Abstract
Diseases are the main problem for barley in Latin America. Spot blotch (caused by Cochliobolus sativus), stripe rust (caused by Puccinia striiformis f. sp. hordei), and leaf rust (caused by Puccinia hordei) are three of the most important diseases that attack the crop in the region. Chemical control of those diseases is both economically and environmentally inappropriate, making the development of durable resistant varieties a priority for breeding programs. However, the availability of new resistance sources is a limiting factor. The objective of this work was to detect genomic regions associated to durable resistance to spot blotch, stripe rust, and leaf rust in Latin American germplasm. Associations between disease severities measured in several environments across the Americas and 1,536 SNPs (belonging to the barley OPA1) in a population of 360 genotypes were used to identify genomic regions associated with disease. Several models for association mapping with mixed models were compared. These models considered either the structure of the population (Q) through PCA analysis, the identity by descent through coancestry information (K), or both. Results show significant marker-trait associations for spot blotch and leaf and stripe rust. Associations are environment specific.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
18.1 Introduction
Barley (Hordeum vulgare, L.) is the fourth most important cereal crop in the world in terms of total world production (FAOSTAT 2008), and malting quality is one of its most relevant traits. The main use of this crop in Latin America is for malt production (German 2004). One of the main limitations for achieving high yields and malt of good quality is the presence of fungal diseases (German 2007).
Fungal diseases affect barley production both directly (i.e., affecting grain weight and germinability) and indirectly (i.e., foliar diseases that affect photosynthesis decreasing yield; Nutter et al. 1985). Spot blotch (caused by Cochliobolus sativus), stripe rust (caused by Puccinia striiformis f. sp. hordei), and leaf rust (caused by Puccinia hordei) are three of the most important diseases that attack the crop in Latin America (Pereyra 1996). Chemical control of those diseases is both economically and environmentally inappropriate, making the development of durable resistant varieties a priority for breeding programs. However, the availability of new resistance sources is a limiting factor.
Genome-wide association mapping (GWA) can be used to detect genomic regions associated to relevant traits (Jannink et al. 2001) and provide targets for MAS in relevant germplasm (Mather et al. 1997). The advantages of GWA include simultaneous assessment of broad diversity, higher resolution for fine mapping, effective use of historical data, and immediate applicability to cultivar development because the genetic background in which QTLs are estimated is directly relevant for plant breeding (Kraakman et al. 2004). Additionally, GWA has been proposed as a promising tool for selfing-species crop improvement and specifically barley breeding (Kraakman et al. 2004, 2006; Hayes and Szücs 2006; Stracke et al. 2009; Waugh et al. 2009; Roy et al. 2010; Bradbury et al. 2011; von Zitzewitz et al. 2011). The objective of this work was to detect genomic regions associated to durable resistance to spot blotch, stripe rust, and leaf rust in Latin American germplasm.
18.2 Materials and Methods
A total of 360 genotypes from ICARDA and national breeding programs were used. Each genotype was evaluated for disease resistance in several environments. Severity of spot blotch infection (in percentage of covered area from 0 to 100%) was evaluated in three environments: EEMAC (northern Uruguay) in 2009 and 2010 and INIA-LE (west Uruguay) in an off-season nursery in 2009–2010. Severity of leaf rust infection (in a scale from 1 to 10) was measured in four environments: EEMAC 2009 and 2010 and INIA-LE 2009 and 2010. Stripe rust was evaluated in two environments: Mexico 2010 and Peru 2011.
All accessions were genotyped for 1,536 SNPs using one Illumina GoldenGate oligonucleotide pool assays (OPAs) developed for the barley CAP. The development and application of the barley OPAs (BOPA1) is described in detail in Close et al. (2009) and Szücs et al. (2009). In this research, we used the consensus SNP map developed by Close et al. (2009), available by downloading the 1.77 version of the barley HarvEST database (2011).
Several models for association mapping with mixed models were compared to use the most appropriate model in each case. Modifications of the general mixed model equation were used:
where Y is the phenotypic vector, X is the molecular marker matrix, β is the unknown vector of allele effects to be estimated, Q is the population structure represented by the scores of the relevant axis of a principal components analysis, υ is the vector of population effects (parameters), Z is a matrix that relates each measurement to the individual from which it was obtained (an identity matrix in our case), u is the vector of random background polygenic effects, and e is the residual errors. Random effects are underlined. We compared the following mainstream models: (1) naïve; a simple test of association (Kruskal–Wallis) with no correction for population structure (Y = X β + e); (2) fixed, a fixed-effects model using population structure as fixed covariate (Y = X β + Q υ + e); (3) kinship, a mixed model including the coancestry matrix among genotypes as a random effect (Y = X β + Zu + e following Parisseaux and Bernardo 2004); (4) Price, a mixed-effects model including population structure but as a random effect (Y = X β + Q υ + e following Price et al. 2006); and (5) Yu, a mixed-effects model including both population structure and coancestry among genotypes (Y = X β + Q υ + Zu + e following Yu et al. 2009 ).
18.3 Results and Discussion
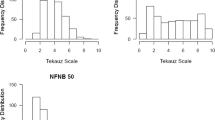
There is not a single best model for all the traits. A mixed model with population structure either as random (Price) or fixed (Yu) effect was the best model for spot blotch (Fig. 18.1a) in all environments. However, a mixed model without population structure and with coancestry information (kinship) was the best model for leaf rust (Fig. 18.1b) in all environments. The Price and Yu models performed relatively well for leaf rust.
We detected several QTL for all the diseases studied (Table 18.1). Eleven significant marker-trait associations were found for leaf rust. Associations were found on chromosomes 2H, 3H, 5H, 6H, and 7H. Nine significant marker-trait associations were found for spot blotch. Associations were found on chromosomes 1H, 3H, 4H, 5H, 6H, and 7H. Only one significant marker-trait association was detected for stripe rust.
Although we expect that the underlying distribution of disease scoring be normal, because of the way these diseases are measured (i.e., in a scale), not all of the traits follow a normal distribution. Additionally, some markers had heterogeneous variances. The most common transformations of the original variable were used to fit final models when the untransformed data did not perform properly. Further study of these traits with more complex models would be beneficial.
18.4 Conclusions
We were able to find significant marker-trait associations for leaf rust, spot blotch, and stripe rust. Although the best correction for population structure was dependent on the trait studied, significant associations were consistent across models. These preliminary results show candidate QTL that should be further studied and compared to previous reports. Evaluations in more environments are being conducted to improve the estimation of QTL. Association mapping is a promising tool to detect candidate QTL to include in marker-assisted selection programs.
References
Bradbury, P., Parker, T., Hamblin, M. T., & Jannink, J. L. (2011). Assessment of power and false discovery rate in genome-wide association studies using the barley CAP germplasm. Crop Science, 51, 52–59.
Close, T. J., Bhat, P. R., Lonardi, S., Wu, Y., Rostoks, N., Ramsay, L., Druka, A., Stein, N., Svensson, J. T., Wanamaker, S., Bozdag, S., Roose, M. L., Moscou, M. J., Chao, S., Varshney, R., Szucs, P., Sato, K., Hayes, P. M., Matthews, D. E., Kleinhofs, A., Muehlbauer, G. J., DeYoung, J., Marshall, D. F., Madishetty, K., Fenton, R. D., Condamine, P., Graner, A., & Waugh, R. (2009). Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics, 10, 582.
FAOSTAT. (2008). Available at http://faostat.fao.org/site/339/default.aspx. Accessed 5 May 2011.
German, S. (2004). Available at http://www.cazv.cz/service.asp?act=print&val=32831 . Breeding malting barley under stress conditions in South America (pp: 140–144). Colonia, Uruguay.
German, S. (2007). Roya de la hoja en cultivos de invierno: Epidemiologia de la enfermedad y comportamiento varietal. Jornada de cultivos de invierno, Young, Serie actividades de difusion 484.
HarvEST. (2011). Available at http://harvest.ucr.edu/HBarley178.exe. Accessed 5 May 2011.
Hayes, P., & Szücs, P. (2006). Disequilibrium and association in barley: Thinking outside the glass. Proceedings of the National Academy of Sciences of the United States of America, 103(49), 18385–18386.
Jannink, J. L., Bink, M. C. A. M., & Jansen, R. C. (2001). Using complex plant pedigrees to map valuable genes. Trends in Plant Science, 6, 337–342.
Kraakman, A. T. W., Niks, N. E., van den Berg, P. M. M. M., Stam, P., & van Eeuwijk, F. A. (2004). Linkage disequilibrium mapping of yield and yield stability in modern spring barley cultivars. Genetics, 168, 435–446.
Kraakman, A. T. W., Martinez, F., Mussiraliev, B., van Eeuwijk, F. A., & Niks, R. E. (2006). Linkage disequilibrium mapping of morphological, resistance, and other agronomically relevant traits in modern spring barley cultivars. Molecular Breeding, 17, 41–58.
Mather, D. E., Tinker, N. A., Laberge, D. E., Edney, M., Jones, B. L., Rossnagel, B. G., Legge, W. G., Briggs, K. G., Irvine, R. B., Falk, D. E., & Kasha, K. J. (1997). Regions of the genome that affect grain and malt quality in a North American two row barley cross. Crop Science, 37, 544–554.
Nutter, E. W., Pederson, V. D., & Foster, A. E. (1985). Effect of inoculations with Cochliobolus sativus at species growth stages on grain yield and quality of malting barley. Crop Science, 25, 933–938.
Parisseaux, B., & Bernardo, R. (2004). Insilico mapping of quantitative trait loci in maize. Theoretical and Applied Genetics, 109, 508–514.
Pereyra, S. (1996). Manejo de enfermedades en cereales de invierno y pasturas, Montevideo, INIA. Serie Tecnica, 74, 110–111.
Price, A. L., Patterson, N. J., Plenge, R. M., Weinblatt, M. E., Shadick, N. A., & Reich, D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics, 38, 904–909.
Roy, J. K., Smith, K. P., Muehlbauer, G. J., Chao, S., Close, T. J., & Steffenson, B. J. (2010). Association mapping of spot blotch resistance in wild barley. Molecular Breeding, 26, 243–256.
Stracke, S., Haseneyer, G., Veyrieras, J.-B., Geiger, H. H., Sauer, S., Graner, A., & Piepho, H.-P. (2009). Association mapping reveals gene action and interactions in the determination of flowering time in barley. Theoretical and Applied Genetics, 118, 259–273.
Szűcs, P., Blake, V. C., Bhat, P. R., Close, T. J., Cuesta-Marcos, A., Muehlbauer, G. J., Ramsay, L. V., Waugh, R., & Hayes, P. M. (2009). An integrated resource for barley linkage map and malting quality QTL alignment. Plant Genome, 2, 134–140.
von Zitzewitz, J., Cuesta-Marcos, A., Condon, F., Castro, A. J., Chao, S., Corey, A., Filichkin, T., Fisk, S. P., Gutierrez, L., Haggard, K., Karsai, I., Muehlbauer, G. J., Smith, K. P., Veisz, O., & Hayes, P. M. (2011). The genetics of winter hardiness in barley: Perspectives from genome-wide association mapping. Plant Genome, 4, 76–91.
Waugh, R., Jannink, J.-L., Muehlbauer, G. J., & Ramsay, L. (2009). The emergence of whole genome association scans in barley. Current Opinion in Plant Biology, 12, 218–222.
Yu, P., Hart, K., & Du, L. (2009). An investigation of carbohydrate and protein degradation ratios, nitrogen to energy synchronization, and hourly effective rumen digestion of barley: Effect of variety and growth year. Journal of Animal Physiology and Animal Nutrition, 93, 555–567.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Zhejiang University Press and Springer Science+Business Media Dordrecht
About this paper
Cite this paper
Gutiérrez, L. et al. (2013). Genome-Wide Association Mapping Identifies Disease-Resistance QTLs in Barley Germplasm from Latin America. In: Zhang, G., Li, C., Liu, X. (eds) Advance in Barley Sciences. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-4682-4_18
Download citation
DOI: https://doi.org/10.1007/978-94-007-4682-4_18
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-4681-7
Online ISBN: 978-94-007-4682-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)