompatible with splicing abilities [30], and is not directly involved in the siRNA /miRNA-triggered RNA degradation process. Based on AGO4’s structure and its interaction with other AGO family proteins and methylase enzymes in particular, we hypothesize that AGO4 has important roles in RNA-mediated DNA methylation or other epigenetic regulation processes. Thus the RNAa mechanism is a fertile field rich with scientific questions that need answers.
5 Conclusion
Recent studies demonstrate that RNAa is an endogenous mechanism of gene regulation, wherein small duplex RNAs including miRNAs activate the gene expression through their interaction with the promoter. Evidence suggests that promoter-guiding effects of small duplex RNAs are primarily regulated through the Argonaute family proteins. It is currently accepted that once small RNAs get bound to the AGO complex, they modify histones and recruit transcription factors to form RNA polymerase II complex on gene promoters. In the past 10 years, RNAa has emerged as an important topic in miRNA research. However, our understanding of RNAa is still limited with many questions still unanswered. These include questions related to the sequence of events that occur during the process of RNAa such as (1) whether the number of seed sequences on the promoters will have different effects on transcription and (2) the number of mechanisms such as histone modifications and recruitment of TFs and/or RNA Pol II that associate with the miRNA binding on the promoter. We expect that the RNA immune precipitation experiments followed by mass spectrometry as well as pulse chase experiments will identify most of the critical proteins associated with conformational changes that lead to the transcriptional activation. Most of the studies on RNAa are employing overexpression methods to increase the levels in miRNA in cells to achieve RNAa. Therefore, it still remains unexplored whether RNAa is an important operational mechanism for transcriptional regulation under normal physiological conditions. Thus more research in this area is required in order to unravel the complexities in RNAa mechanisms, in particular those associated with AGO. Finally, whether this mechanism is found in other cell types and plays a role in diseases other than cancer needs further exploration .
References
Ambros V (2001) microRNAs: tiny regulators with great potential. Cell 107(7):823–826
Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D (2003) MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13(10):807–818
Azlan A, Dzaki N, Azzam G (2016) Argonaute: the executor of small RNA function. J Genet Genomics 43(8):481–494. doi:10.1016/j.jgg.2016.06.002
Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129(4):823–837. doi:10.1016/j.cell.2007.05.009
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Britten RJ, Davidson EH (1969) Gene regulation for higher cells: a theory. Science 165(3891):349–357
Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 298(5595):1039–1043. doi:10.1126/science.1076997
Chaluvally-Raghavan P, Jeong KJ, Pradeep S, Silva AM, Yu S, Liu W, Moss T, Rodriguez-Aguayo C, Zhang D, Ram P, Liu J, Lu Y, Lopez-Berestein G, Calin GA, Sood AK, Mills GB (2016) Direct upregulation of STAT3 by MicroRNA-551b-3p deregulates growth and metastasis of ovarian cancer. Cell Rep 15(7):1493–1504. doi:10.1016/j.celrep.2016.04.034
Dykxhoorn DM, Novina CD, Sharp PA (2003) Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol 4(6):457–467. doi:10.1038/nrm1129
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391(6669):806–811. doi:10.1038/35888
Huang V, Qin Y, Wang J, Wang XL, Place RF, Lin GT, Lue TF, Li LC (2010) RNAa is conserved in mammalian cells. Plos One 5(1). doi:ARTN e8848 10.1371/journal.pone.0008848
Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, Li LC (2012) Upregulation of cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res 40(4):1695–1707. doi:10.1093/nar/gkr934
Huang V, Zheng J, Qi Z, Wang J, Place RF, Yu J, Li H, Li LC (2013) Ago1 interacts with RNA polymerase II and binds to the promoters of actively transcribed genes in human cancer cells. PLoS Genet 9(9):e1003821. doi:10.1371/journal.pgen.1003821
Hutvagner G, Simard MJ (2008) Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9(1):22–32. doi:10.1038/nrm2321
Jain R, Iglesias N, Moazed D (2016) Distinct functions of argonaute slicer in siRNA maturation and heterochromatin formation. Mol Cell 63(2):191–205. doi:10.1016/j.molcel.2016.05.039
Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR (2007) Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol 3(3):166–173. doi:10.1038/nchembio860
Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R (2006) Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A 103(46):17337–17342. doi:10.1073/pnas.0607015103
Lopez P, Wagner KD, Hofman P, Van Obberghen E (2016) RNA activation of the vascular endothelial growth factor gene (VEGF) promoter by double-stranded RNA and hypoxia: role of noncoding VEGF promoter transcripts. Mol Cell Biol 36(10):1480–1493. doi:10.1128/MCB.01096-15
Matsui M, Chu Y, Zhang H, Gagnon KT, Shaikh S, Kuchimanchi S, Manoharan M, Corey DR, Janowski BA (2013) Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res 41(22):10086–10109. doi:10.1093/nar/gkt777
Matsui M, Li L, Janowski BA, Corey DR (2015) Reduced expression of argonaute 1, argonaute 2, and TRBP changes levels and intracellular distribution of RNAi factors. Sci Rep 5:12855. doi:10.1038/srep12855
Matsui M, Prakash TP, Corey DR (2016) Argonaute 2-dependent regulation of gene expression by single-stranded miRNA mimics. Mol Ther 24(5):946–955. doi:10.1038/mt.2016.39
Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T (2004) Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15(2):185–197. doi:10.1016/j.molcel.2004.07.007
Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K (2007) The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 27(10):3769–3779. doi:10.1128/MCB.01432-06
Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408(6808):86–89. doi:10.1038/35040556
Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R (2008) MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A 105(5):1608–1613. doi:10.1073/pnas.0707594105
Place RF, Noonan EJ, Foldes-Papp Z, Li LC (2010) Defining features and exploring chemical modifications to manipulate RNAa activity. Curr Pharm Biotechnol 11(5):518–526
Portnoy V, Huang V, Place RF, Li LC (2011) Small RNA and transcriptional upregulation. Wiley Interdiscip Rev RNA 2(5):748–760. doi:10.1002/wrna.90
Reebye V, Saetrom P, Mintz PJ, Huang KW, Swiderski P, Peng L, Liu C, Liu X, Lindkaer-Jensen S, Zacharoulis D, Kostomitsopoulos N, Kasahara N, Nicholls JP, Jiao LR, Pai M, Spalding DR, Mizandari M, Chikovani T, Emara MM, Haoudi A, Tomalia DA, Rossi JJ, Habib NA (2014) Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology 59(1):216–227. doi:10.1002/hep.26669
Saini HK, Griffiths-Jones S, Enright AJ (2007) Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci U S A 104(45):17719–17724. doi:10.1073/pnas.0703890104
Schurmann N, Trabuco LG, Bender C, Russell RB, Grimm D (2013) Molecular dissection of human argonaute proteins by DNA shuffling. Nat Struct Mol Biol 20(7):818–826. doi:10.1038/nsmb.2607
Turunen MP, Lehtola T, Heinonen SE, Assefa GS, Korpisalo P, Girnary R, Glass CK, Vaisanen S, Yla-Herttuala S (2009) Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism: a novel example of epigenetherapy. Circ Res 105(6):604–609. doi:10.1161/CIRCRESAHA.109.200774
Vaucheret H, Vazquez F, Crete P, Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18(10):1187–1197. doi:10.1101/gad.1201404
Wang X, Wang J, Huang V, Place RF, Li LC (2012) Induction of NANOG expression by targeting promoter sequence with small activating RNA antagonizes retinoic acid-induced differentiation. Biochem J 443(3):821–828. doi:10.1042/BJ20111491
Wang T, Li M, Yuan H, Zhan Y, Xu H, Wang S, Yang W, Liu J, Ye Z, Li LC (2013) saRNA guided iNOS up-regulation improves erectile function of diabetic rats. J Urol 190(2):790–798. doi:10.1016/j.juro.2013.03.043
Wei Y, Li L, Wang D, Zhang CY, Zen K (2014) Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J Biol Chem 289(15):10270–10275. doi:10.1074/jbc.C113.541417
Yue X, Schwartz JC, Chu Y, Younger ST, Gagnon KT, Elbashir S, Janowski BA, Corey DR (2010) Transcriptional regulation by small RNAs at sequences downstream from 3′ gene termini. Nat Chem Biol 6(8):621–629. doi:10.1038/nchembio.400
Acknowledgments
P. Chaluvally-Raghavan is supported by the Liz-Tilberis Career Development Award and the Ann Schreiber Scholar Award from the Ovarian Cancer Research Foundation and funds from the Marsha Rivkin Center for Ovarian Cancer Research. Ramani Ramchandran is supported by endowment funds from the Department of OBGYN at MCW, programmatic support funds from the Department of Pediatrics, Children’s Research Institute, and NIH grants HL123338, HL112639, HL033833, and HL128374.
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Ramchandran, R., Chaluvally-Raghavan, P. (2017). miRNA-Mediated RNA Activation in Mammalian Cells. In: Li, LC. (eds) RNA Activation. Advances in Experimental Medicine and Biology, vol 983. Springer, Singapore. https://doi.org/10.1007/978-981-10-4310-9_6
Download citation
DOI: https://doi.org/10.1007/978-981-10-4310-9_6
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-4309-3
Online ISBN: 978-981-10-4310-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)