Abstract
DNA polymerase δ (Pol δ) is a member of the B-family DNA polymerases and is one of the major replicative DNA polymerases in eukaryotes. In addition to chromosomal DNA replication it is also involved in DNA repair and recombination. Pol δ is a multi-subunit complex comprised of a catalytic subunit and accessory subunits. The latter subunits play a critical role in the regulation of Pol δ functions. Recent progress in the structural characterization of Pol δ, together with a vast number of biochemical and functional studies, provides the basis for understanding the intriguing mechanisms of its regulation during DNA replication, repair and recombination. In this chapter we review the current state of the Pol δ structure-function relationship with an emphasis on the role of its accessory subunits.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
12.1 Introduction
Eukaryotic DNA polymerase δ (Pol δ) belongs to the B-family of DNA polymerases (Pavlov et al. 2006). It addition to its central role in chromosomal DNA replication (Downey et al. 1990; Garg and Burgers 2005), Pol δ is involved in DNA repair and recombination (Baranovskiy et al. 2008; Gibbs et al. 2005; Huang et al. 2000, 2002; Lawrence 2002; Lydeard et al. 2007). In concert with Pol α–primase, it can synthesize both leading and lagging DNA strands in the SV40 system and in yeast strains carrying a catalytically inactive Pol ε (Kesti et al. 1999; Waga et al. 1994). The best studied Pol δs are those from human, the fission yeast Schizosaccharomyces pombe (Sp) and the budding yeast Saccharomyces cerevisiae (Sc). The former two Pol δs are found as four-subunit complexes (Liu et al. 2000; Zuo et al. 1997), while the latter Pol δ is found only as a three-subunit complex (Gerik et al. 1998). A unified nomenclature for the subunits of these three Pol δs has been proposed (MacNeill et al. 2001) and is provided in Table 12.1 along with updated comments on their roles and intersubunit interactions.
Currently published data on intermolecular interactions supports the overall architecture of Pol δ in which the A-, B- and D-subunits interact with each other and the C-subunit interacts only with the B-subunit, while all four subunits somehow interact with PCNA (Fig. 12.1). Progress on the three-dimensional structural characterization of Pol δ was slow for a long time and limited to the crystal structure of the p66 C-terminal peptide with PCNA (Bruning and Shamoo 2004). Initial significant progress was achieved when the crystal structure was solved for a complex of human p50 subunit with the N-terminal p50-interacting domain of the p66 subunit (Baranovskiy et al. 2008). The next notable progress made was the crystal structure determination of the Pol3p catalytic core in ternary complex with a template primer and an incoming nucleotide (Swan et al. 2009). Based on these recent achievements and a vast amount of biochemical, biophysical and functional studies, in this chapter we provide a review rationalizing the structure-function relationship for Pol δ. The first part of the chapter is devoted to the crystal structure of the A-subunit catalytic core and possible structural consequences of cancer-causing mutations, the organization and role of the A-subunit C-terminal domain (CTD), its metal-binding motifs (MBM) and comparisons with CTDs of other B-family members. The second part is devoted to a complex of the B- and C-subunits, its interaction with CTD and the interaction of the C-subunit with PCNA. The final third part is devoted to the least structurally characterized D-subunit, its interactions and functions, and speculation on the possibility of its evolution from the B-subunit.
12.2 Catalytic Subunit (A-subunit)
The purification of eukaryotic Pol δ from the cytoplasm of erythroid hyperplastic bone marrow was first reported in 1976 (Byrnes et al. 1976). Pol δ has been shown to possess both polymerase and 3′–5′ exonuclease activities within a purified single 122 kDa polypeptide (Goscin and Byrnes 1982). This polypeptide is now known as a catalytic subunit of Pol δ and designated as an A-subunit (Table 12.1). The proofreading exonuclease activity significantly enhances the fidelity of the A-subunit (Byrnes 1984; Simon et al. 1991). Cloning and sequencing of Sp, Sc, bovine and human Pol δ A-subunits revealed that, in general, their structures are similar (Chung et al. 1991; Pignede et al. 1991; Zhang et al. 1991). Primary structure analysis indicates that A-subunits could be considered as comprising of less conserved N-terminal sequences, the highly conserved catalytic core and CTD. Until recently, information for understanding the three-dimensional structure of the Pol δ catalytic core was derived mainly from the crystal structures of bacteriophage RB69 as well as its replicating and editing complexes (Franklin et al. 2001; Shamoo and Steitz 1999; Wang et al. 1997). Recently the 2.0 Å resolution crystal structure of Sc Pol3p catalytic core (residues 68–985) in ternary complex with a template primer and an incoming dCTP nucleotide has been reported by Swan and co-authors (Swan et al. 2009). This structure, which is reviewed below, enables precise mapping of functionally important residues and provides the basis for targeted genetic and functional studies.
12.2.1 Crystal Structure of Catalytic Core
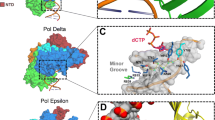
The overall structure of the Pol3p catalytic core replicating complex is shown in Fig. 12.2a. The DNA is located in a clasp comprised of the thumb, palm, fingers and exonuclease domains. The thumb forms extensive contacts with the sugar-phosphate backbones of template primer around the minor groove while the palm interacts with the replicative end of the DNA. The nascent G-dCTP base pair packs against the second α-helix (αP) of the fingers domain. A long β-hairpin of exonuclease domain is extended toward the DNA major groove forming a hydrogen bond with a DNA base and a number of van der Waals contacts with the backbone of a template strand. Finally, the N-terminal domain (NTD), which is wedged between the exonuclease and fingers domains and interacting with the palm domain, contacts only the unpaired segment of the template strand. Two active site residues (Asp608 and Asp764) that catalyze the nucleotidyl transfer reaction are in the palm domain. The exonuclease active site residues Asp321, Glu323 and Asp407 are located close to a single Ca2+. The distance between the polymerase and exonuclease active sites is ~45 Å.
A-subunit structure. (a) Cartoon representation of Sc Pol δ catalytic core (PDB code 3iay) (Swan et al. 2009). The NTD, palm, thumb, fingers, and exonuclease domains are color coded. The side chain atoms of exonuclease and polymerase active site residues are shown as light green and light cyan balls, respectively. The calcium ions are shown as blue balls. The side chain atoms of Sc Pol δ residues corresponding to cancer-causing mutations are shown as red balls. DNA and dCTP molecules are drawn as sticks. The panel (a) was prepared with PyMol software (Delano Scientific). (b) Comparison of human Pol α CTD amino acid sequences with Pol ε, and Pol δ with Pol ζ. The secondary structure elements predicted with Phyre server (Kelley and Sternberg 2009) and metal-coordinating cysteines are marked
Pol3p catalytic core exhibits several notable differences compared with the RB69 Pol. Their structures can be superimposed with a root-mean-square deviation of 2.50 Å for 631 matching α-carbons. The most notable differences are in NTD and the fingers domain. The NTD is significantly larger in Pol3p and contains of an OB-fold and a RNA-binding motif, raising the possibility of RNA involvement in Pol δ function. Unlike the NTD, the fingers domain in Pol3p is comprised of only two shorter and straighter antiparallel α-helices than in RB69 Pol. Another significant difference is in DNA recognition by the long β-hairpin of exonuclease domain. In RB69 Pol structure this β-hairpin does not associate with DNA. Contrarily, in the Pol3p structure it is tightly bound to the template, which is consistent with its role in strand separation. The strand separation is proposed to be facilitated by holding the template strand in place by a β-hairpin that allows the primer strand to separate and migrate to the exonuclease active site (Hogg et al. 2007; Stocki et al. 1995).
12.2.2 Cancer-Causing Mutations
High-fidelity DNA replication is necessary for cell to avoid disease-causing mutations. In the case of Pol δ, DNA replication fidelity is maintained by accurate nucleotide selectivity and exonucleolytic proofreading activity. Furthermore, the DNA mismatch repair pathway contributes to an additional increase of fidelity. Engineering mutations leading to defects in Pol δ fidelity in mice increased the genome instability and accelerated tumorigenesis, supporting the mutator hypothesis for cancer (Albertson et al. 2009; Goldsby et al. 2001, 2002; Venkatesan et al. 2007). Unlike the mutations in mice Pol δ, which were intentionally modeled to destroy specific polymerase functions, the mechanisms of actions of cancer-causing mutations in the human Pol δ A-subunit are not apparent. However, using the structure of the Pol3p catalytic core it became possible to map the location of cancer-causing mutations and provide the rationale for their structure-function relationship (Swan et al. 2009)
Examination of Pol δ mRNA in six colon cancer cell lines (DLD-1, HCT116, SW48, HT29, SW480 and SW620) and seven sporadic human colorectal cancers revealed Arg506His, Arg689Trp and Ser746Ile substitutions located in the catalytic core (Fig. 12.2a – note that in the figure these are numbered according to their location in Sc Pol3p, as described below) (da Costa et al. 1995; Flohr et al. 1999).
Arg506 of p125 is aligned with Arg511 of Pol3p that is located in a long α-helix (αI) opposite the active site of the exonuclease domain. Replacement of this arginine residue by histidine may alter its interactions with the surrounding residues from loop β13β14 and consequently shift the position of the αI helix. Such a shift of the αI helix may cause an allosteric effect on the exonuclease activity of Pol δ. The Arg511His substitution in yeast Pol δ resulted in a small but significant (2.5-fold) increase in the rate of mutagenesis in a MMR-deficient strain (Daee et al. 2010). Because the amino acid sequences and hence local structures are different between the yeast and human Pol δs, a more profound effect of Arg506His substitution is a possibility for human Pol δ.
Arg689 of p125 is the equivalent of Pol3p Arg696 which is located in the second α-helix (αP) of the fingers domain. The Arg696 side chain is sandwiched between the NTD and the fingers domain. Modeling of Trp696 in place of Arg696 shows that, in any allowable conformation, the bulky tryptophan side chain creates significant clashes with the surrounding residues. In order to release the clashes and sterical hindrance introduced by Arg696Trp substitution, it is necessary to have a significant shift of, and/or conformational changes in, the αP helix. Because the αP helix is one of the most important determinants for the selectivity of incoming nucleotide, any shift in its position will inevitably affect the accuracy of base selection. Indeed, the Arg696Trp substitution is lethal in yeast, with the lethality caused by a catastrophic increase in spontaneous mutagenesis attributed to low-fidelity DNA synthesis (Daee et al. 2010).
The effect of Ser746Ile mutation on the catalytic activities of Pol δ is less clear. The equivalent residue in Pol3p is Lys753 which is located in loop αQβ24 at the edge of the palm domain. The Lys753 side chain is completely solvent-exposed. However, it is possible that the local structure of the loop αQβ24 is different in human Pol δ and that Ser746 is less exposed; consequently, its replacement by a bulkier isoleucine may result in conformational changes. Another possibility is that substitution of serine by isoleucine introduces conformational changes in the loop αQβ24 by packing the isoleucine side chain in the nearby hydrophobic pocket. In both such scenarios an allosteric effect of Ser746Ile mutation to the polymerase active site is possible.
Pol δ from highly malignant Novikoff hepatoma cells was found to contain an Arg648Gln substitution (Popanda et al. 1999). Several biochemical characteristics of this polymerase were found to be altered, including a decrease in copying fidelity. The Arg648 equivalent in Pol3p is Arg658, which is located in loop β23αO connecting the fingers and palm domains (Fig. 12.2a). Thus the conformational changes caused by Arg648Gln substitution could allosterically alter the selectivity of incoming nucleotide via the shift of the αO and αP helices.
In summary, an inspection of the structure-function relationship of the reported cancer-causing amino acid replacements in the Pol δ A-subunit catalytic core indicates that all these mutations have a potential to alter its exonuclease or polymerase activities via allosteric effects. That is consistent with the mutator properties of Pol δ in cancer.
12.2.3 C-terminal Domain
Two putative zinc finger motifs (ZnF1 and ZnF2) each with four conserved metal-coordinating cysteines has been identified in the CTD of Pol δs from a variety of sources (Sanchez Garcia et al. 2004). ZnF2 was found to be both necessary and sufficient for binding to the B-subunit in vivo and in vitro. The ZnF2 metal-binding site is critical for the activity of Sc and Sp Pol δs since substitution of ZnF2 cysteines with alanines abolishes B-subunit binding and their in vivo function. Recently Netz and co-authors have established that the cysteines in ZnF2, but not in ZnF1, coordinate the [4Fe-4S] cluster instead of the previously assumed zinc ion (Netz et al. 2011). They also determined that the ZnF1 is required for PCNA-binding and PCNA-mediated Pol δ processivity. However the nature of the metal in ZnF1 motif has not yet been clarified and in further discussions the ZnF1 and ZnF2 will be referenced as the metal-binding motifs 1 and 2 (MBM1 and MBM2).
At present, the only reported CTD structure is the structure of yeast Pol α CTD in complex with its B-subunit (Klinge et al. 2009) (see Chap. 9, this volume). Pol α CTD forms an elongated bi-lobal shape. The two lobes are connected by a three-helix bundle and each lobe contains an MBM. In contrast to what has been observed with the Pol δ CTD, the conserved cysteines in both MBM1 and MBM2 of Pol α CTD are coordinated with zinc ions. The CTD of Pol α forms a stable complex with the B-subunit. However, in case of Pol δ the CTD interaction with the B-subunit is dependent on the stability of the Fe-S cluster. The complex becomes unstable and dissociates at ambient conditions due to loss of iron from the Fe-S cluster. In addition to the differences in MBM2 metal content, the primary structure of Pol δ CTD is nearly two times shorter than in Pol α CTD. The secondary structure prediction of Pol δ CTD using the Phyre server (Kelley and Sternberg 2009) revealed three helices alternated with MBM1 and MBM2 in the order: helix-MBM1-helix-MBM2-helix (Fig. 12.2b). Assuming the resemblance of overall architecture of Pol α and Pol δ CTDs, the predicted helices in Pol δ CTD would be packed as a three-helix bundle flanked by the MBM1 and MBM2 lobes. However, unlike Pol α CTD having central three-helix bundle separate of MBMs, MBM2 in particular, the three-helix bundle of Pol δ CTD appears to comprise a part of its MBMs. In Pol δ the first conserved cysteine residue of MBM1 is located at the C-terminus of the first helix while the first and second conserved cysteine residues of MDM2 are protruding from the second helix, pointing to significant structural differences between the MBMs of Pol α and Pol δ. Contrarily, in Pol α MBMs the metal-coordinating cysteines are located in either loops or β-strands. The discrepancy in structure is especially apparent for MBM2 which results in the different nature of their ligands: Zn2+ in Pol α and [4Fe-4S] cluster in Pol δ.
12.2.4 Similarities Between C-terminal Domains of Pol δ and Pol ζ
Unlike the bulkier CTDs of Pol α and Pol ε, the CTD of Pol ζ appears to have a structure which is highly similar to Pol δ’s CTD (Fig. 12.2b). The alignment of amino acid sequences and secondary structures of the latter two CTDs shows that the length and positions of their helices as well as the positions of metal-coordinating cysteines coincide (Fig. 12.2b). Their similarity raises the possibility of similarity in the structures of their binding partners as well. The CTDs of Pol α, Pol ε and Pol δ are known to bind their respective B-subunits. However, a B-subunit was not found in Pol ζ. The three following explanations can be posited for this. First, Pol ζ does not possess a B-subunit. Second, a B-subunit was not discovered due to low expression levels or other technical complications. Third, Pol ζ shares a B-subunit with Pol δ. The latter case would fit with a plausible model of efficient polymerase switch at replication-blocking lesions during translesion DNA synthesis (TLS). According to such a model, Polδ and Polζ will exchange only their catalytic subunits on a preassembled complex while the shared accessory subunits of Pol δ will be retained by DNA-loaded PCNA. This model is in accordance with previously observed involvement of Pol δ accessory subunits in TLS (Gerik et al. 1998; Gibbs et al. 2005; Huang et al. 2000, 2002; Lydeard et al. 2007).
12.3 B- and C-subunits
The B- and C-subunits of both yeast and mammalian Pol δs have been cloned, purified and characterized (Gerik et al. 1998; Hughes et al. 1999; Mo et al. 2000; Zhang et al. 1995; Zuo et al. 1997). The sequence homology is high among the B-subunits with amino acid sequence similarity of 23–28%. However, the homology is low among the C-subunits. Using the two-hybrid screening, human p66 has been shown to contain p50- and PCNA-binding domains within the 144 N- and 20 C-terminal amino acids, respectively (Pohler et al. 2005). A similar domain arrangement was found also for Pol32p (Johansson et al. 2004). The B-subunit interacts with the A-, C- and D-subunits, as well as PCNA (Lu et al. 2002). The C-subunit also interacts with PCNA (Bruning and Shamoo 2004; Zhong et al. 2006). The parts of the B-subunit responsible for interactions with the D-subunit have not been defined. Interestingly, many essential functions of Pol δ, including the regulation of replication, TLS and BIR, are mediated by the C-subunit (Gerik et al. 1998; Gibbs et al. 2005; Huang et al. 2000, 2002; Lydeard et al. 2007) and thus apparently depend on the interaction between its B- and C-subunits. The crystal structure of human p50 in complex with the N-terminal domain of human p66 is described below.
12.3.1 Crystal Structure of p50•p66N
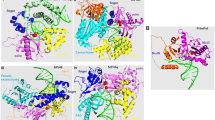
The B-subunit of the human Pol δ is the first among B-subunits of the B-family DNA polymerases whose crystal structure has been solved, providing insights into the overall architecture of B-subunits and their structure-function relationship (Baranovskiy et al. 2008). The structure of p50 was determined in complex with the 144-amino-acids N-terminal domain of the C-subunit (p66N) and the complex hereinafter will be referred as p50•p66N. The structure revealed an extended cashew-shaped molecule with three domains: phosphodiesterase-like (PDE) and oligonucleotide/oligosaccharide-binding (OB) domains in p50, and a winged helix-turn-helix (wHTH) domain in p66N (Fig. 12.3a). The PDE domain is located in the center of the molecule and bound to an OB domain on one side, and a wHTH domain on the other side. The fusion of the PDE and OB domains is a distinct feature of the second subunits of replicative B-family DNA polymerases covering all species from archaea to humans (Aravind and Koonin 1998; Makiniemi et al. 1999).
B- and C-subunit structures. (a) Cartoon representation of p50•p66N (PDB code 3iay) (Baranovskiy et al. 2008). On the left side view the secondary structure elements are color coded and labeled. α-helices, β-strands and coils are red, yellow and green in p50, and cyan, magenta and light pink in p66N. The disordered loops are shown as dotted lines. The expected CTD docking site is highlighted in pink color. On the right side the molecule was rotated to optimize the view of domains. The wHTH, PDE and OB domains are color coded. (b) Cartoon representation of p66 (residues 453–466) in complex with human PCNA (PDB code 1u76) (Bruning and Shamoo 2004). Two orthogonal views are shown. The surface of one PCNA protomer is shown with bound p66 peptide represented as sticks. Both panels were prepared with PyMol software (Delano Scientific)
The PDE domain of p50 is comprised of a two-layer β-sheet with two α-helices flanking on one side, and three α-helices flanking on the other side. However, unlike the catalytically active PDE domains, the B-subunits of the eukaryotic B-family DNA polymerases, including p50, do not contain conserved histidine and aspartate residues involved in metal coordination and catalysis (Aravind and Koonin 1998). Instead, the catalytically inactive PDE domains of eukaryotic polymerases play a central role in protein-protein interactions. They have nearly twice the size and surface area compared with catalytically active PDE domains. For example, comparison of p50 with the structure of a monomeric single subunit archaeal phosphodiesterase protein MJ0936 (Chen et al. 2004) (PDB code 1s3l) highlights significant differences in their structures. The MJ0936 has a compact globular shape with a surface area of 6,579 Å2. In contrast, the PDE domain of p50 has a surface area of 12,858 Å2 and contains many protruding parts and flexible loops. These differences make p50 a perfect docking platform for Pol δ subunits and possibly for other regulatory proteins.
The OB domain is connected with the PDE domain via two linkers, 17 interdomain hydrogen bonds and several scattered interdomain hydrophobic contacts. The OB domain consists of a five-stranded β-barrel (Murzin 1993) wrapped by a bended α-helix on one side. Another side of the β-barrel remains fully accessible and may have a potential for DNA and protein binding as in the case of the OB domains of replication protein A (RPA) (Bochkarev et al. 1997; Bochkareva et al. 2005).
p50-bound p66N is comprised of a four-stranded, anti-parallel β-sheet surrounded by five α-helices and a C-terminal tail, and has a V-shaped structure. It contains a novel variation of the wHTH motif that is often found in dsDNA-binding domains of transcriptional activators and repressors.
12.3.2 p50•p66 Interactions
p66N appears as an extension of p50 because one of the layers of a PDE domain two-layer β-sheet is joined with the β-sheet of p66 via parallel β-strands, resulting in an extended 10-stranded β-sheet (Fig. 12.3a). In addition, one more inter-subunit β-β interaction is observed between the parallel β-strands (the first β-strand of p50 and the last β-strand of p66N). Overall, 37 inter-subunit hydrogen bonds, including 12 main-chain to main-chain hydrogen bonds, inter-subunit hydrophobic interactions covering one larger area and three smaller areas and inter-subunit electrostatic interactions contribute to p50•p66N dimer formation. The latter interactions are formed by the negatively charged p66-interacting surface of p50 and the complementary, positively charged surface of p66. The p50•p66 complex formation produces significantly large (5,398 Å2) surface area buried at the dimer interface
Because of extended inter-subunit surface and a large number of main-chain to main-chain hydrogen bonds, the impact of one or two interacting side chains truncations (for example by substitution to alanine) to the stability of p50•p66 complex is expected to be low. However, amino acid substitution with a bulky side chain at the interface, which results in a sterical hindrance, or substitution with a side chain, which results in a same charge repulsion, would be the efficient ways to prevent the complex formation. Three sites in p50 (Leu217, Gly224 and Glu231) were selected to generate single and double tryptophan substitutions, aiming to disrupt the p50•p66 interactions by sterical hindrance and the interactions were examined by two-hybrid assays. The data confirmed that sterical hindrance causing amino acid substitutions leads to disruption of p50•p66 interaction.
12.3.3 Functional Studies
The crystal structure of Pol δ p50•p66N provides a platform for the explanation of earlier accumulated genetic and functional studies as well as to set up novel structure-based rational genetic and functional studies. Surprisingly, the structure also revealed that in spite of the functional importance, the residues from the p50•p66 dimerization interface belong to the least conserved regions. This significantly complicated the mapping of p50•p66 interacting residues by genetic studies. As result, the earlier genetic studies of p50 orthologs in yeast, Pol31p (Vijeh Motlagh et al. 2006) and Sp Cdc1 (MacNeill et al. 1996) were performed mainly targeting the highly conserved regions. For example, the temperature-sensitive Pol31p mutants (Vijeh Motlagh et al. 2006) are mapped to the secondary structure elements of p50 that are necessary for its proper folding. After inspection of the structure of p50•p66N we know that these temperature-sensitive phenotypes are due to deformations in the overall folding of Pol31. In another example the deletion of the C-terminal 20 residues of Cdc1 abolished its interaction with Cdc27 in yeast two-hybrid assays (MacNeill et al. 1996). Indeed, corresponding residues of p50 provide 7 out of the 12 inter-subunit backbone interactions with p66. It has also become apparent why the deletion of up to 100 N-terminal residues of the Pol32p abolishes its interaction with Pol31p (Johansson et al. 2004).
Sc Polδ was used to verify the functional importance of the interaction between the B- and C-subunits. The Pol31p•Pol32p complex was disrupted by a combination of charge repulsion and sterical hindrance. Two structure-guided Pol31p variants were constructed. In the first Pol31p variant, an Arg-Arg-Gly is inserted in the loop located in the center of the Pol31p•Pol32p interface. In the second Pol31p variant, an Arg-Arg-Gly insertion was combined with two point mutations (Gly242Trp, Asp249Trp). These mutations were introduced into the yeast strain allowing for concomitant detection of induced and spontaneous mutations in three genetic loci (Pavlov et al. 2002). Neither of the mutations led to a growth defect or temperature sensitivity, indicating that the mutations in this essential gene do not affect the general properties of the second subunit. However, in comparison to the parent strain, both mutations led to mild cold-sensitivity, increased sensitivity to hydroxyurea, elevated sensitivity to UV light irradiation and reduced UV mutagenesis. Overall, the mutations behaved as strains with the complete deletion of the third subunit. These experiments confirm that the mutations to affect specifically the interaction between the B- and C-subunits, which is consistent with the structure-predicted functional consequences of the amino acid changes.
In another set of experiments by the MacNeill group, random and directed mutagenesis techniques were used to create a collection of 30 genes encoding mutant Cdc1 proteins (Sanchez Garcia et al. 2009). Each mutant protein was tested for function in fission yeast and for binding to Pol3 and Cdc27 using the two-hybrid system. Mapping of the location of each mutant onto the three-dimensional structure of p50 provided a plausible explanation for in vivo function of each Cdc1 mutant and also identified the amino acid residues and regions within the Cdc1 protein that are essential for interaction with Pol3 and Cdc27. Mutants specifically defective in Cdc1•Cdc27 and Cdc1•Pol3 interactions allowed the identification of Cdc27- and Pol3-binding areas on Cdc1. The Cdc27-binding area of Cdc1 coincides with the p66-binding area of p50. The Cdc1 mutations disrupting Pol3 binding were found mainly to affect the four disordered regions which are localized in close proximity to each other and protruding to the same surface area (Fig. 12.3a). Based on these observations, the disordered area of Cdc1 was predicted to bind the CTD of Pol3. Similar conclusions were obtained by Maloisel group for Sc Pol δ (Brocas et al. 2010). Later, the crystal structure of the Pol α B-subunit in complex with the C-terminal domain of its catalytic subunit has been reported (Klinge et al. 2009). The structure confirmed that in Pol δ, the B-subunit area that was predicted to interact with the CTD matches well with the corresponding interaction area in the Pol α B-subunit.
12.3.4 Crystal Structure of p66•PCNA
The crystal structure of the C-terminal PCNA-interacting protein motif domain (or PIP-box) of p66 (residues 452–466) has been determined in complex with human PCNA (Fig. 12.3b) (Bruning and Shamoo 2004). The structure shows that the binding mode of p66 and site of interaction is similar to the PIP-box structures of FEN1 and p21CIP1 (Bruning and Shamoo 2004; Gulbis et al. 1996). As in these structures, the PIP-box of p66 is comprised of an extended N-terminal region, a central conserved region containing 310-helix, and a region C-terminal to the PIP-box. A PIP-box exists in proteins involved in cell cycle regulation and DNA processing. Sequential and regulated binding of PIP-box-containing proteins to PCNA has been suggested to contribute to the ordering of events during DNA replication and repair (Warbrick 2000).
12.4 D-subunit
The fourth D-subunit of Pol δ was the latest addition to the list of Pol δ subunits. Hurwitz and coauthors purified and characterized the composition of Sp Pol δ (Zuo et al. 1997). Further analysis of the complex revealed four subunits with molecular masses of 125, 55, 54 and 22 kDa (Zuo et al. 2000). Western blot analysis confirmed that the first three subunits correspond to Pol3, Cdc1, and Cdc27, respectively. The identity of the smallest subunit was determined by sequence analysis, indicating that this is identical to Cdm1, which was previously described as a multi-copy suppressor of the temperature-sensitive cdc1-P13 mutant (MacNeill et al. 1996). Unlike Sp Pol δ, Sc Pol δ is comprised of three subunits only and lacks the D-subunit (Gerik et al. 1998).
Shortly after the discovery of D-subunit in yeast Pol δ, Lee and coauthors discovered the D-subunit of mammalian Pol δ exhibiting a 25% identity to Cdm1 (Liu et al. 2000). The D-subunit of human Pol δ, designated as p12, is a 107 amino acid protein with a molecular mass of 12.4 kDa. The four-subunit human Pol δ complex was reconstituted by overexpression of the four subunits in Sf9 insect cells by two groups (Podust et al. 2002; Xie et al. 2002). The purified complex displayed a specific activity comparable with that of the human, bovine, and fission yeast proteins isolated from natural sources. Recently a large-scale production of human Pol δ has been achieved by its overexpression in silkworm larvae, providing a basis for its better structural and functional characterization, especially the role of p12 (Zhou et al. 2011).
12.4.1 D-subunit Structure and Inter-Subunit Interactions
A stable three-subunit Sp Pol δ complex containing Pol3, Cdc1, and Cdm1 was reconstituted by simultaneous expression of corresponding polypeptides in baculovirus-infected insect cells (Zuo et al. 2000). The presence of such a complex pointed to an interaction of Cdm1 with either or both Pol3 and Cdc1 (Zuo et al. 2000). Further clarification of D-subunit interactions was reported in the case of human Pol δ. The yeast two-hybrid and pull-down assays revealed that p12 interacts with both the catalytic subunit and the B-subunit of Pol δ (Li et al. 2006).
The three-dimensional structure of the D-subunit from any source has not been determined. Compared with other Pol δ subunits the human p12 is a relatively small polypeptide. Secondary structure predictions using the Phyre server (Kelley and Sternberg 2009) shows that N-terminal residues may fold as two short β-strands followed by a long stretch of disordered region that is rich in acidic residues and three well-defined helices that are located at C-terminal half. The detailed mapping of p12 regions interacting with the catalytic subunit and the B-subunit as well as determination of its docking areas on the surface of these subunits has not been performed. In addition to linking the Pol δ with PCNA, interactions with p50 and p125 subunits (Li et al. 2006) point to several likely scenarios for the architectural role of p12 within the Pol δ complex. One possible scenario is when p12 binds simultaneously to the p50 and the CTD of p125. However, it may also form a bridge between the p50 and the catalytic core of p125, or even interact with both CTD and the catalytic core of p125 as well as p50. The p50-interacting CTD is flexible relative to the catalytic core of p125 (Jain et al. 2009). Consequently, in the latter two scenarios p12 will reduce the overall flexibility of the Pol δ complex.
As it was mentioned above, Sc Pol δ lacks the D-subunit and functions as a three-subunit complex (Gerik et al. 1998) indicating that some of these subunits may compensate, at least in part, for the function of the D-subunit. Analysis of the amino acid sequences of Sc Pol δ subunits revealed a highly surprising similarity between the amino acid sequences close to the C-terminal part of Pol31 and the human p12 (Tahirov, unpublished data, see Fig. 12.4). This supports the idea that upon the evolution the D-subunit was derived by duplication of the part of B-subunit. In particular, Pol31 contains an insertion that is rich in acidic residues. Such acidic sequences are absent in B-subunits of four-subunit Pol δs but may have migrated to their D-subunits. Another interesting finding is an apparent matching of three tryptophan residues in Pol31 and p12. An evolutionary relationship could be observed upon changing from a lower eukaryotic organism to a higher eukaryotic organism: during these changes, fewer of these tryptophan residues remained in B-subunit and more appeared in D-subunit, but their total number in both subunits remained at least three (Fig. 12.4). For example, in the case of Sp Pol δ only one tryptophan appears in Cdm1 and two remain in Cdc1, and, finally, in the case of human Pol δ all four tryptophan residues appear in p12 and none remain in p50.
Amino-acid sequence alignment of B- and D-subunits of Pol δ from different organisms. Hs (human), Dr (Danio rerio), At (Arabidopsis thaliana), Sp (S. pombe) and Sc (S. cerevisiae). Note that S. cerevisiae does not encode a D-subunit. The aspartate and glutamate residues in Asp/Glu-rich insert of the S. cerevisiae B-subunit Pol31p and in corresponding regions of D-subunits are marked in cyan. The tryptophan residues discussed in Sect. 12.4.1 are marked in magenta
The revealed evolutionary relationship of the B- and D-subunits along with the crystal structure of the B-subunit (Baranovskiy et al. 2008) may help to map the location of Pol31 sequences that “migrated” to form the D-subunit, and hence provide ideas about potential sites where the D-subunit might dock. In particular, the position of the insertion that is rich in acidic residues is mapped to the loop between the α-helices α7 and α8 of B-subunit, and the sequences harboring the tryptophan residues are in an extended loop between the α-helix α8 and β-strand β14 of the B-subunit. Both of these loops protrude towards the B-subunit surface that is proposed to bind the C-terminal domain of the catalytic subunit (Fig. 12.3a) (Sanchez Garcia et al. 2009). In summary, these observations point out that the most likely scenario for D-subunit docking is when the D-subunit binds simultaneously to the B-subunit and the C-terminal domain of the catalytic subunit close to their interaction interface; also, additional interaction with the catalytic core cannot be excluded since p12 alters the catalytic activity of Pol δ (Meng et al. 2009, 2010).
12.4.2 D-subunit Function
Earlier studies with Sp Pol δ showed that cells deleted for the D-subunit are viable, indicating that the D-subunit is non-essential for Sp mitotic growth (Reynolds et al. 1998). Subsequent characterization of the D-subunit and its functional role were extensively pursued using mainly human Pol δ. In initial studies the four-subunit Pol δ and three-subunit Pol δ lacking the p12 subunit were reconstituted by co-expressing the corresponding subunits in insect cells (Podust et al. 2002; Xie et al. 2002). The four-subunit Pol δ activity was comparable to that isolated from natural sources and it efficiently replicated singly primed M13 DNA in the presence of RPA, PCNA, and replication factor C (RFC) and was active in the SV40 DNA replication system (Podust et al. 2002). The DNA polymerizing activity of three-subunit Pol δ was 15-fold lower than that of the four-subunit Pol δ. However, addition of p12 stimulated the activity of the three-subunit Pol δ fourfold on poly(dA)-oligo(dT) template-primer, confirming that the p12 subunit is required to reconstitute fully active recombinant human DNA polymerase (Podust et al. 2002). In further studies, a PCNA-binding motif KRLITDSY (residues 4–11) at the N terminus of p12 was determined by binding assays (Li et al. 2006). The interaction with PCNA was confirmed by site-directed mutagenesis by replacing the p12 residues Ile7, Ser10, and Tyr11 with alanines. Comparison of DNA polymerizing activities of four-subunit Pol δ containing wild-type p12 with Pol δ containing mutated p12 with lost PCNA binding has been performed in the presence of increasing amounts of PCNA using singly primed M13 DNA as the template. The data revealed that Pol δ with mutant p12 is less active than the wild-type Pol δ in the presence of similar concentrations of PCNA. Thus, based on obtained results authors proposed that p12 not only stabilizes Pol δ but also plays an important role in integration of Pol δ with PCNA (Li et al. 2006).
Lee and coauthors have recently discovered that genotoxic agents such as UV and alkylating chemicals trigger a DNA damage response in which the four-subunit Pol δ is converted to a three-subunit Pol δ by rapid degradation of p12 (Meng et al. 2009, 2010; Zhang et al. 2007). The p12 degradation required an active ubiquitin pathway and could be inhibited by proteasome inhibitors. Degradation was regulated by ATR kinase that controls the damage response in S-phase. The three-subunit Pol δ exhibits an increased capacity for proofreading 3′–5′ exonuclease activity compared with four-subunit Pol δ (Meng et al. 2010). In particular, it cleaves single-stranded DNA two times faster and transfers mismatched DNA from the polymerase site to the exonuclease site nine times faster. However, Pol δ extends mismatched primers three times more slowly in the absence of p12.
In parallel studies, Suzuki and coauthors applied the short hairpin RNA (shRNA) to control the levels of p12 in a variety of human cells (Huang et al. 2010a). Reduction of p12 resulted in a marked decrease in colony formation activity by Calu6, ACC-LC-319, and PC-10 cells but increased population of karyomere-like cells. The latter cells retained an ability to progress through the cell cycle suggesting that p12 reduction induces modest genomic instability (Huang et al. 2010a). Furthermore they also found that reduced expression of p12 plays a role in genomic instability in lung cancer (Huang et al. 2010b).
The p12 subunit has been shown to be a target of posttranslational modifications. Analysis of the p12 sequence for putative phosphorylation sites revealed a single potential casein kinase 2 (CK2) phosphorylation site at Ser24 (Gao et al. 2008). Mutation of this serine to aspartate resulted in resistance to phosphorylation, confirming that Ser24 is indeed a target for CK2 phosphorylation. Dephosphorylation of Pol δ subunits, including p12, is catalyzed by the protein phosphatase-1 (PP1) (Gao et al. 2008). In addition to phosphorylation and dephosphorylation, the p12 subunit was found to be target for ubiquitination (Liu and Warbrick 2006). However the regulation of the levels of p12 by the proteasome was found to be through a mechanism that is not dependent upon p12 ubiquitination (Liu and Warbrick 2006). The relation of these post-translational modifications to the above-described Pol δ functions has not been established.
p12 alone and within Pol δ has been reported to interact specifically in vitro and in vivo with BLM (the Bloom’s syndrome protein) and stimulate its DNA helicase activity, and, reciprocally, BLM stimulates Pol δ strand displacement activity (Selak et al. 2008). A region of p12 comprising amino acids 31–60 was responsible for interaction with BLM that indeed includes the disordered acidic region of p12. Interestingly, BLM was found to co-localize with Pol δ and prevent the degradation of p12 when cells were treated with hydroxyurea (HU). These findings for the first time provide a link between BLM and the replicative machinery in human cells and points to the possible involvement of Pol δ in Bloom’s syndrome (BS) (Selak et al. 2008). BS, which is caused by mutation of the BLM gene, is associated with excessive chromosomal instability and a high incidence of cancers of all types.
12.5 Conclusions and Prospects
Decades of research has resulted in the identification and characterization of eukaryotic B-family DNA polymerases. Now it is evident that the accessory subunits of these high-molecular weight complexes are playing crucial roles in regulating their activity. However, deciphering the exact mechanisms of their action remains highly challenging. Among the major hurdles toward achieving this goal is the lack of comprehensive high-resolution three-dimensional structural information, especially for the complexes containing accessory subunits. As we described in this chapter, in the case of Pol δ, partial but significant progress has recently been achieved is this direction. Importantly, the crystal structures were determined for the human Pol δ p50•p60N complex and the Sc Pol δ catalytic core. However, other important pieces of the puzzle related to Pol δ structure and function remain unknown. These include the structure of the D-subunit and its complexes with other subunits of Pol δ, the structure of the CTD with its bound iron-sulfur cluster, the interaction of the CTD with the B-subunit, the structure of the C-subunit C-terminal, the effect of post-translational modifications, the interaction of Pol δ subunits with PCNA, the interaction of Pol δ subunits with other polymerases and regulatory factors, etc. Further collective efforts of structural biologists will be required to find the missing pieces of this puzzle and put them together in order to achieve a more complete understanding of Pol δ.
References
Albertson TM, Ogawa M, Bugni JM, Hays LE, Chen Y, Wang Y, Treuting PM, Heddle JA, Goldsby RE, Preston BD (2009) DNA polymerase ε and δ proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci U S A 106:17101–17104
Aravind L, Koonin EV (1998) Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res 26:3746–3752
Baranovskiy AG, Babayeva ND, Liston VG, Rogozin IB, Koonin EV, Pavlov YI, Vassylyev DG, Tahirov TH (2008) X-ray structure of the complex of regulatory subunits of human DNA polymerase δ. Cell Cycle 7:3026–3036
Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L (1997) Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 385:176–181
Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, Bochkarev A (2005) Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci U S A 102:15412–15417
Brocas C, Charbonnier JB, Dherin C, Gangloff S, Maloisel L (2010) Stable interactions between DNA polymerase δ catalytic and structural subunits are essential for efficient DNA repair. DNA Repair (Amst) 9:1098–1111
Bruning JB, Shamoo Y (2004) Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase δ p66 subunit and flap endonuclease-1. Structure 12:2209–2219
Byrnes JJ (1984) Structural and functional properties of DNA polymerase delta from rabbit bone marrow. Mol Cell Biochem 62:13–24
Byrnes JJ, Downey KM, Black VL, So AG (1976) A new mammalian DNA polymerase with 3′ to 5′ exonuclease activity: DNA polymerase δ. Biochemistry 15:2817–2823
Chen S, Yakunin AF, Kuznetsova E, Busso D, Pufan R, Proudfoot M, Kim R, Kim SH (2004) Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii. J Biol Chem 279:31854–31862
Chung DW, Zhang JA, Tan CK, Davie EW, So AG, Downey KM (1991) Primary structure of the catalytic subunit of human DNA polymerase δ and chromosomal location of the gene. Proc Natl Acad Sci U S A 88:11197–11201
da Costa LT, Liu B, el-Deiry W, Hamilton SR, Kinzler KW, Vogelstein B, Markowitz S, Willson JK, de la Chapelle A, Downey KM et al (1995) Polymerase δ variants in RER colorectal tumours. Nat Genet 9:10–11
Daee DL, Mertz TM, Shcherbakova PV (2010) A cancer-associated DNA polymerase δ variant modeled in yeast causes a catastrophic increase in genomic instability. Proc Natl Acad Sci U S A 107:157–162
Downey KM, Tan CK, So AG (1990) DNA polymerase δ: a second eukaryotic DNA replicase. Bioessays 12:231–236
Flohr T, Dai JC, Buttner J, Popanda O, Hagmuller E, Thielmann HW (1999) Detection of mutations in the DNA polymerase δ gene of human sporadic colorectal cancers and colon cancer cell lines. Int J Cancer 80:919–929
Franklin MC, Wang J, Steitz TA (2001) Structure of the replicating complex of a pol α family DNA polymerase. Cell 105:657–667
Gao Y, Zhou Y, Xie B, Zhang S, Rahmeh A, Huang HS, Lee MY, Lee EY (2008) Protein phosphatase-1 is targeted to DNA polymerase δ via an interaction with the p68 subunit. Biochemistry 47:11367–11376
Garg P, Burgers PM (2005) DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol 40:115–128
Gerik KJ, Li X, Pautz A, Burgers PM (1998) Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J Biol Chem 273:19747–19755
Gibbs PE, McDonald J, Woodgate R, Lawrence CW (2005) The relative roles in vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6–4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics 169:575–582
Goldsby RE, Lawrence NA, Hays LE, Olmsted EA, Chen X, Singh M, Preston BD (2001) Defective DNA polymerase δ proofreading causes cancer susceptibility in mice. Nat Med 7:638–639
Goldsby RE, Hays LE, Chen X, Olmsted EA, Slayton WB, Spangrude GJ, Preston BD (2002) High incidence of epithelial cancers in mice deficient for DNA polymerase δ proofreading. Proc Natl Acad Sci U S A 99:15560–15565
Goscin LP, Byrnes JJ (1982) DNA polymerase δ: one polypeptide, two activities. Biochemistry 21:2513–2518
Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J (1996) Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell 87:297–306
Hogg M, Aller P, Konigsberg W, Wallace SS, Doublie S (2007) Structural and biochemical investigation of the role in proofreading of a β hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J Biol Chem 282:1432–1444
Huang ME, de Calignon A, Nicolas A, Galibert F (2000) Pol32, a subunit of the Saccharomyces cerevisiae DNA polymerase δ, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr Genet 38:178–187
Huang ME, Rio AG, Galibert MD, Galibert F (2002) Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase δ, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics 160:1409–1422
Huang QM, Akashi T, Masuda Y, Kamiya K, Takahashi T, Suzuki M (2010a) Roles of POLD4, smallest subunit of DNA polymerase δ, in nuclear structures and genomic stability of human cells. Biochem Biophys Res Commun 391:542–546
Huang QM, Tomida S, Masuda Y, Arima C, Cao K, Kasahara TA, Osada H, Yatabe Y, Akashi T, Kamiya K, Takahashi T, Suzuki M (2010b) Regulation of DNA polymerase POLD4 influences genomic instability in lung cancer. Cancer Res 70:8407–8416
Hughes P, Tratner I, Ducoux M, Piard K, Baldacci G (1999) Isolation and identification of the third subunit of mammalian DNA polymerase δ by PCNA-affinity chromatography of mouse FM3A cell extracts. Nucleic Acids Res 27:2108–2114
Jain R, Hammel M, Johnson RE, Prakash L, Prakash S, Aggarwal AK (2009) Structural insights into yeast DNA polymerase δ by small angle X-ray scattering. J Mol Biol 394:377–382
Johansson E, Garg P, Burgers PM (2004) The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J Biol Chem 279:1907–1915
Kelley LA, Sternberg MJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371
Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C (1999) DNA polymerase ε catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell 3:679–685
Klinge S, Nunez-Ramirez R, Llorca O, Pellegrini L (2009) 3D architecture of DNA Pol α reveals the functional core of multi-subunit replicative polymerases. EMBO J 28:1978–1987
Lawrence CW (2002) Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair (Amst) 1:425–435
Li H, Xie B, Zhou Y, Rahmeh A, Trusa S, Zhang S, Gao Y, Lee EY, Lee MY (2006) Functional roles of p12, the fourth subunit of human DNA polymerase δ. J Biol Chem 281:14748–14755
Liu G, Warbrick E (2006) The p66 and p12 subunits of DNA polymerase δ are modified by ubiquitin and ubiquitin-like proteins. Biochem Biophys Res Commun 349:360–366
Liu L, Mo J, Rodriguez-Belmonte EM, Lee MY (2000) Identification of a fourth subunit of mammalian DNA polymerase δ. J Biol Chem 275:18739–18744
Lu X, Tan CK, Zhou JQ, You M, Carastro LM, Downey KM, So AG (2002) Direct interaction of proliferating cell nuclear antigen with the small subunit of DNA polymerase δ. J Biol Chem 277:24340–24345
Lydeard JR, Jain S, Yamaguchi M, Haber JE (2007) Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448:820–823
MacNeill SA, Moreno S, Reynolds N, Nurse P, Fantes PA (1996) The fission yeast Cdc1 protein, a homologue of the small subunit of DNA polymerase δ, binds to Pol3 and Cdc27. EMBO J 15:4613–4628
MacNeill SA, Baldacci G, Burgers PM, Hubscher U (2001) A unified nomenclature for the subunits of eukaryotic DNA polymerase δ. Trends Biochem Sci 26:16–17
Makiniemi M, Pospiech H, Kilpelainen S, Jokela M, Vihinen M, Syvaoja JE (1999) A novel family of DNA-polymerase-associated B subunits. Trends Biochem Sci 24:14–16
Meng X, Zhou Y, Zhang S, Lee EY, Frick DN, Lee MY (2009) DNA damage alters DNA polymerase δ to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res 37:647–657
Meng X, Zhou Y, Lee EY, Lee MY, Frick DN (2010) The p12 subunit of human polymerase δ modulates the rate and fidelity of DNA synthesis. Biochemistry 49:3545–3554
Mo J, Liu L, Leon A, Mazloum N, Lee MY (2000) Evidence that DNA polymerase δ isolated by immunoaffinity chromatography exhibits high-molecular weight characteristics and is associated with the KIAA0039 protein and RPA. Biochemistry 39:7245–7254
Murzin AG (1993) OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J 12:861–867
Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ (2011) Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol 8:125–132
Pavlov YI, Newlon CS, Kunkel TA (2002) Yeast origins establish a strand bias for replicational mutagenesis. Mol Cell 10:207–213
Pavlov YI, Shcherbakova PV, Rogozin IB (2006) Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int Rev Cytol 255:41–132
Pignede G, Bouvier D, de Recondo AM, Baldacci G (1991) Characterization of the POL3 gene product from Schizosaccharomyces pombe indicates inter-species conservation of the catalytic subunit of DNA polymerase delta. J Mol Biol 222:209–218
Podust VN, Chang LS, Ott R, Dianov GL, Fanning E (2002) Reconstitution of human DNA polymerase delta using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J Biol Chem 277:3894–3901
Pohler JR, Otterlei M, Warbrick E (2005) An in vivo analysis of the localisation and interactions of human p66 DNA polymerase δ subunit. BMC Mol Biol 6:17
Popanda O, Flohr T, Fox G, Thielmann HW (1999) A mutation detected in DNA polymerase δ cDNA from Novikoff hepatoma cells correlates with abnormal catalytic properties of the enzyme. J Cancer Res Clin Oncol 125:598–608
Reynolds N, Watt A, Fantes PA, MacNeill SA (1998) Cdm1, the smallest subunit of DNA polymerase δ in the fission yeast Schizosaccharomyces pombe, is non-essential for growth and division. Curr Genet 34:250–258
Sanchez Garcia J, Ciufo LF, Yang X, Kearsey SE, MacNeill SA (2004) The C-terminal zinc finger of the catalytic subunit of DNA polymerase δ is responsible for direct interaction with the B-subunit. Nucleic Acids Res 32:3005–3016
Sanchez Garcia J, Baranovskiy AG, Knatko EV, Gray FC, Tahirov TH, MacNeill SA (2009) Functional mapping of the fission yeast DNA polymerase δ B-subunit Cdc1 by site-directed and random pentapeptide insertion mutagenesis. BMC Mol Biol 10:82
Selak N, Bachrati CZ, Shevelev I, Dietschy T, van Loon B, Jacob A, Hubscher U, Hoheisel JD, Hickson ID, Stagljar I (2008) The Bloom’s syndrome helicase (BLM) interacts physically and functionally with p12, the smallest subunit of human DNA polymerase δ. Nucleic Acids Res 36:5166–5179
Shamoo Y, Steitz TA (1999) Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell 99:155–166
Simon M, Giot L, Faye G (1991) The 3′ to 5′ exonuclease activity located in the DNA polymerase δ subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J 10:2165–2170
Stocki SA, Nonay RL, Reha-Krantz LJ (1995) Dynamics of bacteriophage T4 DNA polymerase function: identification of amino acid residues that affect switching between polymerase and 3′ → 5′ exonuclease activities. J Mol Biol 254:15–28
Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK (2009) Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat Struct Mol Biol 16:979–986
Venkatesan RN, Treuting PM, Fuller ED, Goldsby RE, Norwood TH, Gooley TA, Ladiges WC, Preston BD, Loeb LA (2007) Mutation at the polymerase active site of mouse DNA polymerase δ increases genomic instability and accelerates tumorigenesis. Mol Cell Biol 27:7669–7682
Vijeh Motlagh ND, Seki M, Branzei D, Enomoto T (2006) Mgs1 and Rad18/Rad5/Mms2 are required for survival of Saccharomyces cerevisiae mutants with novel temperature/cold sensitive alleles of the DNA polymerase δ subunit, Pol31. DNA Repair (Amst) 5:1459–1474
Waga S, Hannon GJ, Beach D, Stillman B (1994) The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 369:574–578
Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA (1997) Crystal structure of a pol α family replication DNA polymerase from bacteriophage RB69. Cell 89:1087–1099
Warbrick E (2000) The puzzle of PCNA’s many partners. Bioessays 22:997–1006
Xie B, Mazloum N, Liu L, Rahmeh A, Li H, Lee MY (2002) Reconstitution and characterization of the human DNA polymerase δ four-subunit holoenzyme. Biochemistry 41:13133–13142
Zhang J, Chung DW, Tan CK, Downey KM, Davie EW, So AG (1991) Primary structure of the catalytic subunit of calf thymus DNA polymerase δ: sequence similarities with other DNA polymerases. Biochemistry 30:11742–11750
Zhang J, Tan CK, McMullen B, Downey KM, So AG (1995) Cloning of the cDNAs for the small subunits of bovine and human DNA polymerase delta and chromosomal location of the human gene (POLD2). Genomics 29:179–186
Zhang S, Zhou Y, Trusa S, Meng X, Lee EY, Lee MY (2007) A novel DNA damage response: rapid degradation of the p12 subunit of DNA polymerase δ. J Biol Chem 282:15330–15340
Zhong X, Garg P, Stith CM, Nick McElhinny SA, Kissling GE, Burgers PM, Kunkel TA (2006) The fidelity of DNA synthesis by yeast DNA polymerase ζ alone and with accessory proteins. Nucleic Acids Res 34:4731–4742
Zhou Y, Chen H, Li X, Wang Y, Chen K, Zhang S, Meng X, Lee EY, Lee MY (2011) Production of recombinant human DNA polymerase δ in a Bombyx mori bioreactor. PLoS One 6:e22224
Zuo S, Gibbs E, Kelman Z, Wang TS, O’Donnell M, MacNeill SA, Hurwitz J (1997) DNA polymerase δ isolated from Schizosaccharomyces pombe contains five subunits. Proc Natl Acad Sci U S A 94:11244–11249
Zuo S, Bermudez V, Zhang G, Kelman Z, Hurwitz J (2000) Structure and activity associated with multiple forms of Schizosaccharomyces pombe DNA polymerase δ. J Biol Chem 275:5153–5162
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Tahirov, T.H. (2012). Structure and Function of Eukaryotic DNA Polymerase δ. In: MacNeill, S. (eds) The Eukaryotic Replisome: a Guide to Protein Structure and Function. Subcellular Biochemistry, vol 62. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-4572-8_12
Download citation
DOI: https://doi.org/10.1007/978-94-007-4572-8_12
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-4571-1
Online ISBN: 978-94-007-4572-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)