Abstract
This contribution presents methods that can be used to describe and analyse forest structure and diversity with particular reference to CCF management. Despite advances in remote sensing, mapped tree data in large observation windows are very rarely available in CCF management situations. Thus, although we present methods of second order statistics (SOC), the emphasis is on nearest neighbor statistics (NNS). The first section gives a general introduction and lists the objectives of the chapter. Methods of analysing non-spatial structure and diversity are presented in the second section. The third section introduces procedures for analysing unmarked and marked patterns of forest structure and diversity. Relevant R codes are provided to facilitate application of the methods. Examples of measuring differences between patterns and of reconstructing forests from samples are also presented. Finally, in Sect. 4 we discuss some important issues and summarize the main findings of this chapter.
Author contributions Lead author, text and calculations (unless indicated otherwise); co-authors, design of the chapter and contribution to data sets.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Structure is a fundamental notion referring to patterns and relationships within a more or less well-defined system. We recognise structural attributes of buildings, crystals and proteins. Computer scientists design data structures and mathematicians analyse algebraic structures. All these tend to be rigid and complete. There are also open and dynamic structures. Human societies tend to generate hierarchical or networking structures as they evolve and adjust to meet a variety of challenges (Luhmann 1995). Flocks of birds, insect swarms and herds of buffalo exhibit specific patterns as they move. Structure may develop as a result of a planned design, or through a process of self-organization. Structure is an open-ended theme offering itself to interpretation within many disciplines of the sciences, arts and humanities (Pullan and Bhadeshia 2000).
Important theoretical concepts relating to biological structures include self-organisation, structure/property relations and pattern recognition. Self-organisation involves competition and a variety of interactions between individual trees. Selective harvesting of trees in continuous cover forest (CCF) management modifies growing spaces and spatial niches. Physics and materials science have greatly contributed to structural research, e.g. through investigations of structure–property relationships (Torquato 2002). Biological processes not only leave traces in the form of spatial patterns, but the spatial structure of a forest ecosystem also determines to a large degree the properties of the system as a whole. Forest management influences tree size distributions, spatial mingling of tree species and natural regeneration. Forest structure affects a range of properties, including total biomass productions, biodiversity and habitat functions, and thus the quality of ecosystem services. The interpretation of tree diameter distributions is an example of pattern recognition often used by foresters to describe a particular forest type or silvicultural treatment.
“Forest structure” usually refers to the way in which the attributes of trees are distributed within a forest ecosystem. Trees are sessile, but they are living things that propagate, grow and die. The production and dispersal of seeds and the associated processes of germination, seedling establishment and survival are important factors of plant population dynamics and structuring (Harper 1977). Trees compete for essential resources, and tree growth and mortality are also important structuring processes. Forest regeneration, growth and mortality generate very specific structures. Thus, structure and processes are not independent. Specific structures generate particular processes of growth and regeneration. These processes in turn produce particular structural arrangements. Associated with a specific forest structure is some degree of heterogeneity or richness which we call diversity. In a forest ecosystem, diversity does, however, not only refer to species richness, but to a range of phenomena that determine the heterogeneity within a community of trees, including the diversity of tree sizes.
Data which are collected in forest ecosystems have not only a temporal but also a spatial dimension. Tree growth and the interactions between trees depend, to a large degree, on the structure of the forest. New analytical tools in the research areas of geostatistics, point process statistics, and random set statistics allow more detailed research of the interaction between spatial patterns and biological processes. Some data are continuous, like wind, temperature and precipitation. They are measured at discrete sample points and continuous information is obtained by spatial interpolation, for example by using kriging techniques (Pommerening 2008).
Where objects of interest can be conveniently described as single points, e.g. tree locations or bird nests, methods of point process statistics are useful. Of particular interest are the second-order statistics, also known as second-order characteristics (we are using the abbreviation SOC in this chapter) which were developed within the theoretical framework of mathematical statistics and then applied in various fields of research, including forestry (Møller and Waagepetersen 2007; Illian et~al. 2008). Examples of SOCs in forestry applications are Ripley’s K and Besag’s L function, pair and mark correlation functions and mark variograms. SOCs describe the variability and correlations in marked and non-marked point processes. Functional second-order characteristics depend on a distance variable r and quantify correlations between all pairs of points with a distance of approximately r between them. This allows them to be related to various ecological scales and also, to a certain degree, to account for long-range point interactions (Pommerening 2002).
In landscape ecology researchers may wish to analyse the spatial distribution of certain vegetation types in the landscape. Here single trees are often not of interest, but rather the distribution of pixels that fall inside or outside of forest land. Such point sets may be analysed using methods of random set statistics.
Structure and diversity are important features which characterise a forest ecosystem. Complex spatial structures are more difficult to describe than simple ones based on frequency distributions. The scientific literature abounds with studies of diameter distributions of even-aged monocultures. Other structural characteristics of a forest which are important for analysing disturbances are a group which Pommerening (2008) calls nearest neighbor summary statistics (NNSS). In this contribution we are using the term nearest neighbor statistics (abbreviated to NNS). NNS methods assume that the spatial structure of a forest is largely determined by the relationship within neighborhood groups of trees. These methods have important advantages over classical spatial statistics, including low cost field assessment and cohort-specific structural analysis (a cohort refers to a group of reference trees that share a common species and size class, examples are presented in this chapter). Neighborhood groups may be homogenous consisting of trees that belong to the same species and size class, or inhomogenous. Greater inhomogeneity of species and size within close-range neighborhoods indicates greater structural diversity.
The evaluation of forest structure thus informs us about the distribution of tree attributes, including the spatial distribution of tree species and their dimensions, crown lengths and leaf areas. The assessment of these attributes facilitates a comparison between a managed and an unmanaged forest ecosystem. Structural data also provide an essential basis for the analysis of ecosystem disturbance, including harvest events.
The structure of a forest is the result of natural processes and human disturbance. Important natural processes are species-specific tree growth, mortality and recruitment and natural disturbances such as fire, wind or snow damage. In addition, human disturbance in the form of clearfellings, plantings or selective tree removal has a major structuring effect. The condition of the majority of forest ecosystems today is the result of human use (Sanderson et~al. 2002; Kareiva et~al. 2007). The degradation or invasion of natural ecosystems often results in the formation of so-called novel ecosystems with new species combinations and the potential for change in ecosystem functioning. These ecosystems are the result of deliberate or inadvertent human activity. As more of the Earth’s land surface becomes transformed by human use, novel ecosystems are increasing in importance (Hobbs et~al. 2006). Natural ecosystems are disappearing or are being modified by human use. Thus, forest structure is not only the outcome of natural processes, but is determined to a considerable extent by silviculture.
Structure is not only the result of past activity, but also the starting point and cause for specific future developments. The three-dimensional geometry of a forest is naturally of interest to silviculturalists who study the spatial and temporal evolution of forest structures (McComb et~al. 1993; Jaehne and Dohrenbusch 1997; Kint et~al. 2003). Such analyses may form the basis for silvicultural strategies. Important ecosystem functions, and the potential and limitations of human use are defined by the existing forest structures. Spatial patterns affect the competition status, seedling growth and survival and crown formation of forest trees (Moeur 1993; Pretzsch 1995). The vertical and horizontal distributions of tree sizes determine the distribution of micro-climatic conditions, the availability of resources and the formation of habitat niches and thus, directly or indirectly, the biological diversity within a forest community. Thus, information about forest structure contributes to improved understanding of the history, functions and future development potential of a particular forest ecosystem (Harmon et~al. 1986; Ruggiero et~al. 1991; Spies 1997; Franklin et~al. 2002).
Forest structure is not only of interest to students of ecology, but has also economic implications. Simple bioeconomic models have been criticised due to their lack of realism focusing on even-aged monocultures and disregarding natural hazards and risk. Based on published studies, Knoke and Seifert (2008) evaluated the influence of the tree species mixture on forest stand resistance against natural hazards, productivity and timber quality using Monte Carlo simulations in mixed forests of Norway spruce and European beech. They assumed site conditions and risks typical of southern Germany and found superior financial returns of mixed stand variants, mainly due to significantly reduced risks.
The management of uneven-aged forests requires not only a basic understanding of the species-specific responses to shading and competition on different growing sites, but also more sophisticated methods of sustainable harvest planning. A selective harvest event in an uneven-aged forest, involving removal of a variety of tree sizes within each of several species, is much more difficult to quantify and prescribe than standard descriptors of harvest events used in rotation management systems, such as moderate high thinning or clearfelling.
Mortality, recruitment and growth, following a harvest event, are more difficult to estimate in an uneven-aged multi-species hardwood forest than in an even-aged pine plantation. The dynamics of a pine plantation, including survival and maximum density, is easier to estimate because its structure is much simpler than the structure of an uneven-aged multi-species forest. Thus, a better understanding of forest structure is a key to improved definition of harvest events and to the modeling of forest dynamics following that event. A forest ecosystem develops through a succession of harvest events. Each harvest event is succeeded by a specific ecosystem response following those disturbances. Improved understanding of forest structure may greatly facilitate estimation of the residual tree community following a particular harvest event, and the subsequent response of that community.
The objective of this contribution is to present methods that can be used to describe and analyse forest structure and diversity with particular reference to CCF methods of ecosystem management. Despite advances in remote sensing and other assessment technologies, mapped tree data are often not available, except in specially designed research plots. Forest inventories tend to provide tree data samples in small observation windows. Thus, in the overwhelming number of cases, the amount of data and their spatial range is too limited to use SOC methods (Fig. 2.1).
Simplified decision tree indicating methods that may be used depending on available data. The abbreviation DBH/H refers to diameter distributions and diameter height relations which are frequently used to reveal structure. In addition to second order statistics (SOCs), nearest neighbor statistics (NNS) are particularly useful for structural analysis (cf. Pommerening 2008)
Often, tree locations are not measured, but the tree attributes, e.g. species, DBHs, diversity indices and other marks, are established directly in the field. This is a typical case for applying NNS. If, however, mapped data in large observation windows are available, SOCs are often preferred. Such point patterns may depart from the hypothesis of complete spatial randomness (CSR) and marks may be spatially independent or not. Cumulative characteristics such as the L function or the mark-weighted L functions can be used to test such hypotheses. If they are rejected, it makes sense to proceed with an analysis involving SOCs, such as pair correlation functions or mark variograms. If the hypothesis of mark independence is accepted, it suffices to use for example diameter distributions, non-spatial structural indices or NNS.
Because of their practical relevance in CCF, particular emphasis in this chapter will be on analytical tools that do not depend on mapped tree data. Foresters need to be able to analyse the changes in spatial diversity and structure following a harvest event. Their analysis must be based on data that are already available or that can be obtained at low cost.
We will first review non-spatial approaches and then present methods which will facilitate spatially explicit analysis using SOC and NNS, including examples of analysing harvest events and measuring structural differences between forest ecosystems. In some cases implementations of methods are shown using the Comprehensive R Archive Network (R Development Core Team 2011).
2 Non-spatial Structure and Diversity
A first impression about forest structure is provided by the frequency distributions of tree sizes and tree species. Accordingly, this section introduces methods that can be used for describing structure based on tree diameters and diameter/height relations. We will also present approaches to defining residual target structures in CCF systems.
2.1 Diameter Distributions
The breast height diameter of a forest tree is easy to measure and a frequently used variable for growth modeling, economic decision-making and silvicultural planning. Frequency distributions of tree diameters measured at breast height (DBH) are often available, providing a useful basis for an initial analysis of forest structure.
2.1.1 Unimodal Diameter Distributions for Even-Aged Forests
Unimodal diameter frequency distributions are often used to describe forest structure. The 2-parameter cumulative Weibull function is defined by the following equation:
where F(x) is the probability that a randomly selected DBH X is smaller or equal to a specified DBH x. To obtain maximum likelihood estimates of the Weibull pdf, we can use the function fitdistr in package MASS of the Comprehensive R Archive Network (http://cran.r-project.org/). Using the vectors n.trees and dbh.class, the R code for the 2-parameter Weibull function would be: 
The Weibull function may be inverted which is useful for simulating a variety of forest structures characterised by the Weibull parameters:
where P(X > x) = 1 − F(x) is the probability that a randomly selected DBH X is greater than a random number distributed in the interval [0, 1], assuming parameter values a = 30, b = 13.7 and c = 2.6. The following kind of question can be answered when using the inverted Weibull function: what is the DBH of a tree if 50% of the trees have a bigger DBH? Using our dataset, the answer would be: \( x \,{=}\, {30} + {13}{.4} \cdot {\left[ { - \ln \left( {{0}{.5}} \right)} \right]^{{\frac{{1}}{{{2}{.6}}}}}} = {41}{.6}\;{\text{cm}} \). Thus, we can simulate a diameter distribution by generating random numbers in the interval 0 and 1, and calculating the associated DBH’s using Eq. 2.2.
2.1.2 Bimodal Diameter Distributions
There is a great variety of empirical forest structures that can be described using a theoretical diameter distribution. DBH frequencies may occur as bimodal distributions which represent more irregular forest structures. Examples are presented by Puumalainen (1996), Wenk (1996) and Condés (1997). Hessenmöller and Gadow (2001) found the bimodal DBH distribution useful for describing the diameter structures of beech forests, which are often characterised by two distinct subpopulations, fully developed canopy trees and suppressed but shade-tolerant understory trees. They used the general form \( f(x) = g \cdot {f_u}(x) + \left( {1 - g} \right) \cdot {f_o}(x) \) where \( {f_u}(x) \) and \( {f_o}(x) \) refer to the functions for the suppressed and dominant trees, respectively and g is an additional parameter which links the two parts.
Figure 2.2 shows an example of the bimodal Weibull fitted to a beech forest near Göttingen in Germany. The distribution of the shade tolerant beech (Fagus sylvatica) typically shows two subpopulations of trees over 7 cm diameter DBH. In Fig. 2.2, the population of dominant canopy trees is represented by a rather wide range of diameters (18–46 cm), whereas the subpopulation of small (and often old) suppressed and trees has a much narrower range of DBHs. The two subpopulations are usually quite distinct. However, the proportions of trees that belong to either the suppressed or the dominant group may differ, depending mainly on the silvicultural treatment history (Fig. 2.2).
The Weibull model may also be used to describe the diameter distributions in forests with two dominant species (Chung 1996; Liu et~al. 2002), using a mix of species-specific DBH distributions. The diameter structure, however, is increasingly difficult to interpret as the number of tree species increases. This is one of the reasons why so much work has been done on the structure of monocultures. They are easy to describe using straightforward methods.
2.2 Diameter-Height Relations
One of the most important elements of forest structure is the relationship between tree diameters and heights. Information about size-class distributions of the trees within a forest stand is important for estimating product yields. The size-class distribution influences the growth potential and hence the current and future economic value of a forest (Knoebel and Burkhart 1991). Height distributions may be quantified using a discrete frequency distribution or a density function, in the same way as a diameter distribution. However, despite greatly improved measurement technology, height measurements in the field are considerably more time consuming than DBH measurements. For this reason, generalised height-DBH relations are being developed, which permit height estimates for given tree diameters under varying forest conditions (Kramer and Akça 1995, p. 138).
2.2.1 Generalised DBH-Height Relations for Multi-species Forests in North America
Generalised DBH-height relations for uneven-aged multi-species forests in Interior British Columbia, Canada, were developed by Temesgen and Gadow (2003). The analysis was based on permanent research plots. Eight tree species were included in the study: Aspen (Populus tremuloides Michx.), Western Red Cedar (Thuja plicata Donn.), Paper Birch (Betula papyrifera March.), Douglas-Fir (Pseudotsuga menziesii (Mirb.) Franco), Larch (Larix occidentalis Nutt.), Lodgepole Pine (Pinus contorta Dougl.), Ponderosa Pine (Pinus ponderosa Laws.) and Spruce (Picea engelmanii Parry × Picea glauca (Monench) Voss). Five models for estimating tree height as a function of tree diameter and several plot attributes were evaluated. The following model was found to be the best one:
with \( a = {a_1} + {a_2} \cdot BAL + {a_3} \cdot N + {a_4} \cdot G \) and \( c = {a_5} + {a_6} \cdot BAL \). BAL is the basal area of the larger trees (m²/ha); G is the plot basal area (m²/ha); N is the number of trees per ha; a 1 … a 7, b and c are species-specific coefficients listed in Table 2.1.
A generalised DBH-height relation such as the one described above, may considerably improve the accuracy of height estimates in multi-species natural forests. In a managed forest, however, the parameter estimates should be independent of G and BAL because density changes at each harvest event. This affects the parameter estimates, which is not logical.
2.2.2 Mixed DBH-Height Distributions
Unmanaged forests are used as a standard for comparison of different types of managed stands. There are numerous examples showing that virgin beech forests exhibit structures which include more than one layer of tree heights (Korpel 1992). In a managed beech forest, the vertical structure depends on the type of thinning that is applied. In a high thinning, which is generally practised in Germany, only bigger trees are removed while the smaller ones may survive for a very long time, resulting in a typical pattern with two subpopulations with different diameter-height relations. Thus, the population of trees is composed of a mixture of two subpopulations having different diameter-height distributions. Zucchini et~al. (2001) presented a model for the diameter-height distribution that is specifically designed to describe such populations.
A mixture of two bivariate normal distributions was fitted to the diameter-height observations of 1,242 beech trees of a DBH greater than 7 cm in the protected forest Dreyberg, located in the Solling region in Lower Saxony, Germany. The dominant species is beech and the stand is very close to the potentially natural stage. All parameters have familiar interpretations. Let \( f ( {d,h} ) \) denote the bivariate probability density function of diameter and height. The proposed model then is
where α, a parameter in the interval (0, 1), determines the proportion of trees belonging to each of the two component bivariate normal distributions \( {n_1}\left( {d,h} \right) \) and \( {n_2}\left( {d,h} \right) \). The parameters of \( {n_j}\left( {d,h} \right) \) are the expectations \( {u_{{dj}}},{u_{{hj}}} \); the variances \( \sigma_{{dj}}^2\;{\text{and}}\;\sigma_{{hj}}^2 \), and the correlation coefficients, \( {\rho_j},j = 1,2 \). A simple height-diameter curve, as shown in Fig. 2.3 (left), describes how the mean height varies with diameter at breast height, but it does not quantify the complete distribution of heights for each diameter.
Simple height-diameter relation (left) and contour plot of the fitted density function (After Zucchini et~al. 2001)
The larger subpopulation (approximately 80% of the entire population) comprises dominant trees in which the slope of the height-diameter regression is less steep than that for the smaller subpopulation (approximately 20% of the population). This can be seen more distinctly in Fig. 2.4 (right). Mixed DBH-height distributions may be fitted using the “flexmix” package (Leisch 2004) of the Comprehensive R Archive Network.
2.2.3 Generalised DBH-Height Relations in a Spatial Context
Schmidt et~al. (2011, in press) present an approach to modeling individual tree height-diameter relationships for Scots pine (Pinus sylvestris) in multi-size and mixed-species stands in Estonia. The dataset includes 22,347 trees from the Estonian Permanent Forest Research Plot Network (Kiviste et~al. 2007). The distribution of the research plots is shown in Fig. 2.4.
The following model which is known in Germany as the “Petterson function” (Kramer and Akça 1995) and in Scandinavia as the “Näslund function” (Kangas and Maltamo 2002) was used:
where hijk and dijk are the total height (m) and breast height diameter (cm) of the kth tree on the ith plot at the jth measurement occasion, respectively; α, β and γ are empirical parameters. Models are often linearized with the aim to apply a mixed model approach (Lappi 1997; Mehtätalo 2004; Kinnunen et~al. 2007). In addition, the use of generalized additive models (GAM) requires specification of a linear combination of (nonlinear) predictor effects. The Näslund function can be linearized by setting the exponent γ constant (in our case γ = 3, see Kramer and Akça 1995):
A two-level mixed model was fitted, with random effects on stand and measurement occasion levels. Thus, it was possible to quantify the between-plot variability as well as the measurement occasion variability. The coefficients of the diameter-height relation could be predicted not only using the quadratic mean diameter, but also the Estonian plot geographic coordinates x and y. The problem of spatially correlated random effects was solved by applying the specific methodology of Wood (2006) for 2-dimensional surface fitting. Thus, the main focus of this study was to use an approach which is spatially explicit allowing for high accuracy prediction from a minimum set of predictor variables. Model bias was small, despite the somewhat irregular distribution of experimental areas.
3 Analysing Unmarked and Marked Patterns
This section presents methods for analysing unmarked and marked patterns of forest structure and diversity. We show examples of measuring differences between patterns and of reconstructing forests from samples. The emphasis is on nearest neighbor statistics which can be easily integrated in CCF management because they do not require large mapped plots.
3.1 Unmarked Patterns
The two-dimensional arrangement of tree locations within an observation window may be seen as a realisation of a point process. In a point process, the location of each individual tree, i, can be understood as a point or event defined by Cartesian coordinates {x i , y i }. This section deals with unmarked point patterns, using methods for mapped and unmapped tree data. The required code of the Comprehensive R Archive Network is also presented to make it easier for potential users to apply the methods.
3.1.1 Mapped Tree Data Available
Second-order characteristics (SOCs) were developed within the theoretical framework of mathematical statistics and then applied in various fields of natural sciences including forestry (Illian et~al. 2008; Møller and Waagepetersen 2007). They describe the variability and correlations in marked and non-marked point processes. In contrast to nearest neighbour statistics (NNS), SOCs depend on a distance variable r and quantify correlations between all pairs of points with a distance of approximately r between them. This allows them to be related to various ecological scales and also, to a certain degree, to account for long-range point interactions (Pommerening 2002). SOCs may be employed when mapped data from large observation windows are available. Examples are given in this section, but more can be found in the cited literature.
In the statistical analysis, it is often assumed that the underlying point process is homogeneous (or stationary) and isotropic, i.e. the corresponding probability distributions are invariant to translations and rotations (Diggle 2003; Illian et~al. 2008). Although methods have also been developed for inhomogeneous point processes (see e.g. Møller and Waagepetersen 2007; Law et~al. 2009), patterns are preferred in the analysis for which the stationarity and isotropy assumption holds. These simplify the approach and allow a focused analysis of interactions between trees by ruling out additional factors such as, for example, varying site conditions. In this context the choice of size and location of an observation window is crucial.
A rather popular second-order characteristic is Ripley’s K-function (Ripley 1977). λK(r) denotes the mean number of points in a disc of radius r centred at the typical point i (which is not counted) of the point pattern where λ is the intensity, i.e. the mean density in the observation window. This function is a cumulative function. Besag (1977) suggested transforming the K-function by dividing it by π and by taking the square root of the quotient, which yields the L-function with both statistical and graphical advantages over the K-function. The L-function is often used for testing the complete spatial randomness (CSR) hypothesis, e.g. in Ripley’s L-test (Stoyan and Stoyan 1994; Dixon 2002). For example, using the libraries spatstat and spatial of the Comprehensive R Archive Network, the following function Lfn (provided by ChunYu Zhang) calculates the L function and the 99% confidence limits for a random distribution. 
The above functions are implemented in a field experiment where the dominant species is beech (Fagus sylvatica). The plot Myklush Zavadivske covering 1 ha, is located near Lviv in Western Ukraine. The following R code generates the graphics presented in Fig. 2.5.

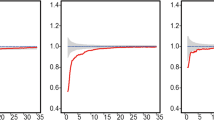
The beech trees in the experimental plot “Myklush_Zavadivske”, show significant regular distribution up to distances of 10 m, which can be seen by the values of the L function well below the lower bound envelope.
Cumulative functions are not always easy to interpret and for detailed structural analysis functions of the nature of derivations are preferred. One of these is the pair correlation function, g(r), which is related to the first derivative of the K function according to the interpoint distance r (Eq. 2.7; see Illian et~al. 2008):
For a heuristic definition consider P(r) as the probability of one point of the point process being at r and another one at the origin o. Let dF denote the area of infinitesimally small circles around o and r and λ dF the probability that there is a point of the point process in a circle of area dF. Then
In this approach, the pair correlation function g(r) acts as a correction factor. For Poisson processes and for large distances when the point distributions are stochastically independent g(r) = 1. In the case of attraction of points g(r) > 1 (leading to a cluster processes) and for inhibition between points g(r) < 1 (resulting in regular patterns).
Figure 2.6 shows an example from the Walsdorf forest in Germany (provided by Arne Pommerening) with beech, Norway spruce and oak. The pair correlation function, ĝ (r), indicates strong clustering of trees at short distances up to approximately 1.5 m. This is followed by a deficit of pairs of trees occurring at distances between 1.5 and 4 m. The most frequent larger intertree distance is 5 m. There are spatial correlations between trees up to 30 m (correlation range). The partial pair correlation function g ij (r), which by analogy to K ij (r) (Lotwick and Silverman 1982) can also be referred to as the intertype pair correlation function, is a tool for investigating multivariate point patterns. Its interpretation is similar to that of g(r). We will not present examples here, mainly because the focus of this section is on unmarked processes. The reader may refer to Penttinen et~al. (1992), Stoyan and Penttinen (2000), Illian et~al. (2008), Kint et~al. (2004) and Suzuki et~al. (2008).
3.1.2 Mapped Tree Data Not Available
Staupendahl and Zucchini (2006) present a brief review of a variety of forest structure indices which were specially developed or adapted for forestry use (cf. Upton and Fingleton 1989, 1990; Biber 1997; Gleichmar and Gerold 1998; Smaltschinski 1998; Gadow 1999). For practical forestry purposes, where complete mappings of populations are normally not feasible, the distance methods are particularly useful. These methods, which are also used for density estimation, are based on nearest-neighbour distances. The measurements made are of two basic types: distances from a sample point to a tree or from a tree to a tree. Again from these methods the T square sampling (Besag and Glaves 1973) and their modifications (Hines and Hines 1979) are particularly useful (Diggle et~al. 1976; Byth and Ripley 1980), especially regarding the test of complete spatial randomness.
Assunção (1994) describes the spatial characteristics of a population of forest trees based on the angle between the vectors joining a particular sample point to its two nearest neighbouring trees. Unaware of Assunção’s work, but with the same intention, Gadow et~al. (1998) also evaluated the angles between neighbouring trees. They did not use a reference point but a reference tree, thus allowing cohort-specific structural analysis. Cohort-specific structure refers to the specific structure in the vicinity of a cohort of reference trees, for example the structure in the vicinity of a given species, or in the vicinity of very large trees. They also considered more than two neighbors, thus extending the neighborhood range. Finally, they proposed that the angles should not be measured exactly, which would be time consuming during field assessment, but classified. They defined a standard angle, for example 72° in the case of four neighbors (Hui and Gadow 2002). The number of observed angles between two neighboring vectors which are smaller than the standard angle, are added up and divided by the total number of angles, as follows:
With four neighbors, W
i
can assume five values: 0.0; 0.25; 0.5; 0.75 and 1.0. The following R-function calculates the uniform angle index: 
The constellations for two reference trees, located in the centre of the square, are shown in Fig. 2.7. The following R code generates the plots for the two groups and calculates the uniform angle indices, using the function uai:

Staupendahl and Zucchini (2006) studied an index considering the three trees nearest to a reference point. Figure 2.8 schematically illustrates the three types of angle-based approach. All three indices measure the angle αij which is the smaller one of the two possible angles formed by the vectors v ij and v ij + 1 joining the reference point (a, c) or reference tree (b) to tree j and tree j + 1 respectively, with j = 1 (a),~~j = 1…4 (b) and j = 1…3 (c). In (b) and (c) v ij is sorted by azimuth into ascending order. To avoid edge effects sample points are chosen from the unshaded subregion with measurements allowed to trees also in the shaded buffer zone.
Schematic representation of the sample unit of three types of angle-based measures of spatial pattern at sample point i within a hypothetical forest: (a) angle test using the two nearest trees to sample point, (b) uniform angle index W using the four nearest trees to a reference tree which is the nearest tree to the sample point and (c) index W p using the three trees nearest to the sample point
A structure unit thus consists of a reference tree, or a reference point, and the n nearest neighbors in the vicinity of the tree or point. A reference tree-based structure unit will reveal spatial patterns in the vicinity of a particular tree cohort (species or size class), which allows more meaningful interpretations.
The treatment of edge trees can affect the estimation of structural indices since they can involve off-plot neighbours. Pommerening and Stoyan (2006) investigated in what circumstances edge-correction methods are necessary, and evaluated different approaches. They proposed a variable buffer zone around the edge of the plot. Only such trees were selected as reference trees, which were located further away from the plot edge than the distance to their n th nearest neighbour.
The expected value of the uniform angle index is the average of an infinite number of realisations of W obtained by repeatedly randomizing orientations of the sample grid:
where \( \hat{W} \) is the estimate of W in the rth simulation. Staupendahl and Zucchini (2006) used sampling simulations in three hypothetical forests with a regular, random and clustered pattern and sample sizes varying between 5 and 50, with 1,000 replications each. They show that the point-based criterion \( {\hat{W}_p} \) is virtually unbiased for the entire range of sample sizes and for different spatial patterns. They conclude that the performance of the neighborhood-based method is comparable to alternative methods that are more costly to implement, and recommend a sample size of 20–30 sample points per compartment. Corral-Rivas et~al. (2010) found that nearest neighbour-based indices enable a categorisation of a spatial pattern of forests with a sensitivity comparable to that of Ripley’s L(r) test, at finer scales. The assessment of the angles can be integrated into routine forest surveys, virtually without additional cost.
3.2 Marked Patterns
We believe that one of the basic requirements of sustainable CCF management is the ability to assess complex forest structure at affordable cost. Therefore, this section presents a brief introduction to methods that can be used to incorporate structural assessment in routine forest inventories. We then give examples of measuring tree size diversity. Finally, we deal with species diversity, again in a spatial context.
3.2.1 Assessment
Traditionally, a forest ecosystem is characterised by area-based attributes (basal area, biomass, number of trees per hectare), mean values and distributions (diameters), and relationships (DBH-height regression). These classical attributes of a forest ecosystem are usually assessed in the field by sampling in field plots of specified shape and size (Kleinn et~al. 2010). Fixed area plots of circular, square or rectangular shape (Fig. 2.9) and sample points using the angle count method are most common. For details refer to standard forest mensuration textbooks (e.g. Van Laar and Akça 2007).
Variables which characterise the spatial structure and diversity of an ecosystem are either included in a rather rudimentary way, or not considered at all, in standard forest sampling schemes. Forest spatial structure is characterised by nearest neighbor statistics (NNS) which include the variables aggregation, species mingling and size differentiation. Aggregation refers to the regularity of tree positions. High aggregation is often described as “clumped”. Species mingling defines the degree of spatial segregation of the tree species in a forest. Low species mingling means high segregation, no matter how many species occur in the forest. Size differentiation measures the degree by which trees of different sizes are spatially mingled. High size differentiation implies that trees of varying size occur in close vicinity of each other. These relationships are explained by three simple diagrams in Fig. 2.10.
Traditional forest sampling concentrates on assessment of forest density, volume and timber products. For characterising spatial structure and diversity in uneven-aged multi-species forest, we require information about the distributions of aggregation, species mingling and size differentiation. A convenient sampling scheme for assessing these variables is distance sampling, which is popular among ecologists despite the potential for bias (Krebs 1999; Nothdurft et~al. 2010). A convenient sampling unit is the n-tree-structure-unit (NSU), which consists of a sample point, or a reference tree which is located closest to the sample point, and its n nearest neighbors (Fig. 2.11). The n-tree-structure-unit is not to be confused with the n-tree-sample where the plot radius is equal to the distance to the nth tree, as proposed by Prodan (1966). In spatial statistics the terms point-related and test-location-related summary characteristics are sometimes used (Pommerening 2008; Illian et~al. 2008). In a circular plot sampling design where the azimuth and distance to the plot centre are known for each tree, the attributes of an n-tree-structure-unit can be calculated using the methods explained in other sections of this chapter. In sampling designs where tree coordinates are not known, the NSU can simply be integrated into the design. Measuring between tree distances is costly and unnecessary. Unbiased estimates of basal areas may be obtained using the angle count method. Thus, using the NSU, it will be possible to assess forest structure and diversity plus all the other traditional variables, without having to measure tree-to-tree distances or tree coordinates.
Examples of point-based structure variables are shown below:
Examples of tree-based structure variables are shown below:
Figure 2.11 shows two neighborhood units, a tree-based one and a point-based one. The four nearest neighbours around the reference point, or the reference tree, are shown. Both units feature the same two species A (one or two trees) and B (three trees). Numbers represent the tree DBHs.
Only one angle α ik is smaller than the standard angle α 0 (which happens to be 72°). The numbers in the diagrams represent each tree’s DBH. The following structure variables may be calculated for a tree-based and a point-based unit respectively:
Hui and Albert (2004) proposed a slightly modified sampling design. Their sampling unit consists of the four trees closest to a sample point. Each of these four trees is a reference tree. This gives four n-tree-structure-units, instead of just one. Thus, the structure of a forest is characterised by the distributions of the variables assessed in all the field sample points. An advantage of the tree-based approach is the fact that forest structure may be analysed cohort-specific (structure in the vicinity of trees of species X with a DBH greater than 50 cm, for example).
Motz et~al. (2010) carried out a study to investigate how tree diversity measures may be estimated as extensions of existing forest resource inventories. They compared the precision of angle count and fixed radius plot sampling with respect to nine representative diversity indices in three different forest types at stand, enterprises and national forest level. Their results indicated that most of the spatially explicit indices are more precisely estimated by fixed radius plots. The superiority of fixed radius plot sampling to angle count sampling increased significantly with increasing diameter differentiation of forests. Basal areas were estimated by angle count sampling with at least the same precision as from fixed radius plots.
The design of a forest inventory system is an optimisation problem (Staupendahl and Gadow 2008; Kleinn et~al. 2010). It should be possible to estimate the target variables with high accuracy, subject to the constraint of limited resources. In an attempt to respond to the need for more flexible forest inventory designs, Gadow and Schmidt (1998) proposed that forest sampling should take place after the trees had been marked for a thinning and before they are harvested. Thus, a harvest event assessment delivers information about the products that will be removed, the structural changes caused by the harvest, and the condition of the forest remaining after the harvest event. The emphasis is on the timing of the inventory. Instead of assessing the entire resource at fixed time intervals, information is gathered where it is needed most. Because of the specific timing, Harvest event assessment is particularly well suited to management control in complex forest structures, such as found in CCF (Puumalainen 1998).
3.2.2 Tree Size Diversity
The examples in the previous sections have shown how the structure of a forest may be defined by the distribution of tree sizes and by the particular relationship between different size variables (DBH-height; DBH-crown width). Such distributions, however, do not reveal the particular spatial arrangement of these variables (Schütz 2002). It requires little imagination to realize that a variety of spatial patterns of tree diameters may be possible for any particular diameter frequency distribution. One might refer to a high degree of “size mingling” when large and small trees occur in close vicinity of each other, or to low size mingling when large and small trees are spatially segregated. Such spatial arrangements may be assessed in the field using sample plots (Fig. 2.13, left) or neighborhood groups of trees (Fig. 2.12, right).
DBH mingling may be assessed in the field using sample plots or neighborhood groups of trees. In order to avoid edge effects, sample points are chosen from the unshaded subregion. References are allowed to trees which are located in the shaded buffer zone, this is known as plus-sampling. Another method to avoid edge effects, known as minus-sampling, considers only those trees as reference trees in the analysis which are located further away from the plot edge than the distance to their nth nearest neighbor (Pommerening and Stoyan 2006)
There are two main categories of edge-correction: plus-sampling and minus-sampling (Illian et~al. 2008). Plus-sampling makes full use of all data within the observation window either by recording objects outside the observation window or by simulating them. Minus-sampling (also referred to as border method) only makes use of an inner sub-set of the observation window. Pommerening (2008) summarised edge-correction methods according to these two categories.
We define the trees in such a group as a “structure unit”. Fixed-area sample plots are preferred for unbiased assessments of area-based variables, e.g. number of trees or basal area per ha. Neighborhood groups are useful if the target information should be independent of density, as in the case of spatial mingling of tree diameters. Each structure unit has certain attributes. The distribution of the DBH mingling values of the sampled structure units will reveal the particular spatial arrangement of tree sizes in a larger forest area. Within a given diameter distribution, greater homogeneity of structure units reveals spatial segregation: trees of the same size are found in close proximity to each other.
Examples of a segregation of tree sizes may be found in a forest with dense groups of saplings within gaps adjoining areas of mature trees. On the other hand, a high degree of spatial mingling may be found in a forest of shade tolerant species where all tree sizes occur in close proximity to each other. Both forests may have the same diameter distribution, but completely different diameter mingling patterns. DBH mingling within a structure unit may be described using a variety of indices (Kint et~al. 2003). Six examples of indices calculated for neighborhood groups are listed in Table 2.2. The distribution of each index may facilitate the analysis of the structural composition of the entire forest and the reader is referred to reviews by Staudhammer and LeMay and McElhinny et~al. (2005).
Sterba and Zingg (2006) found close correlations and specific relationships between the four heterogeneity indices (H′, CV DBH , Gini, Skew) and the two neighborhood indices (T i , U i ), based on data from a great variety of forest types, including coppice forests of different age, even-aged and uneven-aged forests. The four heterogeneity indices can be used to evaluate the evenness of tree sizes within a particular structure unit (a sample plot or a neighborhood group). The two neighborhood indices can be used to evaluate the structural attributes of a cohort, i.e. a specific subpopulation of trees that are similar in some way.
Hyytiäinen and Haigth (2011) employ a specific form of the Shannon index (S) to evaluate habitat quality. They describe the simultaneous species and size richness of a forest as follows:
The weights of species and size diversity are denoted by w
sp
and w
size
, respectively. B is the total basal area, B
u
is the total basal area of trees which belong to species u, and B
v
is the basal area of trees in diameter class v. The number of species is y, the number of diameter classes is z. The following R code groups the basal areas of all trees in the plot Ulaschkiwski29(6) which had been measured in 2009 in the Carpatian mountains, Western Ukraine. The dataframe is Korol_dat and the basal areas are Korol_dat$B:

The following code calculates the total basal area of a particular species, for example for all pine trees: 
The same is done for all the other three species (oak=48483.41, beech=17110.87 and spruce = 5255.89). The total basal area is 180372.7 cm2. The first and 2nd term of Eq. 11 are calculated as follows, assuming weights of 0.6 and 0.4 respectively: 
There is no straight-forward approach to constraining the calculations such that (a) S assumes values between 0 and 1 and (b) S allows a comparison between two arbitrary ecosystems. The forest with the maximum number of species and the greatest diameter variances is not known. However, a practical solution might be to use as reference a fixed number of basal area classes (for example 7, as in our example) and a representative number of species occurring within a region. A negative characteristic of the Shannon-Weaver index is the fact that the value increases with increasing evenness of the relative frequencies. However, rare species are often considered to contribute more to diversity than common species (see Hui et~al. 2011).
3.2.3 Cohort-Specific Structure
Nearest neighbor statistics are particularly useful in the study of cohort-specific structural attributes. We are using the data of two experimental field plots to illustrate the analysis of the diversity of tree sizes in the neighborhood of all trees which belong to a specific cohort of reference trees:
-
1.
Plot Jiaohe1 is located in the Jiaohe forest in Jilin Province of North-Eastern China. The forest is managed by selective harvesting. The plot area is one hectare. The DBHs range from 0.4 to 79.2 cm. The following tree species occur in the plot: Betula costata, Carpinus cordata, Fraxinus mandshurica, Acer mandshurica, Tilia amurensis, Acer mono, Syringa reticulata var. mandshurica, Juglans mandshurica, Abies holophylla, Pinus koraiensis, Ulmus laciniata, Juglans mandshurica, Phellodendron amurense and Ramus davurica. The spatial arrangement of species and tree sizes, and the 5 m buffer zone outside the shaded area, is shown in Fig. 2.13a.
-
2.
Plot 1080 is located in the south-central part of Estonia. The plot shape is circular, the plot radius being 30 m. Three tree species occur in the plot: Pinus sylvestris, Betula pendula and Picea abies. The spatial arrangement of species and tree sizes is shown in Fig. 2.13b. Plot 1080 has no fixed buffer zone. Instead, only those trees are considered in the analysis, which are located further away from the plot perimeter than the distance to their third nearest neighbor (see Pommerening and Stoyan 2006).
The following R code was used to identify the neighbor indices of a specific cohort of trees (tree0) inside the buffer zone (inside) using the nnwhich() function of the spatstat library. The dataframe dat refers to all trees including those located in the buffer zone; dat$X and dat$Y are their coordinates: 
The diameter coefficient of variation (cvd) of the four nearest neighbors increases with decreasing, as well as increasing, size of reference tree (Fig. 2.14).
The lowest cvd values in the neighborhood group are found between DBHs of 10 and 25 cm of the reference tree. It is important to note that this particular analysis does not require mapped datasets. The important advantage of using neighborhood groups is the fact that data can be assessed in routine forest surveys at practically no additional cost. More results of the analysis of the two plots are presented at the end of this section.
3.2.4 Tree Species Diversity
The main structural feature of a forest which only includes one single species, is the distribution and spatial mingling of tree sizes. A multi-species forest is additionally characterized by tree species richness and spatial mingling of tree species. A direct consequence of the large-scale forest destruction, especially since the second half of the twentieth century, is a serious depletion of tree species diversity. Many species have already become extinct or are threatened by extinction. Realising these threats, scientists have been increasing their activities in the area of biodiversity research.
3.2.4.1 Species Richness
Scientists began to use measures of biodiversity during the early years of the twentieth century. Among the first to propose indices of diversity were Fisher et~al. (1943) and Simpson (1949). The Shannon-Weaver index was first applied in studies on community species diversity by Margalef (1957). Whittaker (1972) divided the diversity indices into four spatial scales, the α, β, γ and δ diversity. The literature on biodiversity seems endless and new ideas continue to emerge. Ganeshaiah and Shaanker (2000, 2003) for example, developed the Avalanche index which is based on taxonomic differences between species. Scholes and Biggs (2005) proposed a biodiversity intactness index which measures the decline of populations relative to their presumed pre-modern levels. Recent reviews of biodiversity indices include those by Ferris and Humphrey (1999) and Spanos and Feest (2007). One of the results of this research is the establishment of relationships between the size of sample areas and the number of species.
As expected, the number of tree species increases with increasing assessment area. Hubbell (2001) has shown empirical and theoretical relationships between area size and species number. His theoretical three-phase-curve of species diversity shows that at the local level the number of species increases rapidly with increasing area. At the regional level, the cumulative increase in the number of species is not influenced so much by the relative species frequency, but more by the balance between species formation, spatial distribution and extinction. The continental and intercontinental biogeographic scale produces spatially segregated evolutionary developments. The consequence is another accelerated increase in the species number with increasing area.
The increase of the number of species with increasing sample area is known as the species–area relationship. Lawton (1999) has referred to this relationship as one of the few fundamental laws in ecology. The difference in micro-site conditions generally increases with area and accordingly, the variety of species that can be supported generally increases with area (Williams 1964; Barkman 1989). The species–area relationship is more suitable for assessment of diversity than the mere number of species (Lepě and Stursa 1989). Several models have been proposed to describe this relationship (Monod 1950; de Caprariis et~al. 1976; Gitay et~al. 1991; Buys et~al. 1994; Williams 1995; Tjørve 2003).
These models allow us to determine the minimum species areal, i.e. the smallest area which is required to capture all the species present within a given contiguous region. Using relatively large contiguous sample areas with known tree positions, assessed in different climatic regions with varying tree species abundances, Gadow and Hui (2007) could establish a specific relationship between the maximum number of tree species within a forest (S max ) and the minimum area required to capture all of them. This minimum species areal (A min ) can be directly estimated using a power function (Fig. 2.15). The analysis has shown that, for contiguous forest areas, the form of the species–area relationship is directly defined by the species abundance, the maximum number of tree species. This result confirms assumptions made by Preston (1962), May (1975) and Hubbell (2001).
Relationship between the minimum areal A min and the estimated maximum number of tree species S max . Gadow and Hui (2007) expressed the relationship by \( {A_{{\min }}} = 487.8 \cdot S_{{\max }}^{{0.524}} \)
Near-natural forest management systems with selective harvesting require undisturbed reference areas which are sufficiently large to include the essential features of an unmanaged forest. One of the key features of an unmanaged forest is the tree species distribution. Thus, A min , the smallest area which represents the species richness of the entire population, is of great practical relevance. The minimum tree species areal may provide an important scientific basis for types of selective forest management which attempt to mimic natural processes of forest dynamics. Furthermore, if it is possible to estimate the minimum species area, then by analogy Wehenkel et~al. (2011) propose to estimate the balanced structure area (BSA), which has been defined by Koop (1981) as the minimum contiguous area that includes all tree developmental stages. In the study presented by Wehenkel et~al. (2011), the BSA is the minimum area required for sustainable management in a Mexican multi-sized selection forest. Their analysis has shown that a multi-sized forest represents a balanced structural unit if a specific relationship between harvest and growth can be maintained, using a defined target diameter distribution and disregarding major natural disturbances. Thus, using the BSA as an indicator of demographic sustainability, a range of goods and services may be consistently produced over time, thus realising the vision proposed by Nyland (2002).
Jenssen and Hofmann (2002) have shown relationships between different successional stages of a beech ecosystem and the plant species richness. Using plots of the same size, the average number of species in that particular investigation was increasing from the dense sapling stage to a stage dominated by mature trees. The highest number of plant species was observed in the senescent stage of the trees where the small-scale change between gaps and shaded areas produced a variety of environmental niches. The species diversity of a managed forest is thus greatly influenced by the type of silviculture.
3.2.4.2 Species Spatial Mingling
A useful measure of tree species diversity not only reflects the species richness of a community, but also the particular spatial structure. An important advantage of any index of forest diversity would be easy integration in routine management surveys to facilitate its application in land-use planning and monitoring. To develop more effective variables which reflect not only species richness and evenness, but also include a spatially relevant structural component, is an important challenge for science (Pretzsch 2003; Xia 2007).
The literature on biodiversity is extensive and new ideas continue to emerge (for an exhaustive review of biodiversity indices refer to Ferris and Humphrey (1999) and Spanos and Feest (2007)). Statistical methods for analysing beta diversity include principle component analysis and redundancy analysis (Legendre and Gallagher 2001). Some studies presented sophisticated and advanced evaluations of beta diversity, even in mega-diverse tropical forests (e.g. Condit et~al. 2002). Many of these approaches require tree coordinates which are normally not available, thus presenting a major limitation regarding practical use. If costly assessment of tree coordinates is not required for diversity assessment, spatially relevant biodiversity analyses could be possible, based on data from routine forest surveys.
Hui and Albert (2004) defined the spatial relationship between a particular reference tree and its n nearest neighboring trees as the forest spatial structure unit. Theoretically, n could be any reasonable number. However, based on a series of field studies, they found that the optimum group size of such a spatial structure unit consisted of a reference tree and its four nearest neighboring trees. Pommerening (2006) found that the optimum number of nearest neighbours is that which allows the best spatial reconstruction of a given forest from sampled NNS (see details below).
The species spatial mingling within a structure unit is equal to the proportion of neighbors which do not belong to the same species as the reference tree. The previous work only considered the spatial mingling of the reference tree by calculating the proportion of neighbors which do not belong to the same species as the reference tree. The spatial mingling index, which was described by Gadow (1993) and used by Füldner (1995) and Pommerening (2002), is defined as follows:
where n is the number of nearest neighbors considered, v ij =1 if the jth neighboring tree is not of the same species as the i-th reference tree and v ij =0 otherwise. The distribution of the M i values, in conjunction with the species proportions within a given tree population, allows a detailed study of the spatial diversity within a forest. However, the number of different tree species in the structure unit was not taken into account, and this was a shortcoming of the original mingling index.
A logical improvement of the mingling index would be to consider in Eq. 3.6 not only the spatial mingling, but also the number of tree species. This can be achieved by multiplying M i with S i /n max where S i is the number of tree species in the neighborhood of reference tree i, including tree i, and n max is the maximum number of species in this structure unit i. In our special case n max = 5, which was found to represent a good compromise between scientific assertion and practicability. The reasons for n max = 5 were presented in several previous studies which evaluated different numbers of neighbors under field conditions (Gadow and Hui 2002; Hui and Albert 2004; Hui et~al. 2007). A greater number of neighbors allows a more detailed and differentiated analysis. The smaller the number of neighbors, the lower is the cost of assessment. The most convenient number of neighbors from the practical point of view would be 1, because the one nearest neighbor is very easy to identify in the field. The assessment effort increases with an increasing number of neighbors. The cost of identifying a fifth neigbour increases sharply (because of the need to evaluate many tree-to-tree distances) and the cited field studies have shown that four neighbors (n max = 5) represents the best compromise.
The spatial diversity status (MS i ) of a particular tree species is determined by the relative species richness within the structure unit i and the degree of mingling of the reference tree, and may be expressed as follows:
where the M i are the species mingling values, as defined above (refer to previous work, e.g. Füldner 1995; Pommerening 2002) and n max is the maximum number of species in the structure unit. Equation 2.13 thus measures the tree species richness as well as an important spatial characteristic of a structure unit. A reference tree of a common species is more likely to have neighbors of the same species, which is reflected by low MS i values (Fig. 2.16, left). On the other hand, a rare species is likely to produce a high proportion of high MS i values (Fig. 2.16, right). Thus, MS i is especially sensitive to rare tree species. Again, an important practical advantage, considering the assessment effort, is the fact that it is not necessary to measure tree coordinates in the field.
A useful statistic reflecting the status of each individual tree species in the community is the species average spatial status (MS sp ), which is the average value of the MS i for each tree species in the community:
where N sp is the number of trees of species sp in the community. The commonly used diversity indices are represented by a single statistic which combines the species richness and evenness (the ratio of observed to maximum richness). Assuming additivity, the tree species spatial diversity of a tree population may be expressed as the sum of the average spatial diversity states of the different tree species. This sum is conveniently expressed by the TSS criterion proposed by Hui et~al. (2011):
where s is the number of tree species. TSS assumes a maximum value of 1 when each species is represented by one tree, in which case all MS sp are equal to 1. When there is only one species of N trees in the community, the species richness is a minimum, and the TSS value is zero. The TSS variable thus represents the sum of the average spatial mingling values of all tree species in the community. It is a measure, not only of tree species richness, but also of tree species spatial diversity within a given ecosystem. Table 2.3 presents the MS sp values for the 21 tree species in Jiaohe1 and the three species in plot 1008.
The average MS sp value for the 17 tree species in Jiaohe1 is 0.66, and the TSS value is 11.22. The average MS sp value for the 3 tree species in 1080 is 0.31. Due to its absolute dominance, Pinus sylvestris in plot 1080 has a very low MS sp value. The TSS value in 1080 is only 0.92, which is the result of the comparatively small number of species. The average diameter coefficient of variation is 0.75 in Jiaohe1 and 0.33 in 1080. Both, the TSS value and the average diameter coefficient of variation indicate a high spatial diversity in Jiaohe1. The spatial diversity of tree species and sizes is much lower in 1080.
TSS is sensitive to rare species and to variations in community structure, including species spatial isolation and spatial mingling. For these reasons, the TSS criterion is more effective in measuring tree species diversity than the commonly used indices. It allows detailed interpretation of forest spatial diversity and of forest structural modifications following selective thinnings in CCF systems. A particular advantage of the TSS index is the fact that its assessment, which is based on neighborhood relations, can be easily integrated in routine forest management surveys at practically no additional cost.
3.2.4.3 Expected Values of NNS
Expected values of spatial indices may be of interest to students involved in comparative analysis of forest structures. According to Lewandowski and Pommerening (1997) expected mingling (EM), can be calculated as
where s is the number of species, p is the number of trees in the observation window and p i refers to the number of trees of species i.

Jiaohe forest in north-eastern China (Photo: Klaus von Gadow)
Expected mark diameter differentiation is not as straightforward as expected mark mingling. Using tree diameters, DBH, as example marks Pommerening (1997, p. 18) proposed to sort DBH in ascending order, i.e. i < j ⇒ DBH i ≤ DBH j . As a result, the index set J of a given forest is obtained. Then the auxiliary measure R is defined as
The expected mark differentiation for tree DBH, E T, may now be calculated as
Details about the derivation of Eqs. 2.17 and 2.18 can be found in Pommerening (1997).
3.3 Assessing Differences Between Ecosystems
CCF management involves regular modification of forest structure through harvest events. Foresters need to assess the impact of ecosystem modification caused by a particular harvest event (Puumalainen 1998). To be able to do this, they need to describe the structural changes caused by the tree removals. Harvest events modify the tree diameter distributions, the species distributions and spatial patterns. A major objective of many CCF systems is to mimic natural ecosystem dynamics. Therefore, it is often desirable to compare the current state of a particular ecosystem with some ideal state, such as an unmanaged virgin forest. Accordingly, this chapter introduces methods to quantify differences between forest structures.
3.3.1 Describing Harvest Events with Linguistic Variables
A harvest event is a silvicultural activity which modifies ecosystem structure. A selective thinning, typical in CCF systems, is usually described by foresters using some adjective which defines the weight and the type of ecosystem modification. The European forestry literature abounds with definitions of specific harvest events (Kramer 1988, p. 180). A “moderate low thinning” removes the understorey trees, and the adjective “moderate” refers to the amount removed (Fig. 2.17).
A “heavy high thinning” involves the removal of all competitors of some specially identified trees which are believed to produce future value (Schober 1991). The expected change of forest structure based on a combination of two adjectives is shown in Fig. 2.17. Harvest events are described by a great variety of linguistic variables, including “selective thinning” (German: Auslesedurchforstung; see Schädelin 1942; Leibundgut 1978, p. 116; Abetz 1976; Johann 1982), “plenter thinning” (German: Plenterdurchforstung; see Borggreve 1891; Schütz 1989), “qualitative group selection” (German: qualitative Gruppendurchforstung; see Kato and Mülder 1983), “structural thinning” (German: Strukturierende Durchforstung; see Reininger 1987, p. 142). The “variable retention systems” In North America are defined by expressions like “strip shelterwood”, “irregular shelterwood”, “group retention” or “group selection” (Maguire et~al. 2006).
The use of simple verbal expressions for describing complex structural modifications creates confusion, especially in multi-species forests. The linguistic variables plenter thinning and selective thinning, for example, may refer to virtually identical harvest events and ecosystem modification. On the other hand, one particular linguistic variable, like plenter thinning, may be interpreted in completely different ways. For a more detailed analysis of the confusion created by silvicultural terminology refer to Füldner and Gadow (1994). To describe the structural changes caused by different harvest events is particularly challenging in uneven-aged multi-species forests which are selectively managed in a CCF system. Figure 2.18 presents an example of two harvest events and the corresponding structural changes. The example demonstrates that one would need many adjectives to differentiate between the two harvest events.
3.3.2 Simple Numerical Variables
Johann (1982) proposed the A-thinning index (Eq. 2.19) which defines a critical distance cd ij between tree i and a given neighbour j. This critical distance is defined by the thinning intensity parameter A. Any neighbouring tree j which is located closer to tree i than the critical distance cd ij is removed. Equation 2.19 shows that, apart from the thinning intensity parameter A the index uses the height diameter ratio of tree i and the diameter of the neighbouring tree j. The A-thinning index is thus sensitive to the h/d ratio of tree i: Trees with a greater h/d ratio are more heavily released than those with a lower h/d ratio. The A-values may range from 4 to 8. Higher values indicate decreasing thinning intensity. Johann (1982) recommended values of 4, 5 and 6 for even-aged pure Norway spruce forests which he considered to be synonymous with heavy, moderate and light release. A-values of 4 and 6 are frequently used values in thinning experiments (Hasenauer et~al. 1996; Pretzsch 2002).
The A-index has been used to simulate thinnings in spruce monocultures where silviculture is geared to clearfelling. The index may be difficult to adapt to realistic situations in CCF.
A practical variable for defining the weight of a thinning in CCF is the portion of the basal area (m2/ha) removed during a harvest event. We denote this quantity as rG (Murray and Gadow 1993):
Correspondingly, the removed portion of the number of trees per ha may be denoted as rN. The ratio of these two quantities, NG-ratio, describes the type of thinning:
Both rG and NG can be related to the change of the parameters of the Weibull distribution (Staupendahl and Puumalainen 2000). In an uneven-aged multi-species forest, the thinning weight (rG) and type (NG) may be calculated separately for each species or species group. Figure 2.19 presents an example.
The harvest event in 1999 removed more beech trees (slightly more than 10% of total basal area) than other deciduous species (5% of total basal area) while the NG ratios of about 0.6 indicate a high thinning (removal of predominantly bigger trees) in both groups. The structural modification caused by the 2004 harvest event was similar regarding the “other deciduous” group, but different in beech where the NG ratio was approaching unity, thus indicating removal of smaller beech trees, on average, when compared with the 1999 event. Only 16% of the basal area was removed in 1999, and 20% in 2004.
3.3.3 Removal Preferences in a Spatial Context
Sometimes we wish to know the spatial context of the trees that were removed during a harvest event: did the harvest event preferably target the dominant or the suppressed trees in the vicinity of their immediate neighbors? Where trees selected for removal located preferably in groups composed of one species or in mixed groups? To be able to do this, we can describe the neighborhood constellations of all removed trees and compare that with the neighborhoods of all trees before the harvest. An example of a variable that can be used is the species mingling (the proportion of the n nearest neighbors of a particular reference tree that are not of the same species as the reference tree). Another example is the Dominance (the proportion of the n nearest neighbors of a particular reference tree that are smaller than the reference tree). Considering four neighbors, each of these two variables can assume five values. The relative proportion of the removed reference trees divided by the proportion of all reference trees (before the harvest) within a structural class i and j is a measure of the removal preference (Pr ij ) within a given combination of structural classes:
Table 2.4 presents the removal preferences in the Lensahn experiment during the 2004 harvest event, using the criteria Mingling and Dominance for beech and other deciduous species. The removed beech trees had occurred within a broad array of spatial constellations. They had been suppressed as well as dominant individuals. The highest removal preference (Pr ij = 6.37) refers to neighborhood groups in which the removed beech was (a) the smallest tree among its four nearest neighbors (U = 0.00) and (b) surrounded by three neighbors that were not beech trees (M = 0.75). The other deciduous species were removed with high preference if they were co-dominant (U = 0.75) and surrounded by three out of four neighbors of a different species (M = 0.75).
3.3.4 Spatially Explicit Simulation of Harvest Events
Numerous tools have been developed to facilitate the prediction of tree growth in uneven-aged multi-species forests. Fairly advanced growth models are available in many regions where forest ecosystems are selectively harvested. To be able to evaluate the specific dynamics of a selectively managed ecosystem, harvest event models are indispensible. In our experience, developing a model that predicts the structural modification caused by a harvest event is a rather challenging task, requiring effective algorithms that translate silvicultural prescriptions into tree selection algorithms resulting in complex forest structural modifications (Albert 1999; Hessenmöller 2002). It is not always possible to describe a harvest event using an index or a combination of indices that describe the weight and the type of tree removals. Continuous cover forestry is characterised by a wide range of specific harvesting types, including gap removals and Z-tree release thinnings. A Z tree in Germany is known as a frame tree in the UK, in reference to their role as representing the basic frame of a stand of trees. Frame trees are selected for their outstanding vitality, stem quality, stability and crown morphology. They are released from competition during a thinning by removal of their immediate competitors. Z-tree release thinning is widely practiced in Germany and simulation allows a more detailed and meaningful quantitative analysis of a harvest event. Examples were presented by Albert (1999, 2001; Fig. 2.20).
Harvest event simulation on the experimental plot Frankenberg 131a. Identification of frame trees (•) and removed trees (+) based on the rule “remove the two most competitive neighbors of each frame tree” (Albert 1999)
Albert (2001) presented a simulation of silvicultural alternatives to support the decision process in management planning. Management alternatives were generated by applying different thinning concepts, thinning intervals and changing the silvicultural objectives. The decision to identify a tree as a frame tree was based on the attributes crown vitality, stem quality, and distance to the nearest frame tree. The simulations demonstrated a realistic approach to harvest event prediction.
In his PhD thesis, Hessenmöller (2002) simulated the structural modifications caused by different harvest events. Each harvest event was described by a set of rules published in silvicultural textbooks. The selective thinnings were also characterised by different removal intensities to see if the removal probabilities would be affected by the thinning weight. Figure 2.21 shows a simulated selective and gap thinning for two types of spatial distribution.
Four examples of harvest event simulations presented by Hessenmöller (2002) for an aggregated and a random forest. The left column shows the removed (black dots) and remaining trees (circles). The right column just shows the remaining trees
A harvest event in CCF management modifies the tree size distribution, the species distribution and the spatial pattern. Foresters, who need to assess the impact of ecosystem modification caused by a particular harvest event, have to be able to describe structural changes caused by the tree removals. Mason et~al. (2005) have shown that 2nd order statistics may also be usefully employed for analysing harvest events.
3.3.5 Comparing Ecosystems
As mentioned before, major objective of many CCF systems is to mimic natural ecosystem dynamics. Therefore, it is often desirable to compare the current state of a particular ecosystem with some ideal state, such as an unmanaged virgin forest. Accordingly, this section very briefly introduces a method based on the Gini coefficient (G), which permits a quantitative analysis of differences between forest structures. The Gini measures the area between the Lorenz curve (red colored in Fig. 2.22) and a hypothetical line of absolute homogeneity, expressed as a percentage of the maximum area under the line. The Gini coefficient is the ratio of the area between the Lorenz Curve and the line of absolute equality (numerator) and the whole area under the line of absolute equality (denominator). Based on Fig. 2.22, the Gini Coefficient is thus equal to the area C divided by the area of the triangle 0AB.
It is possible to use the Gini coefficient to measure the degree of structural regularity, i.e. the homogeneity of tree frequencies in the m x m cells. In a situation of perfect homogeneity, the Lorenz curve would overlap the line of perfect equality and the Gini coefficient would be equal to zero. In the theoretical situation of one cell containing all the trees, the Lorenz curve would coincide with the axes and the Gini coefficient would equal unity.
Two 50 × 50 m experimental plots located in Mexico are used to demonstrate the method. The study areas are located in the high range of the Sierra Madre Occidental, in the north-eastern region of the state of Durango, within the geographical coordinates 24º 30′ 59″ – 25º 30′ 20″ N and 106º 25′ 00″ – 105º 52′ 21″ W. Uneven-aged and highly semi-natural pine-oak stands are the predominant forest types, often mixed with Pseudotsuga, Arbutus and Juniperus (Wehenkel et~al. 2011). The forest covering about 30,350 ha in total is owned and managed by a community (locally known as ejido). The ejido “San Diego de Tezains” belongs to the municipality of Santiago. The forest is mainly managed by selective removals, i.e. as a CCF system, according to the Método Mexicano de Ordenación de Bosques Irregulares (MMOBI; see Torres 2000). The following R code subdivides the area into one hundred 5 × 5 m subplots and counts the number of trees in each cell (Fig. 2.23).

The Gini coefficient is calculated using the library reldist and the following code: 
The following code generates a plot of the trees in the cell grids (Fig. 2.23): 
This approach may be useful if we are interested in measuring the difference in structural homogeneity between two forests, e.g. a virgin forest and a managed one.
As mentioned before, the spatial statistics literature abounds with descriptions of point processes. But many of these are merely of academic interest. In the real world, despite advances in remote sensing (especially laser scanning) mapped tree coordinates are just not available. Thus it is reasonable to suggest that in the study of managed forests, which represent almost all that is left of the original “virgin” ecosystems, the most significant challenge is to develop methods for analysing spatial structures for situations where mapped tree data are not available. Forest structures tend to be complex in CCF systems. Meaningful structural analysis can be performed at affordable cost using the neighborhood methods described in several sections of this chapter.
3.4 Reconstruction and Simulation
Several authors have proposed methods which can be used to reconstruct an entire forest based on the structural information of a sample (Pretzsch 1997; Lewandowski and Gadow 1997; Hui et~al. 2003; Pommerening 2006; Pommerening and Stoyan 2008). The objective of reconstructing a forest in this way may be to improve monitoring of silvicultural operations, to create realistic ecosystems as a basis for spatially explicit analysis and visualisation, or to use more advanced growth models based on known neighborhood constellations and competition status. A reconstruction is considered successful if there is a close resemblance between the real forest and the artificial one. It is usually impossible in practice to measure all the tree positions, the reproduction must thus be achieved with limited information. The information typically available to foresters involves distributions of neighborhood relations obtained during a routine forest inventory. A precondition to obtaining such information is the application of assessment techniques, such as described above. The simulation generates a reproduction of a real forest, based on a sample of the structural variables described in the previous section.
Pretzsch (1997) presented a method for producing a spatially explicit forest based on information about the manner in which two different tree species mingle (e.g., in clusters, rows or groups). He used empirical functions to estimate the distance to the nearest neighbor and a set of probability functions combining an inhomogenous Poisson process and hardcore process. Tree size mingling was not considered and the species are limited to two.
According to Lewandowski and Gadow (1997), a reproduction of a forest is considered to be perfect, if each tree in the real stand has a counterpart in the reproduced one, with exactly the same distances to its three nearest neighbors, and if all the species-mingling values and all size-differentiation values in the reproduction occur with the same frequencies as those in the real forest. Their simulation consisted of four separate phases, during which tree positions are generated and shifted and position attributes exchanged, until the distributions of the structural variables of the reproduction are close enough to those of the real stand. The initial positioning of the trees is done in phase 0. The positions may be allocated either at random or in some predefined manner. The denser the stand, the less important is the manner in which initial coordinates are generated. Phase 1 consists of 3 cyclic subphases: optimizing the distances to the first, the second and the third neighbor. The subphases are repeated until all the distances agree with those of the sample from the real forest. The purpose of phase 2 is to optimize the mingling. This is achieved by successively exchanging two trees of different species (swapping trees of the same species would not change the mingling value).Footnote 1 The swap is retained if it reduces the difference between the real and the simulated distributions of the species-mingling values, otherwise it is repealed. Phase 2 terminates when the difference between the two distributions of the mingling values cannot be further reduced. The diameter differentiation was optimized in phase 3. Again, a pairwise swapping of trees of the same species was done with the aim of attaining a distribution of the size-differentiation values, which closely resembles that of the original forest. The algorithm was later refined by Pommerening (2006) who reversed the analysis and thus enabled the synthesis of forest structure from the indices derived. He investigated this idea with a simulation model that uses the concept of cellular automata. The rules according to which the spatial pattern of tree positions developed in the stand matrix were deduced directly from the distributions of the structural indices of the input data. Pommerening used different combinations of indices to assess and simulate the structure of four sample stands. Simulations using species specific distributions of indices and limiting the number of neighbours to three or four were most successful at reconstructing the original stand structure.
Hui et~al. (2003) proposed a method of reconstruction based on the uniform angle index (UAI) described in a previous section of this chapter. They generated tree coordinates for random, clumped and regular distributions, based on the sampled UAI.
Pommerening and Stoyan (2008) presented a method for synthesizing spatial point patterns from nearest neighbor summary statistics sampled in small circular subwindows. They used a stochstic optimisation technique based on simulated annealing and conditional simulation. The success of their reconstruction was tested by comparing tree point patterns reconstructed from sample data with the known patterns in three structurally different forests. Their validation has shown that it is possible to successfully reconstruct complex forest structures from NNS sample data.
Simulation is a useful technique for generating and analysing particular spatial patterns. The following R code generates a random forest, with 100 tree positions in a 100 × 100 m square plot: 
The number of trees in this example has been specified as a constant. Alternatively, the number of points may be specified as a realisation of a Poisson-distributed random variable with the parameters μ = λ × a × b, where λ is the point density and a and b are the sides of a rectangular observation window (x ≤ random number × a; y ≤ random number × b). The results of three separate runs using the above code are shown in Fig. 2.24.
If the tree positions are regarded as constant, tree attributes (marks) can be assigned to them at random. To generate a random forest, the tree attributes are permuted, i.e. randomly reassigned to the observed tree locations (Hartung 1985, p. 96). There are N! ways in which the observed values of one attribute can be assigned to the N tree positions. It is assumed that each of these N! assignments has the same probability 1/N!. Since the tree attributes are not modified, the distributions of tree species and diameters in the random forest are identical to that of the real forest. Lewandowski and Pommerening (1997) applied this method to generate 1,000 random forests with the associated 1,000 spatial patterns using random permutation. Using the same approach, Schröder (1998) found a surprising agreement in the mean observed and expected DBH differentiation (T i ) of different tree species cohorts in the Knysna natural forest in South Africa, using only the nearest neighbor.
There are many ways to simulate clustered forests. The following code (provided by Walter Zucchini) is useful because of its intuitive logic and simplicity: 
Labelling the positions by generating species, independently and in a given proportion, tree diameters and other attributes, may be done using the code shown above (Fig. 2.24).
The simulation technique enables us to generate random structures which can be used as a basic reference. Some CCF management approaches claim to mimic developments in a natural ecosystem. Deviations from randomness may be helpful when analysing virgin or random forests in relation to managed forests.
4 Discussion
The objective of this contribution was to present methods that can be used to analyse forest structure and diversity with particular reference to CCF methods of ecosystem management. Trees are sessile and once established, their locations are fixed. Each population of trees exhibits a very specific structure and diversity at a given point of time. However, this condition is not static. Tree growth, regeneration and mortality represent important structuring processes. Specific structures generate particular processes of growth and regeneration. These processes in turn produce particular patterns and constellations. Thus, structure and processes are mutually dependent. Associated with a specific forest structure is some degree of heterogeneity or richness. In a forest ecosystem, such diversity does however not only refer to species richness, but to a range of phenomena that determine the heterogeneity within a community of trees. Most important on the macro scale is the diversity of tree dimensions.
Despite advances in remote sensing, mapped tree data are usually not available in the practice of CCF management. Forest inventories tend to provide tree data samples in small observation windows. Thus, in the overwhelming number of cases, nearest neighbor statistics (NNS) are a realistic choice. For this reason, a variety of NNS methods are presented in this chapter, including unmarked as well as market patterns and their use in structural analysis and description. NNS methods assume that the spatial structure of a forest is largely determined by the relationship within neighborhood groups of trees. These methods have important advantages over classical spatial statistics, including low cost field assessment and cohort-specific structural analysis.
Trees are objects which can be conveniently described by their locations and attributes. Thus, methods of point process statistics are useful when mapped data are available. Especially important are the second-order statistics (SOCs), which were developed within the theoretical framework of mathematical statistics and then applied in various fields of research, including forestry. Examples of SOCs in forestry applications are Ripley’s K and Besag’s L function, pair correlation functions and mark variograms. SOCs describe the variability and correlations in marked and non-marked point processes. Functional second-order characteristics depend on a distance variable r and quantify correlations between all pairs of points with a distance of approximately r between them. This allows them to be related to various ecological scales and also, to a certain degree, to account for long-range point interactions.
The analysis of forest structure informs us about the distribution of tree attributes, including the spatial distribution of tree species and their dimensions, crown lengths and leaf areas. The assessment of these attributes facilitates a comparison between a managed and an unmanaged forest ecosystem. In CCF it is especially important to evaluate structural modifications caused by harvest events. For this reason, analytical tools that describe structure and diversity are needed. The structure of a forest is the result of natural processes and human disturbance. Human disturbance in the form of selective tree removal in CCF has a major effect and forest structure is determined to a considerable extent by silviculture.
Important ecosystem functions, and the potential and limitations of human use are defined by the existing forest structures. The vertical and horizontal distributions of tree sizes determine the distribution of micro-climatic conditions, the availability of resources and the formation of habitat niches and thus, directly or indirectly, the biological diversity within a forest community.
Forest structure is not only of interest to students of ecology, but has also economic implications. Uneven-aged multi-species forests were found to be more resistant against natural hazards, and sometimes may generate superior financial returns. However, the management of uneven-aged forests requires not only a basic understanding of the species-specific responses to shading and competition on different growing sites, but also more sophisticated methods of sustainable harvest planning to ensure long-term productivity. A selective harvest event in an uneven-aged forest, involving removal of a variety of tree sizes within each of several species, is much more difficult to quantify and prescribe than standard descriptors of harvest events used in rotation management systems. Thus, the methods presented in this chapter may be helpful in assisting foresters, not only in analysing complex structures, but also in predicting the dynamics of an uneven-aged multi-species forest. A better understanding of forest structure is a key to improved definition of harvest events and to the modeling tree growth and recruitment following that event.
Notes
- 1.
A total enumeration would be a fairly hopeless endeavour. For example, in a forest containing 47 trees (with 20 trees of species 1, 15 of species 2 and 12 of species 3), there are
$$ \left( \begin{array}{l} 47 \\ 20 \end{array} \right)*\left( \begin{array}{l} 27 \\ 15 \end{array} \right)*\left( \begin{array}{l} 12 \\ 12 \end{array} \right) = 1.697*{10^{{20}}} $$different ways in which the species can be assigned to the available positions.
References
Abetz P (1976) Kann und soll die Standraumregulierung in Fichtenbeständen programmiert werden? Forst und Holz 31:117–119
Albert M (1999) Analyse der eingriffsbedingten Strukturveränderung und Durchforstungsmodellierung in Mischbeständen. Dissertation Universität Göttingen, Hainholz Verlag, 201 p
Albert M (2001) Predicting the selection of elite trees in mixed- species stands – a rule-based algorithm for silvicultural decision support systems. Allgemeine Forst und Jagd Zeitung 173(9):153–161
AssunÇÃO R (1994) Testing spatial randomness by means of angles. Biometrics 50:513–537
Barkman JJ (1989) A critical evaluation of minimum area concept. Vegetatio 85:89–104
Besag J (1977) Contribution to the discussion of Dr Ripley’s paper. J R Stat Soc Ser B 39:193–195
Besag JE, Glaves JT (1973) On the detection of spatial pattern in plant communities. Bull Int Stat Inst 45:153–158
Biber P (1997) Analyse verschiedener Strukturaspekte von Waldbeständen mit dem Wachstumssimulator SILVA 2. Vortrag anläßlich der Jahrestagung 1997 der Sektion Ertragskunde im Deutschen Verband Forstlicher Forschungsanstalten. Tagungsbericht, pp 100–120
Borggreve B (1891) Die Holzzucht [Growing Timber]. Berlin
Buys MH, Maritz JS, Boucher C, Van Der Walt JJA (1994) A model for species-area relationships in plant communities. J Veg Sci 5:63–66
Byth K, Ripley BD (1980) On sampling spatial patterns by distance methods. Biometrics 36:279–284
Chung D-J (1996) Konkurrenzverhältnisse und Struktur natürlicher Pinus densiflora – Quercus variabilis – Mischwälder in Korea. Diss., Fak. f. Forstw. u. Waldökologie, Universität Göttingen
Clark PJ, Evans FC (1954) Distance to nearest neighbor as a measure of spatial relationships in populations. Ecology 35:445–453
Condés S (1997) Simulación de parcelas arboladas con datos del 2 Inventario Forestal Nacional. Tesis Doctoral. Tesis Doctoral Escuela Técnica Superior de Ingenieros de Montes de Madrid, 616 pp
Condit R, Pitman N, Leigh EG Jr, Chave J, Terborgh J, Foster RB (2002) Beta-diversity in tropical forest trees. Science 295(5555):666–669
Corral-Rivas JJ, Wehenkel C, Castellanos BH, Vargas LB, Diéguez-Aranda U (2010) A permutation test of spatial randomness: application to nearest neighbour indices in forest stands. J Forest Res 15:218–225
de Caprariis P, Lindemann RH, Collins CM (1976) A method for determining optimum sample size in species diversity studies. Math Geol 8(5):575–581
R Development Core Team (2011) R: a language and environment for statistical computing. R Foundation for statistical computing, Vienna, Austria. http://www.R-project.org
Diggle PJ (2003) Statistical analysis of spatial point patterns, 2nd edn. Arnold, London, 159 p
Diggle PJ, Besag JE, Glaves JT (1976) Statistical analysis of spatial point patterns by means of distance methods. Biometrics 32:659–667
Dixon PM (2002) Ripley's K Function. John Wiley & Sons, Ltd: 342 p
Donnelly KP (1978) Simulations to determine the variance and edge effect of total nearest-neighbour distance. In: Hodder I (ed) Simulation methods in Archaeology. Cambridge University Press, London, pp 91–95
Eyre FH (1980) Forest cover types of the US and Canada. Society of American Foresters, Washington, DC
Ferris R, Humphrey JW (1999) A review of potential biodiversity indicators for application in British forests. Forestry 72(4):313–328
Fisher RA, Corbet SA, Williams CB (1943) The relation between the number of species and the number of individuals in a random sample of an animal population. J Anim Ecol 12:42–58
Franklin JF, Spies TA, Van Pelt R, Carey AB, Thornburgh DA, Berg DR, Lindenmayer DB, Harmon ME, Keeton WS, Shaw DC, Bible K, Chen JQ (2002) Disturbances and structural development of natural forest ecosystems with silvicultural implications, using Douglas-fir forests as an example. Forest Ecol Manage 155(1–3):399–423
Füldner K (1995) Strukturbeschreibung von Buchen-Edellaubholz-Mischwäldern. Cuvillier Verlag, Göttingen
Füldner K, Gadow Kv (1994) How to define a thinning in a mixed deciduous beech forest. Proceedings of the IUFRO conference: mixed stands – research plots, measurements and results, models, Lousa, Portugal, pp 31–42
Gadow Kv, Hui GY (2002) Characterizing forest spatial structure and diversity. Proceedings of the Sustainable Forestry in Southern Sweden (SUFOR) conference “Sustainable Forestry in Temperate Regions”, Lund, April 7–9
Gadow Kv (1993) Zur Bestandesbeschreibung in der Forsteinrichtung. Forst und Holz 48(21):602–606
Gadow Kv (1999) Waldstruktur und Diversität. Allgemeine Forst und Jagdzeitung 170(7):117–122
Gadow Kv, Hui GY (2007) Can the tree species-area relationship be derived from prior knowledge of the tree species richness? Forest Stud/Metsanduslikud Uurimused 46:13–22
Gadow Kv, Schmidt M (1998) Periodische Inventuren und Eingriffsinventuren. Forst und Holz 53(22):667–671
Gadow Kv, Hui GY, Albert M (1998) Das Winkelmass – ein Strukturparameter zur Beschreibung der Individualverteilung in Waldbeständen. Centralblatt für das Gesamte Forstwesen 115:1–10
Ganeshaiah KN, Shaanker UR (2000) Measuring biological heterogeneity of forest vegetation types: avalanche index as an estimate of biological diversity. Biodiver Conserv 9:953–963
Ganeshaiah KN, Shaanker UR (2003) A decade of diversity. University of Agricultural Sciences Bangalore/India. Internal report, 100 p
Gitay H, Rexburg SH, Wilson JB (1991) Species-area relation in a New Zealand tussock grassland, with implications for nature reserve design and for community structure. J Veg Sci 2:113–118
Gleichmar W, Gerold D (1998) Indizes zur Charakterisierung der horizontalen Baumverteilung. Forstwissenschaftliches Centralblatt 117:69–80
Harmon ME, Franklin JF, Swanson F, Sollins P, Gregory SV, Lattin JD, Anderson NH, Cline SP, Aumen NG, Sedell JR, Lienkaemper GW, Cromack Jr, Cummins KW (1986) Ecology of coarse woody debris in temperate ecosystems. In: MacFadey A, Ford ED (eds) Advances in ecological research, vol 15, pp 133–302
Harper JL (1977) Population biology of plants. Academic, London
Hartung J (1985) Statistik. Lehr- und Handbuch der angewandten Statistik. R. Oldenbourg Verlag, München Wien, 806 p
Hasenauer H, Leitgeb E, Sterba H (1996) Der A-Wert nach Johann als Konkurrenzindex für die Abschätzung von Durchforstungseffekten. Allgemeine Forst und Jagdzeitung 167:169–174
Heck CR (1904) Freie Durchforstung. Springer, Berlin
Hessenmöller D (2002) Modelle zur Wachstums- und Durchforstungssimulation im Göttinger Kalkbuchenwald. Doctoral Disserttion, University of Göttingen, 134 p
Hessenmöller D, Gadow Kv (2001) Beschreibung der Durchmesserverteilung von Buchenbeständen mit Hilfe der bimodalen Weibullfunktion. Allgemeine Forst und Jagdzeitung 172(3):46–50
Hines WGS, Hines RJO (1979) The Eberhardt index and the detection of nonrandomness of spatial point distributions. Biometrika 66:73–80
Hobbs RJ, Arico S, Aronson J, Baron JS, Bridgewater P, Cramer VA, Epstein PR, Ewel JJ, Klink CA, Lugo AE, Norton D, Ojima D, Richardson DM, Sanderson EW, Valladares F, Vilà M, Zamora R, Zobel M (2006) Novel ecosystems: theoretical and management aspects of the new ecological world order. Global Ecol Biogeogr 15:1–7
Hubbell SP (2001) The unified neutral theory of biodiversity and biogeography. Princeton University Press, Princeton/Oxford, 375 p
Hui GY, Albert M (2004) Stichprobensimulationen zur Schätzung nachbarschaftsbezogener Strukturparameter in Waldbeständen. Allgemeine Forst und Jagdzeitung 175(10):199–209
Hui GY, Gadow Kv (2002) Das Winkelmass – Herleitung des optimalen Standardwinkels. Allgemeine Forst und Jagdzeitung 173(10):173–177
Hui GY, Albert M, Gadow Kv (1998) Das Umgebungsmaß als Parameter zur Nachbildung von Bestandesstrukturen. Forstwissenschaftliches Centralblatt 117:258–266
Hui GY, Albert M, Chen BW (2003) Reproduktion der Baumverteilung im Bestand unter Verwendung des Strukturparameters Winkelmaß. Allgemeine Forst und Jagdzeitung 174:109–116
Hui GY, Gadow Kv, Hu YB, Xu H (2007) Structure-based forest management. China Forestry Publishing House, Beijing, 200 pp (in Chinese, with English abstract)
Hui GY, Zhao XH, Zhao ZH, Gadow Kv (2011) Evaluating tree species spatial diversity based on neighborhood relationships. Forest Science 57(4):292–300 http://www.safnet.org/publications/forscience/index.cfm
Please update page numbers in Reference “Hyytiäinen and Haigth (2011).” Hyytiäinen K, Haigth RG (2011) Optimizing continuous cover forest management. In: Pukkala T, Gadow von K (eds) Economics of continuous cover forestry. Springer, Dordrecht, pp XXX
Illian J, Penttinen A, Stoyan H, Stoyan D (2008) Statistical analysis and modelling of spatial point patterns. Wiley, Chichester, 534 p
Jaehne S, Dohrenbusch A (1997) Ein Verfahren zur Beurteilung der Destandesdivertität. Summary: a method to evaluate forest stand diversity. Forstwissenshaftliches Centralblatt 116:333–345
Jenssen M, Hofmann G (2002) Die Quantifizierung ökologischer Potentiale der Phytodiversität und Selbstorganisation der Wälder. Beiträge zur Forstwirtschaft und Landschaftsökolgie 37, pp 18–27
Johann K (1982) Der„ A-Wert – “ein objektiver Parameter zur Bestimmung der Freistellungsstärke von Zentralbäumen. [The “A-thinning index” – an objective parameter for the determination of release intensity of frame trees]. Tagungsbericht der Jahrestagung 1982 der Sektion Ertragskunde im Deutschen Verband Forstlicher Forschungsanstalten in Weibersbrunn, pp 146–158
Kangas A, Maltamo M (2002) Anticipating the variance of predicted stand volume and timber assortments with respect to stand characteristics and field measurements. Silva Fennica 36(4):799–811
Kareiva P, Watts S, McDonald R, Boucher T (2007) Domesticated nature: shaping landscapes and ecosystems for human welfare. Science 316(5833):1866–1869
Kato F, Mülder D (1983) Qualitative Gruppendurchforstung der Buche. Allgemeine Forst und Jagdzeitung 154:139–145
Kinnunen J, Maltamo M, Päivinen R (2007) Standing volume estimates of forests in Russia: how accurate is the published data. Forestry 80(1):53–64
Kint V, Meirvenne MV, Nachtergale L, Geuden G, Lust N (2003) Spatial methods for quantifying forest stand structure development: A comparison between nearest- neighbour indices and variogram analysis. Forest Sci 49(1):36–49
Kint V, Marc Mv, Lieven N, Guy G, Noel L (2004) Spatial methods for quantifying forest stand structure development: A comparison between nearest-neighbor indices and variogram analysis. Forest Sci 49(1):36–49
Kiviste A, Laarmann D, Hordo M, Sims A (2007) Factors influencing tree mortality on the basis of permanent forest sample plot data. Paper presented at the International Workshop on disturbance regimes in a changing environment, 03–06 October 2007 in Tukums, Latvia
Kleinn Ch, Ståhl G, Fehrmann L, Kangas A (2010) Issues in forest inventories as an input to planning and decision processes. Allgemeine Forst und Jagdzeitung 181(11/12):205–210
Knoebel BR, Burkhart HE (1991) A bivariate distribution approach to modelling forest diameter distributions at two points of time. Biometrics 47:241–253
Knoke T, Seifert T (2008) Integrating selected ecological effects of mixed European beech–Norway spruce stands in bioeconomic modelling. Ecol Model 210:487–498
Koop H (1981) Vegetatiesstructuur en dynamiek van twee natuurlijke bossen: het Neuenburger en Hasbrucher Urwald, Verslagen van landbouwkundige onderzoekingen 904 PUDOW Wageningen
Koop H (1989) Forest dynamics. Springer, New York
Korpel S (1992) Ergebnisse der Urwaldforschung für die Waldwirtschaft im Buchen-Ökosystem. Allgemeine Forstzeitschrift 47(21):1148–1152
Kramer H (1988) Waldwachstumslehre. Paul Parey, Hamburg
Kramer H, Akça A (1995) Leitfaden zur Waldmesslehre. Sauerländer, JD, Frankfurt a. Main, p 298
Krebs ChJ (1999) Ecological methodology, 2nd edn. Addison Wesley Longman, Inc, Menlo Park, 620 p
Lappi J (1997) A longitudinal analysis of height/diameter curves. Forest Sci 43(4):555–570
Law R, Illian J, Burslem DFRP, Gratzer G, Gunatilleke CVS, Gunatilleke IAUN (2009) Ecological information from spatial patterns of plants: insights from point process theory. J Ecol 97:616–628
Lawton JH (1999) Are there general laws in ecology? Oikos 84:177–192
Legendre P, Gallagher ED (2001) Ecologically meaningful transformations for ordination of species data. Oecologia 129:271–280
Leibundgut H (1978) Die Waldpflege. Paul Haupt, Bern und Stuttgart
Leisch F (2004) FlexMix: a general framework for finite mixture models and latent class regression in R. J Stat Softw 11(8). http://www.jstatsoft.org/v11/i08/
Lepě J, Stursa J (1989) Species-area curve, life history strategies, and succession: a field test of relationships. Vegetatio 83:249–257
Lewandowski A, Pommerening A (1997) Zur Beschreibung der Waldstruktur – Erwartete und beobachtete Artendurchmischung. Forstwissenschaftliches Centralblatt 116:129–139
Lewandowski A, Gadow Kv (1997) Ein heuristischer Ansatz zur Reproduk- tion von Waldbesta ̈nden [A heuristic method for reproducing forest stands]. Allgemeine Forst und Jagdzeitung 168:170–174
Liu C, Zhang L, Davis CJ, Solomon DS, Gove JH (2002) A finite mixture model for characterizing the diameter distributions of mixed-species forest stands. Forest Sci 48(4):653–661
Lotwick HW, Silverman BW (1982) Methods for analysing spatial processes of several types of points. J R Stat Soc Ser B 44:406–413
Luhmann N (1995) Social systems. Stanford University Press, Stanford
Maguire DA, Mainwaring D, Halpern CB (2006) Stand dynamics after variable retention harvesting in mature Douglas-fir forests of western North America. Allgemeine Forst und Jagdzeitung 177:120–131
Margalef R (1957) Information theory in ecology. Genet Syst 3:36–71
Mason B, Kerr G, Pommerening A, Edwards C, Hale S, Ireland D, Moore R (2005) Continuous cover forestry in British conifer forests. In: Forest Research (2005). Annual report and accounts 2003–2004, United Kingdom, pp 38–53
May RM (1975) Patterns of species abundance and diversity. In: Cody ML, Diamond JM (eds) Ecology and evolution of communities. Belknap Harvard University, Cambridge, pp 81–120
McComb WC, Spies TA, Emmingham WH (1993) Douglas-fir forests: managing for timber and mature forest habitat. J Forest 91(12):31–42
McElhinny C, Gibbons P, Brack C, Bauhus J (2005) Forest and woodland stand structural complexity: Its definition and measurement. Forest Ecol Manage 218:1–24
Mehtätalo L (2004) A longitudinal height-diameter model for Norway spruce in Finland. Can J Forest Res 34:131–140
Mitscherlich G, Ganssen R (1951) Die Ergebnisse zweier Buchendurchforstungsversuche in höheren Lagen des Schwarzwaldes. Allgemeine Forst und Jagdzeitung 123:1–15
Moeur M (1993) Crown width and foliage weight of northern Rocky Mountain conifers, USDA, Forest Service, Intermountain Forest and Range Experiment Station, Ogden, Utah, Research Paper INT-283, p 14
Møller J, Waagepetersen RP (2007) Modern statistics for spatial point processes. Scand J Stat 34:643–684
Monod J (1950) La technique de culture continue, the´orie et applications. Annales de l’Institut Pasteur 79:390–410
Motz K, Sterba H, Pommerening A (2010) Sampling measures of tree diversity. Forest Ecol Manage 260:1985–1996
Murray DM, Gadow K (1993) A flexible yield model for regional timber forecasting. Southern J Appl Forest 17(2):112–115
Nothdurft A, Saborowski J, Nuske RS, Stoyan D (2010) Density estimation based on k-tree sampling and point pattern reconstruction. Can J Forest Res 40:953–967
Nyland RD (2002) Silviculture – concepts and applications, 2nd edn. McGraw-Hill, New York
O’Hara KL, Gersonde RF (2004) Stocking control concepts in uneven-aged silviculture. Forestry 77(2):131–143
Penttinen A, Stoyan D, Henttonen HM (1992) Marked point process in forest statistics. Forest Sci 38:806–824
Pielou EC (1961) Segregation and symmetry in two-species populations as studied by nearest-neighbour relations. J Ecol 49:255–269
Pommerening A (1997) Eine Analyse neuer Ansätze zur Bestandesinventur in strukturreichen Wäldern [An analysis of new approaches towards stand inventory in structure-rich forests]. PhD dissertation, Faculty of Forestry and Forest Ecology, University of Göttingen, Cuvillier Verlag Göttingen, 187 pp
Pommerening A (2002) Approaches to quantifying forest structures. Forestry 75:305–324
Pommerening A (2006) Evaluating structural indices by reversing forest structural analysis. Forest Ecol Manage 224:266–277
Pommerening A (2008) Analysing and modelling spatial woodland structure. Habilitationsschrift (DSc dissertation), University of Natural Resources and Applied Life Sciences, Vienna, Austria
Pommerening A, Stoyan D (2006) Edge-correction needs in estimating indices of spatial forest structure. Can J Forest Res 36:1723–1739
Pommerening A, Stoyan D (2008) Reconstructing spatial tree point patterns from nearest neighbour summary statistics measured in small subwindows. Can J Forest Res 38:1110–1122
Preston FW (1962) The canonical distribution of commonness and rarity: Part I. Ecology 43:185–215
Pretzsch H (1995) Perspektiven einer modellorientierten Waldwachstumsforschung. Forstwissenschaftliches Centralblatt 114:188–209
Pretzsch H (1997) Analysis and modeling of spatial stand structures. Methodological considerations based on mixed beech-Larch stands in Lower Saxony. Forest Ecol Manage 97:237–253
Pretzsch H (2002) Grundlagen der Waldwachstumsforschung. [The priciples of forest growth science.]. Parey Buchverlag, Berlin
Pretzsch H (2003) Diversität und Produktivität von Wäldern (Diversity and Productivity of Forests). Allgemeine Forst und Jagdzeitung 174(5/6):88–97
PRODAN, M (1966) Forest Biometrics. Oxford: Pergamon Press, 447 p
Pullan W, Bhadeshia H (eds) (2000) Structure in science and art. Cambridge University Press, Cambridge
Puumalainen (1996) Die Beta-Funktion und ihre analytische Parameterbestimmung für die Darstellung von Durchmesserverteilungen. Georg-August-Universität Göttingen. Working Paper 15–96
Puumalainen J (1998) Optimal cross-cutting and sensitivity analysis for various log dimension constrains by using dynamic programming approach. Scandinavian J Forest Res 13:74–82
Puumalainen J, Gadow Kv, Korhonen K (1998) Flexible Ergebnisberechnung einer Eingriffsinventur. [Flexible calculation of a thinning inventory]. Forstarchiv 69:145–150
Reininger H (1987) Zielstärken-Nutzung oder die Plenterung des Altersklassenwaldes. Österreichischer Agrarverlag, Wien
Richter J (1994) Neue Aspekte der Fichtendurchforstung. Allgemeine Forst und Jagdzeitung 49(12):632–637
Ripley BD (1977) Modelling spatial patterns (with discussion). J R Stat Soc Ser B 39:172–212
Ruggiero LF, Jones LC, Aubry KB (1991) Plant and animal habitats associations in Douglas-fir forests of the Pacific Northwest: an overview. In: Ruggiero LF, Aubry KB, Carey AB, Huff HM (tecn cords) Wildlife and vegetation of unmanage Douglas-fir forest. USDA Forest Service, GTR-PNW-285, pp 447–462
Sanderson EW, Jaiteh M, Levy MA, Redfrod KH, Wannebo AV, Woolmer G (2002) The human footprint and the last of the wild. Bioscience 52:891–904
Schädelin W (1942) Die Auslesedurchforstung als Erziehungsbetrieb höchster Wertleistung, 3rd edn, Bern
Schmidt M, Kiviste A, Gadow Kv (2011) A spatially explicit height-diameter model for Scots Pine in Estonia. Eur J Forest Res. doi:10.1007/s10342-010-0434-8
Schober R (1991) Eclaircies par le haut et arbres d’avenir. Revue Forestière XLIII:385–401
Scholes RJ, Biggs R (2005) A biodiversity intactness index. Nature 434(3):45–49
Schreuder HT, Gregoire TG, Wood GB (1993) Sampling methods for multiresource forest inventory. Wiley, New York, 446 p
Schröder J (1998) Beschreibung von Bestandesstrukturen im Knysna-Wald, Südafrika. Diploma thesis, Faculty of Forestry, University Göttingen, 37 p
Schütz J-Ph (1989) Der Plenterbetrieb. Lecture Notes Waldbau III, ETH Zürich, p 54
Schütz J-P (2002) Silvicultural tools to develop irregular and diverse forest structures. Forestry 75:329–338
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Shimatani K, Kubota Y (2004) Quantitative assessment of multispecies spatial pattern with high species diversity. Ecol Res 19:149–163
Simpson EH (1949) Measurement of diversity. Nature 163:688
Smaltschinski Th (1998) Charakterisierung von Baumverteilungen. Forstwissenschaftliches Centralblatt 117:355–361
Spanos KA, Feest A (2007) A review of the assessment of biodiversity in forest ecosystems. Manage Environ Qual 18(4):475–486
Spies TA (1997) Forest stand structure, composition, and function. In: Creating a forestry for the 21st century: the science of ecosystem management. Oxford University Press
Staudhammer CL, LeMay VM (2001) Introduction and evaluation of possible indices of stand structural diversity. Can J Forest Res 31:1105–1115
Staupendahl K, Puumalainen J (2000) Modellierung des Einflusses von Durchforstungen auf die Durchmesserverteilung von gleichaltrigen Fichtenreibeständen. Forstwissenschaftliches Centralblatt 116(4):249–262
Staupendahl K, Gadow Kv (2008) Eingriffsinventuren und Dynamisches Betriebswerk – Instrumente der Operativen Planung im Forstbetrieb. Forstarchiv 79:16–27
Staupendahl K, Zucchini W (2006) Estimating the spatial distribution in forest stands by counting small angles between nearest neighbours. Allgemeine Forst und Jagdzeitung 177(8/9):160–168
Sterba H (1981) Natürlicher Bestockungsgrad und Reinecke’s SDI. Centralblatt für das gesamte Forstwesen 98(2):101–116
Sterba H, Zingg A (2006) Abstandsabhängige und abstandsunabhängige Bestandesstrukturbeschreibung. Allgemeine Forst und Jagdzeitung 177(8/9):169–176
Stöcker G (2002) Analyse und Vergleich von Bestandesstrukturen naturnaher Fichtenwälder mit Lorenz-Funktionen und Gini-Koeffizienten. Centralblatt für das gesamte Forstwesen 119: 12–39
Stoyan D, Penttinen A (2000) Recent applications of point process methods in forestry statistics. Stat Sci 15:61–78
Stoyan D, Stoyan S (1994) Fractals, random shapes and point fields. In: Methods of geometrical statistics, Wiley, Chichester, UK, 394 p
Suzuki SN, Kachi N, Suzuki J-I (2008) Development of a local size-hierarchy causes regular spacing of trees in an even-aged Abies forest: analyses using spatial autocorrelation and the mark correlation function. Ann Bot 102:435–441
Temesgen H, Gadow Kv (2003) Generalized height-diameter models–an application for major tree species in complex stands of interior British Columbia. Eur J Forest Res 123(1):45–51
Tjørve E (2003) Shapes and functions of species-area curves: a review of possible models. J Biogeogr 30:827–835
Torquato S (2002) Random heterogeneous materials. Microstructure and macroscopic properties. Interdisciplinary applied mathematics 16. Springer, New York
Torres RJM (2000) Sostenibiladad del volumen de cosecha calculado con el método de ordenación de montes. Madera y Bosques 6(2):57–72
Upton G, Fingleton B (1989) Spatial data analysis by example, vol 2: categorical and directional data. Wiley, Chichester, 416 p
Upton G, Fingleton B (1990) Spatial data analysis by example, vol 1: point pattern and quantitative data, 2nd edn. Wiley, Chichester, 410 p
Van Laar A, Akça A (2007) Forest mensuration. Springer, Dordrecht, 418 p
Wehenkel C, Corral-Rivas JJ, Hernandez-Díaz JC, Gadow Kv (2011) Estimating Balanced Structure Areas in multi-species forests on the Sierra Madre Occidental, Mexico. Ann Forest Sci 68. doi:10.1007/s13595-011-0027-9
Wenk G (1996) Durchmesserverteilungen im Buchenplenterwald. Tagungsband des Deutschen Verbandes Forstlicher Versuchsanstalten. Sektion Ertragskunde, Mai 1996 in Neresheim
Westphal C, Tremer N, v Oheimb G, Hansen J, Gadow Kv, Härdtle W (2006) Is the reverse J-shaped diameter distribution universally applicable in European virgin beech forests? Forest Ecol Manage 223:75–83
Whittaker RH (1972) Evolution and measurement of species diversity. Int Assoc Plant Taxon 21:213–251
Williams CB (1964) Patterns in the balance of nature. Academic, London
Williams MR (1995) An extreme-value function model of the species incidence and species-area relationship. Ecology 76:2607–2616
Wood SN (2006) Generalized additive models: an introduction with R. Chapman & Hall/CRC, Boca Raton, p 391
Xia FC (2007) Study on the plant biodiversity and spatial patterns of broad-leaved Korean Pine forest in Changbai Mountain. Doctoral dissertation, Beijing Forestry University, Beijing
Zucchini W, Schmidt M, Gadow Kv (2001) A model for the diameter-height distribution in an uneven-aged beech forest and a method to assess the fit of such models. Silva Fennica 35(2):168–183
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media B.V.
About this chapter
Cite this chapter
Gadow, K.v. et al. (2012). Forest Structure and Diversity. In: Pukkala, T., von Gadow, K. (eds) Continuous Cover Forestry. Managing Forest Ecosystems, vol 23. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-2202-6_2
Download citation
DOI: https://doi.org/10.1007/978-94-007-2202-6_2
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-2201-9
Online ISBN: 978-94-007-2202-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)