Abstract
Initiation of DNA synthesis in eukaryotic replication depends on the Pol α-primase complex, a multi-protein complex endowed with polymerase and primase activity. The Pol α-primase complex assembles the RNA-DNA primers required by the processive Pol δ and Pol ε for bulk DNA synthesis on the lagging and leading strand, respectively. During primer synthesis, the primase subunits synthesise de novo an oligomer of 7–12 ribonucleotides in length, which undergoes limited extension with deoxyribonucleotides by Pol α. Despite its central importance to DNA replication, little is known about the mechanism of primer synthesis by the Pol α-primase complex, which comprises the steps of initiation, ‘counting’ and hand-off of the RNA primer by the primase to Pol α, followed by primer extension with dNTPs and completion of the RNA-DNA hybrid primer. Recent biochemical and structural work has started to provide some insight into the molecular basis of initiation of DNA synthesis. Important advances include the structural characterisation of the evolutionarily related archaeal primase, the elucidation of the mechanism of interaction between Pol α and its B subunit and the observation that the regulatory subunit of the primase contains an iron-sulfur cluster domain that is essential for primer synthesis.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
9.1 Introduction
In the semi-conservative model of DNA replication each parental strand acts as a template for the synthesis of a complementary DNA chain. DNA polymerases are responsible for synthesising the novel strands of nucleic acid accurately and efficiently. However, polymerisation depends on the presence of an existing oligonucleotide primer annealed to the template that is extended by addition of deoxynucleotides to the 3′ hydroxyl group of the primer.
In order to be able to begin duplication of their genome, all organisms have evolved a specialised DNA-dependent RNA polymerase, termed primase, which is endowed with the unique ability to initiate DNA synthesis de novo (Frick and Richardson 2001; Kuchta and Stengel 2010). Primases can assemble from the ribonucleotide pool of the cell the short RNA primers that are utilised by the DNA polymerase for processive synthesis. The antiparallel arrangement of the parental strands and the obligate 5′–3′ direction of synthesis of DNA polymerase imply that DNA synthesis needs to be primed repeatedly on the lagging strand. Thus, primase activity is constantly required at the replication fork.
Priming DNA synthesis is a universal requirement of DNA replication and consequently primases are present in all kingdoms of life. Certain important features of primase activity, such as the tendency to initiate with a purine base and to synthesise RNA primers of between 5 and 10 nucleotides, appear to be universal. However, prokaryotic and eukaryotic primases differ radically in structural organisation and mechanism of primer synthesis. Whereas bacterial primase is a single polypeptide, in eukaryotic cells the primase is a heterodimer of catalytic or small subunit (PriS) and regulatory or large subunit (PriL). In addition, in eukaryotic cells the primase is normally associated in a specific, constitutive complex with DNA polymerase α (Pol α) and its B subunit (Muzi-Falconi et al. 2003). The resulting heterotetramer forms the Pol α-primase complex, the multi-subunit protein assembly that initiates DNA synthesis in eukaryotic replication. In the complex, the RNA primer synthesised by the primase undergoes limited extension with deoxynucleotides by Pol α. The resulting RNA-DNA primer is then utilised for processive synthesis by Pol ε and Pol δ on the leading and lagging strands, respectively.
The first reports describing the isolation of primase activity and the biochemical characterisation of the Pol α-primase complex in cell extracts of eukaryotic organisms date to almost 30 years ago (Conaway and Lehman 1982; Hubscher 1983; Kaufmann and Falk 1982; Plevani et al. 1984; Yagura et al. 1982). Despite extensive experimental efforts, our understanding of the molecular steps of RNA primer initiation, completion and limited elongation with dNTPs by the Pol α-primase complex is still surprisingly limited. In this chapter, I will review the current state of knowledge concerning the structural information currently available for the Pol α-primase complex and describe how the present data inform our understanding of the mechanism of primer synthesis in eukaryotic replication.
9.2 Primase
9.2.1 Prim Fold of the Catalytic Subunit
The eukaryotic primase is a heterodimer of catalytic or small subunit (PriS) and regulatory or large (PriL) subunit. Most of our knowledge of its three-dimensional architecture comes from crystallographic analysis of evolutionarily related archaeal primases, which show clear sequence similarity and subunit organisation with their eukaryotic counterparts.
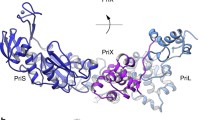
To date, two crystal structures for the isolated catalytic subunit of archaeal primases have been reported (Augustin et al. 2001; Ito et al. 2003) (Fig. 9.1a). The crystallographic analysis showed that the core structural elements of the catalytic subunit fold in a novel DNA-dependent RNA polymerase domain, the ‘prim’ fold, that has at its core two flanged beta sheets accommodating the active site and surrounded by alpha helices on the outside. Overall, the structure of the catalytic subunit has a flat, slab-like appearance with a very exposed active site, entirely different from the classic ‘right-hand-like’ fold of DNA polymerases. The structure of the catalytic subunit is completed by a smaller, species-specific domain that can occupy different positions on the rim of the core ‘prim’ fold (Fig. 9.1a).
Structure of the archaeal/eukaryotic primase. (a) Ribbon diagrams of the catalytic subunits of archaeal primases from Sulfolobus solfataricus (Sso, PDB 1zt2) and Pyrococcus furiosus (Pfu, PDB 1 g71) are shown in grey. In the middle of the panel, a superposition of the two structures is shown, coloured according to structure similarity: parts of the structure of the catalytic subunits that are similar are drawn in red, whereas divergent regions are drawn in blue. The colouring scheme highlights the elements of secondary structure that constitute the ‘prim’ fold. (b) Ribbon representation of the crystal structure of the Sulfolous solfataricus primase in its heterodimeric form. The catalytic subunit (PriS) is drawn is green and the large subunit (PriL) in orange. The PriL polypeptide present in the crystal is a C-terminally truncated version that lacks the Fe-S cluster domain (see Fig. 9.2)
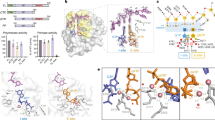
Although the archaeal/eukaryotic primase catalytic subunit fold is novel, the mechanism of nucleotide polymerization is likely to be similar to the general enzymatic mechanism of nucleic acid synthesis. Thus, a triad of aspartate residues have been identified that are invariant across archaeal and eukaryotic primases, and which are deemed to be necessary for catalytic activity (Augustin et al. 2001; Ito et al. 2003) (Fig. 9.2a). It therefore appears that convergent evolution has driven primases to adopt the same mechanism of catalysis, involving two divalent metal ions, proposed for other DNA and RNA polymerases (Yang et al. 2006). The position of the active site was confirmed by diffusion of uridine triphosphate (UTP) in the Pyrococcus horikoshii primase crystals (Ito et al. 2003) (Fig. 9.2a).
Active site architecture of the archaeal/eukaryotic primase. (a) Close-up view of the archaeal primase from Pyrococcus horikoshii (PDB 1v34) with a molecule of UTP bound in the active site. The side chains of the aspartate residues forming the putative catalytic triad are also shown. (b) Structure of the conserved C-terminal domain of the large subunit of the yeast primase. The [Fe4S4] cofactor and the cysteine ligands are also shown in ball-and-stick representation. (c) Superimposition of the yeast PriL-CTD on the active site of cryptochrome Cry3 from A. thaliana, bound to FAD and ssDNA. The PriL-CTD is drawn as a thin tube in blue, the DNA photolyase in green. FAD and ssDNA are shown as sticks, coloured according to element type. (d) A possible model for the essential role of the PriL-CTD in initiation of RNA primer synthesis. In the model, the PriL-CTD is an integral part of the active site and assists the catalytic subunit PriS in binding the two initial ribonucleotides at the initiation site on the template DNA
A highly conserved feature of the catalytic subunit is the presence of a zinc-binding motif situated on the same side of the catalytic subunit as the active site; its putative functional role remains uncertain but its proximity to the active site suggests an involvement in the mechanism of catalysis (Fig. 9.1a). Intriguingly, bacterial primases also contain a zinc-binding domain that has been implicated in binding single-stranded DNA (Corn et al. 2005). It is tempting to speculate that the zinc-binding motif in archaeal and eukaryotic primases might fulfill comparable functional roles. However, in contrast to what observed in the archaeal primase, the zinc- binding motif of the bacterial primase is located in a separate domain of the polypeptide chain.
The prim fold is probably of ancient evolutionary derivation as it has been found in a number of different genetic contexts (Iyer et al. 2005), including multifunctional enzymes with combined primase and helicase activities, encoded in the genome of the bacterium Bacillus cereus (McGeoch and Bell 2005) and in a plasmid of the archaeon Sulfolobus islandicus (Lipps et al. 2003), as well as in certain multi-functional bacterial enzymes active in non-homologous end joining pathway of DNA repair in mycobacteria (Brissett et al. 2007).
9.2.2 The Archaeal/Eukaryotic Primase Is an Iron-Sulfur Protein
The archaeal/eukaryotic primase is a constitutive heterodimer of two subunits, PriS and PriL. The role of the non-catalytic polypeptide in primase activity has remained mysterious for a long time, however it is known that PriL performs an essential role in the priming reaction (Zerbe and Kuchta 2002). Indeed, PriL is an essential gene in yeast (Foiani et al. 1989).
Recent crystallographic and biochemical analysis of the archaeal/eukaryotic primase has made important progress in understanding the role of PriL in RNA primer synthesis. The structure of a truncated form of the heterodimeric primase from the thermophilic archaeon S. solfataricus, lacking the conserved C-terminal portion of PriL, illuminated the relative special arrangement of the two subunits (Lao-Sirieix et al. 2005) (Fig. 9.1b): PriL is a mainly helical polypeptide that extends from the narrow edge of the slab-shaped catalytic subunit and is better understood as a structural ‘arm’ that serves to position its conserved C-terminal domain (PriL-CTD) in the correct position for catalysis. Indeed, the PriL-CTD is essential in the initiation reaction of the heterodimeric primase, but not for its polymerase activity, pointing to a crucial function of the PriL-CTD in the initial step of di-nucleotide synthesis.
Surprisingly, it was found that PriL-CTD contains a [Fe4S4] cofactor (Klinge et al. 2007; Weiner et al. 2007). The iron-sulfur centre appears to have a structural role in maintaining the C-terminal sequence of PriL in its correct three-dimensional shape (Agarkar et al. 2011; Sauguet et al. 2010; Vaithiyalingam et al. 2010) (Fig. 9.2b). At this stage, functional roles of the Fe-S cofactor depending on its redox status remain speculative but cannot be ruled out.
The molecular mechanism underlying PriL-CTD’s involvement in primer synthesis remains unclear, but an intriguing clue as to how this might happen comes from the unexpected structural similarity between PriL-CTD and the active site of DNA photolyase/cryptochrome family of DNA repair enzymes (Sauguet et al. 2010) (Fig. 9.2c). The mode of binding of single-stranded (ss) DNA and flavin adenine dinucleotide (FAD) observed in the co-crystal structure of the DASH cryptochrome 3 from Arabidposis thaliana (Pokorny et al. 2008) suggests that the PriL-CTD could adopt a similar mode of interaction with the template DNA and ribonucleotides during de novo RNA synthesis. In particular, the spacial relationship between FAD and the extruded, cross-linked pyrimidine dimer observed in the active site of DNA photolyase suggests a possible arrangement for the pairing of the first dinucleotide of the RNA primer onto template DNA during the initiation step catalysed by the primase. Thus, the PriL-CTD might participate in RNA primer synthesis by assisting the catalytic subunit PriS in the simultaneous binding of the two initial RNA nucleotides and by promoting dinucleotide base-pairing with template DNA at the initiation site (Fig. 9.2d).
9.3 DNA Polymerase α
9.3.1 Catalytic Activity
In contrast to prokaryotic and bacteriophage replication, where the primer is composed exclusively of RNA, in eukaryotic replication the primer is a hybrid RNA-DNA molecule. The polymerase responsible for synthesising the deoxy-nucleotide portion of the primer is DNA polymerase α (Pol α), a member of the B-family of DNA polymerases and the first DNA polymerase to be detected in mammalian cells. The role of Pol α in priming synthesis is to extend the RNA primer synthesised by the primase with deoxy-nucleotides, in order to assemble an RNA-DNA oligonucleotide of about 20–25 nucleotides. According to current models, transfer of the 3′-hydroxyl of the RNA primer between active sites of the primase and Pol α takes place via an intra-molecular hand-off (Copeland and Wang 1993; Eki et al. 1991; Kuchta et al. 1990; Sheaff et al. 1994). However, the nature of the molecular switch that regulates the transfer is still unknown. The mature RNA-DNA primer is then utilised by DNA Polymerase δ and ε for processive synthesis on the lagging and leading strands, respectively (Stillman 2008) (see Chaps. 12 and 13). Upon completion of lagging strand synthesis and joining of the Okazaki fragments, the primer is excised by the combined action of Pol δ and PCNA, which displace the 5′ end of the downstream fragment, and the enzymatic activities of the Fen1 nuclease and the Dna2 nuclease-helicase, which excise 5′ flaps of ssDNA (Burgers 2009).
A common feature of all three replicative polymerases – α, δ and ε – is the presence of a conserved region that extends past the polymerase fold to a cysteine-rich C-terminal region (CTD) that is necessary for DNA replication and cell viability. A conserved pattern of eight cysteine residues indicated that the CTD of the polymerase likely binds two metal ions, such as Zn2+. Biochemical evidence implicates this region of the polymerase in the interaction with the primase (Mizuno et al. 1999).
9.3.2 Structure of the B Subunit and Its Interaction with Pol α
The three major replicative DNA polymerases – Pols α, δ and ε – share unifying features of their subunit organisation that reveal a clear evolutionary relationship (Fig. 9.3a) (Johansson and MacNeill 2010) (see also Chap. 12 and 13). Of their different cohorts of accessory subunits, only the so-called B subunit is present in all three polymerase assemblies and is clearly conserved in eukaryotic organisms (Aravind and Koonin 1998; Makiniemi et al. 1999). Interestingly, an orthologue of the B subunit has also been found in archaeal organisms as the single accessory polypeptide of a replicative polymerase.
Structure of the yeast Pol α CTD – B subunit complex. (a) Cartoons of the three multi-subunit replicative DNA polymerases α, δ and ε, highlighting the conserved B subunit (blue) and Pol CTDs (orange). (b) Crystal structure of the Pol α CTD – B complex. The B subunit is shown as a orange ribbon, and the Pol α CTD as blue ribbon. The position and extend of the oligonucleotide/oligosaccharide (OB) and inactive nuclease domains are shown, as well as the location of the two zinc atoms
Reflecting their high degree of conservation, the catalytic and B subunits are the only indispensable polymerase components. A large body of experimental evidence has highlighted the functional importance of the CTD interaction with the B subunit (Dua et al. 1998; Mizuno et al. 1999; Sanchez Garcia et al. 2004). Thus, a heterodimer of catalytic and B subunit represents the conserved functional core of the three replicative polymerases.
Recent crystallographic evidence has demonstrated the mode of interaction between the CTD of yeast Pol α and its B subunit (Klinge et al. 2009) (Fig. 9.3b). The B subunit fold derives from the intimate association of an N-terminal oligonucleotide/oligosaccharide (OB) domain with an inactive C-terminal phosphoesterase domain. The CTD of yeast Pol α adopts an elongated bilobal shape reminiscent of an asymmetrically proportioned saddle. Each lobe contains a zinc-binding module: the lobe with the four N-terminal cysteine ligands (Zn-1) is larger and includes additional secondary structure elements as well as irregular coil structure; the lobe with the four C-terminal cysteine ligands is smaller and formed entirely by the zinc-binding module (Zn-2). The two lobes are connected by a three-helix bundle that represents the central portion or ‘backbone’ of the saddle-shaped CTD.
The two zinc-binding motifs bear a clear structural relationship to each other; in both cases, metal-binding results from the ‘handshake’ interaction of two β ribbons, each providing a pair of cysteine ligands for the tetrahedral coordination of the zinc atom. However, the crystal structure indicates that the two metal-binding motifs take on distinct functional roles. The Zn-2 motif is an integral part of the Pol α CTD – B subunit interface, whereas the Zn-1 motif is removed from the interface and more exposed to solvent, possibly poised for interactions with other protein factors or DNA.
The large surface area (4,500 Å2) buried at the interface between the Pol α CTD and the B subunit indicates a tight association between the two polypeptides, supporting the notion that the B subunit performs a function that requires its constitutive association to Pol α. The presence of a phosphodiesterase domain juxtaposed to an OB domain suggests that the evolutionary ancestor of the B subunit might have possessed an enzymatic function, perhaps required alongside the polymerase activity of Pol α. In support of this hypothesis, some nuclease activity appears to have been retained by the archaeal orthologue of the eukaryotic B subunit (Jokela et al. 2004; Shen et al. 2004). Thus, it appears that, in the course of evolution of the replication machinery, the B subunit has morphed from a nuclease into a protein scaffold element responsible for mediating multiple and concomitant protein-protein interactions within the eukaryotic replisome (Baranovskiy et al. 2008; Klinge et al. 2009).
9.4 Towards a Concerted Mechanism for Primer Synthesis by the Pol α-Primase Complex
As already mentioned, the nucleic acid primer that begins synthesis in eukaryotic replication is a hybrid RNA-DNA molecule. Virtually nothing is known about the molecular switch that coordinates the hand-off of the nascent primer between active sites of primase and Pol α. However, given the high frequency of initiation events at the replication fork and the necessity to synchronize priming of synthesis with fork progression and processive synthesis of bulk DNA, it seems reasonable to assume that a coordinated, intra-molecular mechanism of primer transfer between primase and Pol α must exist. Indeed, experimental evidence exists in support of such a model (Copeland and Wang 1993; Eki et al. 1991; Kuchta et al. 1990; Sheaff et al. 1994).
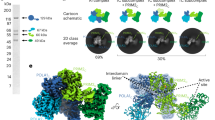
The physical association of the primase and polymerase polypeptides provides a physical basis for their functional coupling. Recent electron microscopy reconstructions of the 3D-architecture of the Pol α- B subunit (Klinge et al. 2009) and of the Pol α-primase complex (Nunez-Ramirez et al. 2011) shows that it is organised as a dumbbell-shaped particle with flexibly connected lobes (Fig. 9.4a, b). The catalytic domain of Pol α resides in one lobe, whereas the other lobe is formed by the interaction of the Pol α CTD, the B subunit and the two primase subunits. The high degree of mobility observed in the relative orientation of the two catalytic lobes might be functionally relevant for the hand-off mechanism of the RNA primer between primase and Pol α (Fig. 9.4c). Thus, it is possible to envisage the process of RNA–DNA primer synthesis as requiring a series of transitions between specific conformational conformations of the Pol α-primase, leading in turn to RNA primer synthesis, transfer to and extension with dNTPs by Pol α (Fig. 9.4c). Confirmation of such mechanism will require the structural characterization of the Pol α-primase complex in its key intermediate states during primer synthesis.
Structure of the accessory B subunit and its interaction with Pol α. (a) Ribbon model of the Pol α CTD – B complex structure is fitted into the EM reconstruction of the Pol α – B complex. The catalytic domain of the archaeal polymerase from T. gorgonarius (PDB 2VWJ) has also been fitted in the EM density. Two rotated views of the EM reconstruction are shown, represented as white transparent density. The Pol α CTD and the catalytic domain of the archaeal polymerase are shown in blue and the B subunit in orange. (b) Collection of 2D reference-free averages of the Pol α-primase. Each row shows a set of averages where the Pol lobe is oriented according to the same view. The conformational flexibility of the primosome is highlighted by a cartoon outline showing the superposition of all averages in a row. Scale bar represents 15 nm. (c) Hypothetical diagram of the steps of RNA–DNA primer synthesis, based on a conformational rearrangement of the Pol and Prim lobes during primer transfer
9.5 Outlook
Elucidating in detail the molecular mechanisms by which the Pol α-primase complex initiates nucleic acid synthesis is essential to our comprehension of DNA replication. Despite its central role in the process of genomic duplication, how the complex assembles the RNA–DNA oligonucleotides that prime DNA synthesis remains surprisingly obscure. The evidence discussed here represents a hopeful sign that rapid progress will be made in providing a structural basis for the enzymatic activity of the of Pol α-primase complex. Indeed, crystallographic models of archaeal or eukaryotic derivation for almost all the proteins or protein domains of the Pol α-primase complex are now available. The challenge for the future will be to integrate our current structural knowledge in a complete picture of the necessary enzymatic steps underlying RNA–DNA primer synthesis in eukaryotic replication.
References
Agarkar VB, Babayeva ND, Pavlov YI, Tahirov TH (2011) Crystal structure of the C-terminal domain of human DNA primase large subunit: implications for the mechanism of the primase-polymerase α switch. Cell Cycle 10(6):926–931
Aravind L, Koonin EV (1998) Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res 26(16):3746–3752
Augustin MA, Huber R, Kaiser JT (2001) Crystal structure of a DNA-dependent RNA polymerase (DNA primase). Nat Struct Biol 8(1):57–61
Baranovskiy AG, Babayeva ND, Liston VG, Rogozin IB, Koonin EV, Pavlov YI, Vassylyev DG, Tahirov TH (2008) X-ray structure of the complex of regulatory subunits of human DNA polymerase δ. Cell Cycle 7(19):3026–3036
Brissett NC, Pitcher RS, Juarez R, Picher AJ, Green AJ, Dafforn TR, Fox GC, Blanco L, Doherty AJ (2007) Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science 318(5849):456–459
Burgers PM (2009) Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem 284(7):4041–4045
Conaway RC, Lehman IR (1982) A DNA primase activity associated with DNA polymerase alpha from Drosophila melanogaster embryos. Proc Natl Acad Sci USA 79(8):2523–2527
Copeland WC, Wang TS (1993) Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J Biol Chem 268(35):26179–26189
Corn JE, Pease PJ, Hura GL, Berger JM (2005) Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol Cell 20(3):391
Dua R, Levy DL, Campbell JL (1998) Role of the putative zinc finger domain of Saccharomyces cerevisiae DNA polymerase ε in DNA replication and the S/M checkpoint pathway. J Biol Chem 273(45):30046–30055
Eki T, Enomoto T, Masutani C, Miyajima A, Takada R, Murakami Y, Ohno T, Hanaoka F, Ui M (1991) Mouse DNA primase plays the principal role in determination of permissiveness for polyomavirus DNA replication. J Virol 65(9):4874–4881
Foiani M, Santocanale C, Plevani P, Lucchini G (1989) A single essential gene, PRI2, encodes the large subunit of DNA primase in Saccharomyces cerevisiae. Mol Cell Biol 9(7):3081–3087
Frick DN, Richardson CC (2001) DNA primases. Annu Rev Biochem 70:39–80
Hubscher U (1983) The mammalian primase is part of a high molecular weight DNA polymerase α polypeptide. EMBO J 2(1):133–136
Ito N, Nureki O, Shirouzu M, Yokoyama S, Hanaoka F (2003) Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis. Genes Cells 8(12):913–923
Iyer LM, Koonin EV, Leipe DD, Aravind L (2005) Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res 33(12):3875–3896
Johansson E, MacNeill SA (2010) The eukaryotic replicative DNA polymerases take shape. Trends Biochem Sci 35:339–347
Jokela M, Eskelinen A, Pospiech H, Rouvinen J, Syväoja JE (2004) Characterization of the 3′ exonuclease subunit DP1 of Methanococcus jannaschii replicative DNA polymerase D. Nucleic Acids Res 32(8):2430–2440
Kaufmann G, Falk HH (1982) An oligoribonucleotide polymerase from SV40-infected cells with properties of a primase. Nucleic Acids Res 10(7):2309–2321
Klinge S, Hirst J, Maman JD, Krude T, Pellegrini L (2007) An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat Struct Mol Biol 14(9):875–877
Klinge S, Nunez-Ramirez R, Llorca O, Pellegrini L (2009) 3D architecture of DNA Pol α reveals the functional core of multi-subunit replicative polymerases. EMBO J 28(13):1978–1987
Kuchta RD, Stengel G (2010) Mechanism and evolution of DNA primases. Biochim Biophys Acta 1804(5):1180–1189
Kuchta RD, Reid B, Chang LM (1990) DNA primase. Processivity and the primase to polymerase α activity switch. J Biol Chem 265(27):16158–16165
Lao-Sirieix SH, Nookala RK, Roversi P, Bell SD, Pellegrini L (2005) Structure of the heterodimeric core primase. Nat Struct Mol Biol 12(12):1137–1144
Lipps G, Rother S, Hart C, Krauss G (2003) A novel type of replicative enzyme harbouring ATPase, primase and DNA polymerase activity. EMBO J 22(10):2516–2525
Makiniemi M, Pospiech H, Kilpelainen S, Jokela M, Vihinen M, Syvaoja JE (1999) A novel family of DNA-polymerase-associated B subunits. Trends Biochem Sci 24(1):14–16
McGeoch AT, Bell SD (2005) Eukaryotic/archaeal primase and MCM proteins encoded in a bacteriophage genome. Cell 120(2):167–168
Mizuno T, Yamagishi K, Miyazawa H, Hanaoka F (1999) Molecular architecture of the mouse DNA polymerase α-primase complex. Mol Cell Biol 19(11):7886–7896
Muzi-Falconi M, Giannattasio M, Foiani M, Plevani P (2003) The DNA polymerase α-primase complex: multiple functions and interactions. ScientificWorldJournal 3:21–33
Nunez-Ramirez R, Klinge S, Sauguet L, Melero R, Recuero-Checa MA, Kilkenny M, Perera RL, Garcia-Alvarez B, Hall RJ, Nogales E, Pellegrini L, Llorca O (2011) Flexible tethering of primase and DNA Pol α in the eukaryotic primosome. Nucleic Acids Res 39(18):8187–8199
Plevani P, Badaracco G, Augl C, Chang LM (1984) DNA polymerase I and DNA primase complex in yeast. J Biol Chem 259(12):7532–7539
Pokorny R, Klar T, Hennecke U, Carell T, Batschauer A, Essen LO (2008) Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc Natl Acad Sci USA 105(52):21023–21027
Sanchez Garcia J, Ciufo LF, Yang X, Kearsey SE, MacNeill SA (2004) The C-terminal zinc finger of the catalytic subunit of DNA polymerase δ is responsible for direct interaction with the B-subunit. Nucleic Acids Res 32(10):3005–3016
Sauguet L, Klinge S, Perera RL, Maman JD, Pellegrini L (2010) Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PLoS One 5(4):e10083
Sheaff RJ, Kuchta RD, Ilsley D (1994) Calf thymus DNA polymerase α-primase: “communication” and primer-template movement between the two active sites. Biochemistry 33(8):2247–2254
Shen Y, Tang XÄ, Yokoyama H, Matsui E, Matsui I (2004) A 21-amino acid peptide from the cysteine cluster II of the family D DNA polymerase from Pyrococcus horikoshii stimulates its nuclease activity which is Mre11-ike and prefers manganese ion as the cofactor. Nucleic Acids Res 32(1):158–168
Stillman B (2008) DNA polymerases at the replication fork in eukaryotes. Mol Cell 30(3):259–260
Vaithiyalingam S, Warren EM, Eichman BF, Chazin WJ (2010) Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proc Natl Acad Sci 107(31):13684–13689
Weiner BE, Huang H, Dattilo BM, Nilges MJ, Fanning E, Chazin WJ (2007) An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J Biol Chem 282(46):33444–33451
Yagura T, Kozu T, Seno T (1982) Mouse DNA replicase. DNA polymerase associated with a novel RNA polymerase activity to synthesize initiator RNA of strict size. J Biol Chem 257(18):11121–11127
Yang W, Lee JY, Nowotny M (2006) Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell 22(1):5
Zerbe LK, Kuchta RD (2002) The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry 41(15):4891–4900
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Pellegrini, L. (2012). The Pol α-Primase Complex. In: MacNeill, S. (eds) The Eukaryotic Replisome: a Guide to Protein Structure and Function. Subcellular Biochemistry, vol 62. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-4572-8_9
Download citation
DOI: https://doi.org/10.1007/978-94-007-4572-8_9
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-4571-1
Online ISBN: 978-94-007-4572-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)