Abstract
Iron is an essential element for nearly all organisms on earth including most bacteria, which have to acquire iron to maintain growth. Iron is an important cofactor of many enzymes, serving as a cofactor in electron carrying proteins, and is also important for RNA and DNA metabolism. Although iron is required for growth, high concentrations can be toxic as excess iron promotes generation of free radicals via the Fenton reaction, radicals that damage DNA, proteins, and the cell membrane (Touati, 2000). At the beginning of life on earth, iron was readily available and soluble. However, as our planet matured the levels of oxygen in the atmosphere increased, resulting in dramatically reduced iron solubility, exacerbating the toxic effects associated with this element. Consequently, bacteria had to develop sophisticated mechanisms to scavenge iron from dilute environmental sources and in parallel regulate tightly cellular iron homeostasis. It is interesting to note that as life on earth continues to evolve, the role of iron as an essential element is maintained. Although the microbial growth requirement for iron has been known for many years, it was discovered only recently that this metal serves also as a signal for bacterial biofilm development. In this chapter, we will review the most recent findings concerning iron regulation of biofilm formation within the more general context of the relationship between iron and bacteria in the environment.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Iron is an essential element for nearly all organisms on earth including most bacteria, which have to acquire iron to maintain growth. Iron is an important cofactor of many enzymes, serving as a cofactor in electron carrying proteins, and is also important for RNA and DNA metabolism. Although iron is required for growth, high concentrations can be toxic as excess iron promotes generation of free radicals via the Fenton reaction, radicals that damage DNA, proteins, and the cell membrane (Touati, 2000). At the beginning of life on earth, iron was readily available and soluble. However, as our planet matured the levels of oxygen in the atmosphere increased, resulting in dramatically reduced iron solubility, exacerbating the toxic effects associated with this element. Consequently, bacteria had to develop sophisticated mechanisms to scavenge iron from dilute environmental sources and in parallel regulate tightly cellular iron homeostasis. It is interesting to note that as life on earth continues to evolve, the role of iron as an essential element is maintained. Although the microbial growth requirement for iron has been known for many years, it was discovered only recently that this metal serves also as a signal for bacterial biofilm development. In this chapter, we will review the most recent findings concerning iron regulation of biofilm formation within the more general context of the relationship between iron and bacteria in the environment.
2 Iron Acquisition and Regulation in Bacteria
In nature, under aerobic conditions the soluble form of iron (Fe2+) is scarce (10−8 M) and most iron is found in the insoluble ferric form (Fe3+), and therefore unavailable for use by biological systems. Thus bacteria have had to evolve sophisticated and versatile iron acquisition systems to promote Fe3+ uptake and its conversion to Fe2+.
2.1 Iron Uptake by Siderophores
One mechanism bacteria utilize to scavenge Fe3+ from the environment involves secretion and uptake of siderophores (iron carriers in Greek), which are low molecular weight compounds (<1,000 Da) that chelate iron. They are produced by bacteria and released into the surrounding environment under limiting iron conditions. Siderophores have high affinity for Fe3+, dissociation constants being in the 1022–1050 M−1 range. Typically, the siderophore backbone comprises amino acids, particularly the non-protein d-amino acids ornithine and citrulline (Pohlmann and Marahiel, 2008), and an iron binding moiety. Siderophores are categorized according to their iron binding moiety: (i) hydroxamate, (ii) catechol, or (iii) hydroxyacid (Orsi, 2004).
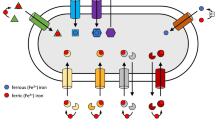
Due to their molecular weight, siderophores are not able to diffuse freely through general porins present in the outer membrane of Gram negative bacteria (which can only transport solutes smaller than 600 Da) and thus require special energy-dependent outer membrane-bound receptors and uptake systems for their transport. In Gram negative bacteria, the cell wall necessitates two uptake stages, initially across the outer membrane into the periplasm and then from the periplasm across the inner membrane into the cytoplasm. Similar to other outer membrane receptors, siderophore receptors form a β-barrel structure comprising 22 transmembranal β-sheets, which are connected by large extracellular and short periplasmic loops. The barrel is “corked” by a globular domain derived from the first 160 amino acids of the N-terminal sequence. Ferri-siderophores bind with high affinity to the external loops and thus trigger a conformational change in the region that mediates contact between the TonB Box, a highly conserved section of the “cork” domain, and the periplasmic part of TonB protein. TonB protein resides within a “TonB complex” (TonB:ExbB:ExbD), which is integrated in the inner membrane and uses the proton gradient as an energy source. The conformational change in the TonB box region induced by the Ferri-siderphore binding to external loops “energizes” TonB such that in turn, Ton B induces a conformational change in the siderophore receptor that promotes transport of the Ferri-siderophore into the periplasm. Once in the periplasm, the Ferri-siderophore is bound by specialized periplasmic binding proteins (PBP) in order to prevent the production of potentially damaging reactive oxygen species via the Fenton reaction. PBPs are classified into nine groups according to sequence similarity (Tam and Saier, 1993; Claverys, 2001). Class 8 PBPs are responsible for Ferri-siderophore binding, with a different PBP binding each category of siderophore. For example, in Escherichia coli FhuD binds to hydroxymate and FepB to catachol type siderophores (Sprencel et al., 2000). Transport of the Ferri-siderophore from the periplasm to the cytoplasm is mediated by an inner membrane transporter of the ATP-binding cassette (ABC) class, a multi-protein complex. Upon delivery of the Ferri-siderophore-PBP complex to the ABC transporter, cytoplasmic ATP is hydrolyzed and the Ferri-siderophore alone is moved into the cytoplasm (Fig. 1). In Gram positive bacteria, which have a different cell wall than Gram negative bacteria, siderophore uptake is much simpler. Siderophores are recognized by membrane-anchored binding proteins and then transported directly into the cytoplasm by ABC transporter systems (Andrews et al., 2003) (Fig. 2).
Once inside the cell, iron has to be released from the siderophore in order to be available for assimilation. There are currently two known mechanisms by which this is achieved (Miethke and Marahiel, 2007):
-
1.
Reduction – Fe3+ is reduced to Fe2+ either enzymatically by cytoplasmic or inner membrane-bound reductases (Schröder et al., 2003) or via a nonenzymatic reaction using intracellular free electron donors, such as NADPH or NADH (Wandersman and Delepelaire, 2004). Since siderophores have a much lower affinity for the Fe2+ ion, this ferrous ion is quickly sequestered by iron binding proteins in the cell. Subsequently, the apo-siderophore is believed to be “recycled,” although a siderophore recycling system has yet to be identified.
-
2.
Degradation – The siderophore is degraded, resulting in release of Fe3+ into the cytoplasm where it is reduced to Fe2+ by cytoplasmic reductases. Examples of proteins that degrade siderophores include Fes, a cytoplasmic esterase that degrades the E. coli siderophore enterobactin (Larsen et al., 2006; Raymond et al., 2003) and Yuil, a cytoplasmic trilactone hydrolase responsible for degrading the Bacillus subtilis siderophore bacillibactin (Miethke et al., 2006).
Many bacteria can scavenge iron from the environment using, in addition to their own, siderophores produced by other bacteria. For example, the Gram positive bacterium Staphylococcus aureus produces the siderophores staphylopherrin and aureochelin but can also utilize the siderophore enterobactin synthesized by the Gram negative bacterium E. coli (Sebulsky and Heinrichs, 2001). In this way, certain bacteria have the ability to “steal” iron from other microorganisms, perhaps providing them with a competitive advantage in some environments.
2.2 Non-Siderophore Iron Uptake
Some bacteria are capable of non-siderophore iron uptake. For example, E. coli expresses ferric citrate receptors that allow them to use ferric citrate as an iron source (e.g., FecA). This transport system also uses TonB to promote movement of ferric citrate into the periplasm (Schröder et al., 2003; Braun and Herrmann, 2007).
During infection, pathogenic bacteria encounter extremely low iron concentrations since most iron is unavailable, bound within organic compounds (heme) or to carrier proteins such as transferrin and lactoferrin. Accordingly, pathogenic bacteria have evolved special mechanisms to survive in such an iron-deprived environment. Some pathogenic bacteria exploit host iron-binding proteins, for example, Neisseria meningitidis, Neisseria gonorrhoeae, and Moraxella catarrhalis can utilize lactoferrin or transferrin as iron sources. Generally, the host iron-binding protein in complex with iron atoms are bound by special bacterial membrane receptors and free iron is transported into the cytoplasm using periplasmic binding proteins and an ABC transporter (Ekins et al., 2004; Miller et al., 2008). However, even in the absence of bacterial receptors to bind particular host iron-binding proteins, iron can still be obtained from these host proteins via their degradation. Examples of enzymes expressed by pathogenic bacteria that degrade host iron-binding proteins include a serine protease from B. subtilis and an alkaline protease from Pseudomonas aeruginosa (Kim et al., 2006; Park et al., 2006).
Some pathogenic bacteria make use of heme as an iron source. Certain Gram negative bacteria release heme-binding proteins, hemophores, which capture free heme and enable its uptake via special receptor systems. Such systems have been identified in P. aeruginosa, Pseudomonas fluorescens, Yersinia pestis, Yersinia enterolitica, Haemophilus influenzae, and Serratia marcescens (Cescau et al., 2007). Due to their high affinity for heme, (e.g., Ka = 5.3 × 1010 M−1 for HasA from S. marcescens), the hemophores can even scavenge heme from within organic compounds, such as hemopexin and hemoglobin (Cescau et al., 2007; Wolff et al., 2008). To date, hemophores have been identified only in Gram negative bacteria. However, recent research has revealed that the Gram positive bacterium Bacillus anthracis secretes two proteins, IsdX1 and IsdX2, which remove heme molecules from hemoglobin (Maresso et al., 2008) and thus, may be considered hemophores. Nonetheless, some Gram positive bacteria express special membrane receptors that bind heme-containing proteins or free heme, which facilitate exploitation of host heme even in the absence of hemophores. For example the Gram positive pathogen S. aureus expresses the haptoglobin-binding protein, HarA (Dryla et al., 2003).
2.3 Iron Starvation Sigma Factor
Iron uptake is an energetically “expensive” process that can bear a “costly price” due to the toxic affects associated with this element. Consequently, bacterial iron homeostasis is strictly regulated. One mechanism by which bacteria coordinate gene expression with iron concentration involves extracytoplasmic function (ECF) sigma factors. ECF sigma factors are a subclass of the σ70 sigma factors (Brooks and Buchanan, 2008). Under normal conditions ECF sigma factors are held by the “antisigma factor” close to the inner membrane, and thus inactivated. The antisigma factor is an integral inner membrane protein, the periplasmic part of which is bound to siderophore receptors. When an iron-loaded siderophore binds the siderophore receptor, an event indicative of low iron conditions, this triggers the antisigma factor to recruit the RNA polymerase (RNAP) machinery to the inner membrane and then release it in complex with the ECF sigma factor. Subsequently, this RNAP–ECF sigma factor complex binds specific promoters and directs their expression in line with limited iron conditions, ensuring a rapid transcriptional response to iron starvation (IS). One of the most studied iron starvation ECF sigma factors is PvdS from the human pathogen P. aeruginosa. When iron-loaded pyoverdine binds its cognate receptor FpvA, the sigma factor PvdS in complex with RNAP is released from the antisigma factor FpvR. Then PvdS guides RNAP to PvdS-dependent promoters, which contain a conserved sequence called the iron starvation box (IS Box) (Visca et al., 2002). The genes under PvdS regulation can be divided into two classes. The first class includes genes responsible for iron acquisition, such as pyoverdine biosynthesis genes and the gene encoding a transferrin degrading enzyme, AprA (Shigematsu et al., 2001). The second class are genes commonly referred to as virulence factors, such as the proteolyic enzyme prpL (Wilderman et al., 2001) and endotoxin A (ToxA) (Hunt et al., 2002). In summary, these environmentally responsive transcription factors enable bacteria to monitor the presence of specific siderophores and respond to the level of iron in the environment.
2.4 Fur and Srna Regulation
An important regulator responsible for iron homeostasis inside the cell is the Ferric Uptake Regulon (Fur) protein. Fur is a dimeric metaloprotein that acts as intracellular iron “sensor.” When iron concentrations surpass a certain threshold, iron binds to the Fur protein and activates it. Activated fur binds to the 19 nucleotide sequence, GATAATGATAATCATTATC, known as the “Fur box,” typically located between −10 and −35 in promoter regions (Rudolph et al., 2006). Fur box binding represses gene expression from that promoter. Fur homologs are found in diverse bacteria. Exceptionally, in N. meningitidis the Fur box is located upstream to the −35 region in a few promoters and Fur binding activates instead of repressing (Delany et al., 2004). The genes repressed by Fur are very diverse and depend on the bacterial type, and even bacterial strain. In P. aeruginosa for example, Fur is an essential protein and is known to repress directly genes involved in iron uptake, such as the pyoverdine operon, heme uptake systems, proteases, and toxins (Vasil, 2007).
In addition, Fur binding to the Fur Box can mediate indirectly activation of gene expression via regulation of sRNAs. sRNAs are short noncoding RNAs, typically 50–500 nucleotides long. sRNAs base pair with a target mRNA, forming an RNA–RNA complex that is recognized and degraded by RNaseE (Pichon and Felden, 2007). In E. coli the sRNA responsible for iron regulation is RyhB, which is expressed constantly. Under iron replete conditions, Fur represses RyhB transcription, resulting in enhanced expression of RyhB target mRNAs, previously degraded due to RyhB. Many RhyB-dependent transcripts encode “nonessential iron-binding proteins,” their expression is desirable only when iron is readily available as these proteins lower the intracellular iron pool and accordingly increase the cellular demand for iron (Jacques et al., 2006). Other RhyB-dependent transcripts are either directly or indirectly involved in cellular metabolism. For example, RhyB influences directly cellular metabolism by regulating the expression of genes involved in the TCA cycle such as sdhABCD (succinate dehydrogenase complex), frdABCD (fumarate reductase) and acnAB (the stationary phase aconitase and exponential phase aconitase, respectively). RhyB indirectly regulates metabolism by affecting expression of Fe–S cluster synthesis enzymes and genes involved in oxidative stress such as sodB (Masse et al., 2005). In P. aeruginosa there are two sRNAs involved in iron metabolism, PrrF1 and PrrF2 (Wilderman et al., 2004). Notably, these same sRNAs were demonstrated recently also to control quorum sensing, a cell density-dependent signaling process, via inhibition of Pseudomonas quinolone quorum sensing signal (PQS) biosynthesis (Oglesby et al., 2008). As quorum sensing is an important social behavior required for successful biofilm formation in P. aeruginosa, this finding hints at the complex relationship between iron and biofilm formation (Davies et al., 1998).
3 Biofilms and Corrosion
The biofilm lifestyle is a protected mode of growth that facilitates bacterial survival in hostile environments. It is now well recognized that microbial cells undergo profound changes during the transition from free-living to matrix-embedded communities (Whiteley et al., 2001; Hall-Stoodley and Stoodley, 2005). Biofilm development occurs in a series of complex but discrete and tightly regulated steps (O’Toole et al., 2000; Hall-Stoodley and Stoodley, 2005): (i) microbial attachment to the surface; (ii) growth, and aggregation of cells into microcolonies; (iii) maturation; and (iv) dissemination of progeny cells for new colony formation.
Bacterial biofilms, which represent the primary colonizers of surfaces in the environment, play an important role in corrosion. Corrosion is an electrochemical process that results in deterioration of a metal due its interaction with the environment (Hamilton, 1985). Corrosion-related costs exceed 276 billion dollars a year in the USA alone (Koch et al., 2001). Rusting of iron materials in the presence of oxygen and water is probably the most familiar and common form of corrosion. A reaction on the metal surface couples iron oxidation (anodic reaction) to reduction of the metal (cathodic reaction). The ferrous ion (Fe2+) is oxidized further to ferric ion (Fe3+) that forms amorphous solid Fe(OH)3 under neutral conditions. Bacterial biofilms influence corrosion by altering the chemistry around a metal. This process is defined as biocorrosion or microbiologically influenced corrosion (Jones and Amy, 2000). Most biocorrosion research has focused on sulfate-reducing bacteria (SRB) that reduce sulfate to sulfide. In the absence of oxygen, sulfide products (such as FeS) act as strong cathodes and accelerate the oxidation of Fe(O) (Lee et al., 1995; Hamilton, 1998, 2003).
The extracellular polymeric matrix, an integral part of biofilms, may also contribute to biocorrosion. The matrix of certain bacterial species can bind metals via anionic functional groups such as phosphate, sulfate, and carboxyl groups (Rohwerder et al., 2003). In particular, the affinity of the matrix for multivalent cations such as Ca2+, Mg2+, and Fe3+ can be very strong, sometimes shifting the standard reduction potentials of the metals. For example, Fe2+/Fe3+ redox potentials vary with different ligands, from +1.2 to −0.4 V. Thus, metals bound to the matrix can act as electron “shuttles” and facilitate novel redox reactions, for example, direct electron transfer from iron or FeS. In the presence of oxygen or another suitable electron acceptor, such a redox reaction depolarizes the cathode and promotes corrosion (reviewed in [Beech and Sunner, 2004]). Chan and colleagues have reported recently that bacterial exopolymers, most notably acidic polysaccharides, can serve as a template for assembling FeOOH crystals. The observed mineralization was shown to result from the contact between the extracellular polymeric substance and oxidized iron, via ferric iron binding with carboxylic groups. This oxidation of ferrous ions and mineralization was noted to incur proton release and a decrease in extracellular pH, which the authors suggested should increase the proton motive force and encourage metabolic energy generation by the cells (Chan et al., 2004). Since the presence of ferrous metal and iron oxyhydroxide in the biofilm matrix enhances oxidation of the ferrous ion, overall the cathodic reaction is promoted and accordingly, corrosion.
Microbial iron respiration is also considered to play an important role in biocorrosion and under some conditions inhibits biocorrosion (reviewed by Lee and Newman (2003). During initial biofilm formation on the metal surface, oxygen is available and is consumed by aerobic respiration. This promotes localized anodic and cathodic reactions, which accelerate electrochemical corrosion. But as oxygen is depleted due to bacterial growth, respiration promotes reduction of the ferric ion, resulting in diffusion of ferrous ion into the surrounding fluid. In this way, under static conditions, iron respiration creates a protective shield of ferrous ions that serve to “soak up” any of the limited oxygen that might be diffusing into the fluid. Any ferrous ions oxidized back to ferric ions are reduced again by respiration. Therefore, the biofilm serves to create locally an anoxic environment that inhibits biocorrosion. Notably, this anoxic environment is exquisitely sensitive to fluid movements. Under flow conditions, Fe2+ ions become diluted and oxygen readily available, similar to the situation during initial biofilm formation, which accelerates corrosion on the surface (Dubiel et al., 2002).
The discovery that biofilms exert a protective affect and inhibit corrosion under certain conditions was unexpected (reviewed in Zuo, 2007). Three mechanisms have been proposed:
-
1.
Removal of corrosive-promoting agents due to bacterial physiological activities – The most obvious corrosive agent is oxygen. Potekhina et al. have shown that biofilm-forming bacteria restrict corrosion under aerobic conditions by utilizing oxygen during respiration (Potekhina et al., 1999). Similarly, Jayaraman and colleagues demonstrated that thicker biofilms protect the surface better, showing that the protection required a viable biofilm and was not due to cellular metabolites secreted by the biofilm, concluding that oxygen consumption and the consequent anoxic environment close to the metal surface reduces corrosion (Jayaraman et al., 1998).
-
2.
Secretion of antimicrobial agents that inhibit growth of corrosion causing bacteria – Jayaraman et al. genetically engineered B. subtilis to express antimicrobials and found that biofilms of this transgenic strain inhibit the growth of corrosion causing SRBs and reduce corrosion rates (Jayaraman et al., 1999a). Furthermore, biofilms of Brevibacillus brevis that naturally secrete gramicidin S have been shown to inhibit SRB colonization and reduce corrosion of mild steel and stainless steel (Jayaraman et al., 1999b). The advantage of this approach to reducing corrosion is that the antimicrobial agents are produced within the biofilm and consequently, do not face the diffusion barriers of the biofilm matrix. Moreover, it is presumed that the limited diffusion through the biofilm matrix ensures a relatively high local concentration of the antimicrobial agent in the biofilm (Jayaraman et al., 1999a).
-
3.
Generation of a layer by the biofilm that protects the surface from corrosion – Bacillus licheniformis biofilms were shown to produce a sticky protective layer of γ-polyglutamate on metal surfaces that reduce corrosion by 90% (Ornek et al., 2002).
Natural biofilms comprise various microbial species with diverse metabolic traits making the study of biocorrosion and biofilm physiology a challenging task. Nonetheless, recent advances in genomic analyses (e.g., metagenomics) and innovative molecular tools should progress our understanding of the mechanisms that promote microbe-induced corrosion, facilitating development of novel approaches to control biocorrosion.
4 Biofilm Formation and Iron Regulation
Research from the last decade has revealed several key cellular processes to be important for biofilm formation including: cell–cell communication (Davies et al., 1998), surface motility (O’Toole and Kolter, 1998), and extracellular polysaccharide production (Matsukawa and Greenberg, 2004). Despite this insight, our current understanding of how bacteria regulate biofilm formation is still limited. In the last few years, an expanding body of work suggests that iron is a major player in the regulation and formation of biofilms. Most studies concerning iron regulation of biofilms have focused on bacterial pathogens as maintaining low free iron concentrations is one of the first lines of defense of the innate immune system. In the following section we will review, species by species, the recent literature describing the effects of iron on biofilm formation.
4.1 Staphylococcus Aureus
S. aureus, a nonmotile Gram positive coccus, is one of the most frequently isolated pathogens associated with nosocomial infections (Johnson et al., 2005). The ability of S. aureus to form biofilms on biotic as well as abiotic surfaces, such as medical devices, contributes greatly to its pathogenicity and capacity to colonize new hosts (Beenken et al., 2004). Staphylococcal biofilms are influenced by various environmental cues such as osmotic stress, anaerobic growth, glucose availability (Johnson et al., 2008), and iron, the latter essential for survival of S. aureus. Accordingly, the bacterium possesses several different iron uptake pathways. It produces three siderophores, termed staphylopherrin A and B and aureochelin (Maresso and Schneewind, 2006). The uptake of these iron-carrying molecules into the bacteria is hypothesized to be mediated by four ABC transporters found in the cytoplasmic membrane of S. aureus. With respect to regulation of iron uptake, the systems identified to date include Fur and a Fur homolog termed perR (Maresso and Schneewind, 2006).
The effects of both replete and restricted iron conditions on biofilm formation by S. aureus have been studied. S. aureus strains display increased biofilm formation in iron-depleted conditions. Addition of 50 µM Fe2(SO4)3 to the growth medium represses biofilm formation. To investigate the involvement of Fur in iron-regulated biofilm formation, an S. aureus Newman strain and an isogenic NewmanDfur::tet mutant were assayed for biofilm formation in restricted and replete iron media (Johnson et al., 2005). The fur mutant, like the wild-type strain, was observed to display reduced biofilm formation in the presence of iron, indicating that Fur is not required to repress biofilm formation in iron replete conditions. However, the fur mutant displayed a fourfold decrease in biofilm production relative to the wild type strain in low iron conditions following a 24 h growth period. This finding suggests that Fur regulates positively biofilm formation under restricted iron conditions (Johnson et al., 2005). But when the strains were grown for a shorter period of time (6 h) in low iron conditions, the fur mutant exhibited more biofilm production than the wild type, pointing to a negative regulatory role for Fur initially in low iron conditions (Johnson et al., 2005). Summarily, Fur appears to have a complex role in the regulation of biofilm formation in iron-depleted conditions. Fur-dependent regulation may be direct, perhaps via binding the promoters of genes involved in biofilm formation, or it may be indirect, via intermediary molecules, like small RNAs (Johnson et al., 2005).
The polymeric N-acetylglucosamine polysaccharide (PNAG) is considered critical for biofilm formation by S. aureus (Cramton et al., 1999). PNAG is synthesized by the products of the ica operon, which comprises the icaR regulatory gene as well as the icaADBC biosynthesis genes (Götz, 2002). Biofilm formation assays show that in low iron conditions a Newman ica mutant produces 93% less biofilm than a wild-type strain (Johnson et al., 2008). However, since wild-type strains grown in iron-rich (low biofilm production) versus iron-poor (high biofilm production) media (Johnson et al., 2005) produce the same amount of PNAG, the aforementioned effect of the ica locus on biofilm formation in low iron is likely mediated by factors other than PNAG. Indeed, this reasoning led to the discovery of two S. aureus secreted proteins, Eap and Emp (Johnson et al., 2005). The levels of these two proteins are reduced significantly in surface protein extracts prepared from the ica mutant grown in low iron media, compared to their levels in extracts taken from the wild-type strain grown similarly. Eap and Emp are bacterial adhesins, noncovalently linked to the S. aureus cell surface. In general, host cell injury, such as a tissue wound or a vessel wall injury, exposes various adhesive glycoproteins including fibronectin, fibrinogen, collagen, and vitronectin, which promote attachment of other eukaryotic cells (Hussain et al., 2001). However, in addition, these exposed glycoproteins serve as substrates for bacterial adhesins such as Eap and Emp. Notably, the interaction between bacterial adhesins and exposed host glycoproteins is considered important for bacterial tissue colonization and biofilm formation inside the host (Hussain et al., 2001). Therefore, it is perhaps not surprising that two bacterial adhesions (Emp and Eap) are regulated by the ica locus and appear to be involved in low iron-induced biofilm formation by S. aureus. Additionally, recent studies have shown that Fur regulates positive expression of these adhesins, which could explain the repressed biofilm formation of the fur mutant observed after sustained growth in low iron conditions (Johnson et al., 2008).
4.2 Staphylococcus Epidermidis
Another biofilm forming bacterium belonging to the Staphylococcus genus is Staphylococcus epidermidis. This nonmotile Gram positive bacterium is a common cause of foreign body-associated infections, such as infections of prosthetic valves, pacemakers, and cerebrovascular shunts. The formation of S. epidermidis biofilms (or “slime”) is largely dependent on production of a polysaccharide called PIA. PIA synthesis is regulated by icaR (Vuong et al., 2005). One study demonstrated that iron-depleted conditions, generated by the addition of the iron chelator EDDA, promote slime production by most clinically relevant strains of S. epidermidis and are associated with the presence of extracellular polysaccharides such as PIA (Deighton and Borland, 1993). The induced PIA production observed in low iron conditions, and concomitant increase in biofilm formation, appears to be related to the functionality of TCA cycle. According to Vuong et al. (2005), a reduction in available iron hinders the functionality of TCA cycle enzymes such as aconitase and fumarase. Consequently, the TCA cycle is disturbed and metabolites are shunted into PIA production (Vuong et al., 2005).
4.3 Pseudomonas Aeruginosa
P. aeruginosa is a motile Gram negative, opportunistic pathogen, associated with nosocomial infections. It is resistant to many types of antibiotic treatments and is able to induce an array of diseases in various hosts. P. aeruginosa is a major player in the chronic lung infections suffered by cystic fibrosis patients and is partially responsible for the high mortality rates of these subjects (Williams et al., 2007). So far, two siderophores have been identified in P. aeruginosa, pyoverdine and pyochelin. The gene products of the pvd locus are responsible for synthesis of pyoverdine, which is effective at acquiring iron from transferrin and lactoferrin (Poole and McKay, 2003). The gene products of two separate operons, pchDCBA and pchEFGHI, determine synthesis of pyochelin, which has a much lower affinity for iron than pyoverdine, but is still effective at acquiring iron from transferrin. A specific receptor transports each of these siderophores, fpvA for pyoverdine and fptA for pyochelin. Pyochelin expression is regulated directly by Fur while the regulation of pyoverdine is mediated by the pvdS sigma factor which is, in turn, regulated by Fur (Poole and McKay, 2003). Additionally, P. aeruginosa can utilize a range of heterologous siderophores originating from various bacteria or fungi and exploit natural iron chelators, like citrate or desferrioxamine, explaining its capacity to thrive in metabolically diverse environments (Poole and McKay, 2003).
In the presence of low iron conditions induced by an iron chelator, like lactoferrin, P. aeruginosa forms an irregular biofilm, characterized as a thin layer of cells. In the absence of lactoferrin, that is, in the presence of sufficient iron concentrations, a normal mushroom-shaped biofilm is formed, comprising a thick layer of cells (Singh et al., 2002). While researching the role of iron in P. aeruginosa aberrant biofilm formation, a novel phenomenon was observed, namely that iron limitation promotes a form of surface motility called twitching (Singh et al., 2002; Patriquin et al., 2008). Twitching is mediated by type IV pili and reflects the spreading of P. aeruginosa over the surface of a substratum during the initial stages of biofilm formation. It is believed that iron limitation causes the cells to “wander” constantly across the surface, thus disrupting their ability to settle and form structured communities (Singh et al., 2002; Singh, 2004). Twitching is also thought to be involved in generating the “stalk” arrangement beneath the mushroom caps of structured biofilms (Klausen et al., 2003) (Fig. 3).
The role of iron in Pseudomonas aeruginosa biofilm formation. (a) In high iron conditions cells attach normally and multiply but do not remain attached to the glass surface and biofilms do not form. (b) In replete (normal) iron conditions (1–100 μM) bacteria attach, multiply and develop into microcolonies that mature into structured mushroom-like biofilms. (c) Low iron conditions promote the twitching phenomenon, cells attach and multiply but daughter cells move away from the point of replication disrupting the formation of structured biofilms.
To investigate the role of iron in biofilm development, several mutant strains of P. aeruginosa PAO1 have been constructed: pyoverdine mutant (PAO.1DpvdA), pyochelin mutant (PAO1pchA:TcR+), and a pyoverdine pyochelin double mutant (pvdA,pchA). Biofilm formation by each of these mutant strains was analyzed carefully in the absence of lactoferrin (replete iron conditions). The pyochelin mutant, like the wild-type strain, produces a normal mushroom-like biofilm. In contrast, the pyoverdine mutant forms an abnormal biofilm, similar to the biofilm formed by wild type in the presence of lactoferrin. However, after exposure to pyoverdine-conditioned medium, the pyoverdine mutant was found to produce a normal biofilm. The pyoverdine pyochelin double mutant also produces an abnormal biofilm, even in the presence of very high iron concentrations. Nevertheless, addition of ferric dicitrate or desferrioxamine, two natural iron chelators that can be exploited by P. aeruginosa, to the growth medium of the pyoverdine mutant was observed to allow normal biofilm development. Taken together, these results evidence that ongoing iron acquisition from the environment is essential for P. aeruginosa to be able to develop a normal mushroom-shaped biofilm (Banin et al., 2005). But iron-regulated biofilm development seems to proceed through “check points.” One check point is that in response to low iron twitching is stimulated. Another, seemingly master Fur-dependent check point is indicated by the finding that a Fur mutant produces a normal biofilm even in the presence of lactoferrin (Banin et al., 2005).
Recent findings suggest a relationship between the iron regulon and the quorum sensing regulon (Bollinger et al., 2001; Cornelis and Aendekerk, 2004; Lequette et al., 2006). This connection highlights the importance of iron in biofilm production since quorum sensing is known to play a key role in biofilm development (Davies et al., 1998; Singh et al., 2000). P. aeruginosa possesses two HSL-mediated quorum sensing systems. The first one consists of LasR, a transcriptional regulator, and LasI, a protein responsible for the synthesis of the las system autoinducer, 3-oxo-C12-HSL. The second quorum sensing system comprises the transcriptional regulator RhlR, and rhlI, a gene responsible for the synthesis of the rhl system autoinducer, C4-HSL. These las and rhl systems regulate the expression of several virulence factors, including alkaline protease, rhamnolipids, elastase, phospholipase C, pyocyanin, as well as biofilm formation (Duan and Surette, 2007). A connection between the iron and quorum sensing regulons was discovered when it was observed that a mutation in the rhlI gene of the P. aeruginosa K2589 strain causes a reduction in twitching and is characterized by normal biofilm formation despite low iron conditions (Patriquin et al., 2008). Specifically, limiting iron conditions induce rhlI expression, resulting in increased amounts of the autoinducer signal C4-HSL, which in turn promotes twitching and affects biofilm formation.
Iron levels influence not only establishment but also maintenance of P. aeruginosa biofilms. High iron concentrations perturb biofilm formation and promote dissociation of a preformed biofilm (Musk et al., 2005; Yang et al., 2007). Specifically, Musk et al. reported that iron salts (ammonium ferric citrate, ferric chloride, ferric sulfate, and ferrous sulfate), at iron concentrations >100 μM, inhibit P. aeruginosa biofilm formation without any effect on growth. This inhibition is not due to reduced adhesion of cells to the surface as initial biofilm formation, the first 10 h of development, is unaffected. Rather the excess iron seems to disrupt the later stages of biofilm development, such that very few cells are adhering to the surface by 48 h (Musk et al., 2005) (Fig. 3). Yang et al. found that elevated iron conditions reduce the amount of extracellular DNA (ecDNA), an important component of biofilm matrices (Yang et al., 2007). Further study implicated that this fluctuation in ecDNA levels is mediated by a third quorum sensing system found in P. aeruginosa, the pqs system (Yang et al., 2007). Briefly, the autoinducer of the pqs system is 2-heptyl-3-hydroxy-4-quinolone, commonly referred to as PQS (Pesci et al., 1999). Two mutant strains deficient in PQS production, mutated at the pqsA and pqsR loci, display iron-independent reductions in ecDNA, supporting that PQS regulates ecDNA production but importantly, revealing that the regulatory effect of iron concentration on ecDNA production is mediated by the pqs system. Further evidence for this link between quorum sensing and iron-regulated biofilm maintenance is provided by the finding that biofilms grown in flow cell chambers with low iron conditions express pqs highly, whereas biofilms grown in high iron conditions express pqs poorly (Yang et al., 2007).
4.4 Escherichia Coli
E. coli is a motile Gram negative bacterium belonging to the family Enterobacteriaceae. Certain strains of E. coli are involved in urinary tract infections (UTIs). Two phenotypic traits of E. coli contribute to colonization of the urinary tract, namely fast growth and superior biofilm formation (Roos et al., 2006; Hancock et al., 2007). E. coli possesses three iron uptake systems, a low affinity aerobactin system and two high affinity systems, yersiniabactin (Ybt) and enterobactin. Synthesis of the Ybt siderophore and its receptor FyuA is dependent on proteins encoded within the high pathogenicity island (HPI). Microarray analysis of the urinary tract isolate E. coli VR50 has shown that during biofilm growth HPI is among the most upregulated gene clusters (Hancock et al., 2008), supporting that iron plays a role in biofilm formation. In order to investigate further a role for iron in biofilm formation, using human urine as the growth medium, E. coli VR50 and a fyuA deletion mutant, VR50fyuA, were grown in a flow chamber system and their biofilm formation monitored. The fyuA deletion mutant displays impaired biofilm formation, with respect to biomass and biofilm structure, forming small, scattered patches of biofilm whereas the wild type strain covers the entire surface of the glass slide. Biofilm formation by another urinary tract E. coli strain 83972 was also examined. A mutation in the fyuA gene of this strain also results in significantly reduced biofilm formation, specifically, a 53% reduction in microtitre plates when compared to the parent strain (Hancock et al., 2008). Taken together, these data indicate that FyuA expression affects biofilm formation and support the premise that iron regulates biofilm formation by UTI E. coli strains. However, it was necessary to demonstrate explicitly that the role of fyuA in biofilm formation relates to its ability to transport iron into the cell via Ybt-Fe binding, for the position of FyuA in the outer membrane raises the possibility that FyuA contributes to biofilm formation by affecting cellular adhesiveness. Therefore, to clarify the role of FyuA in biofilm formation Hancock et al. examined the affect of iron addition to the growth medium on biofilm formation by fyuA deletion mutant strains and found it sufficient to promote biofilm formation. Moreover, three UTI wild-type strains were shown to produce less biofilm in the presence of iron chelators and more biofilm in the presence of added iron. Summarily, these observations corroborate the premise that iron serves as an important signal for biofilm formation by UTI E. coli strains (Hancock et al., 2008).
4.5 Vibrio Cholerae
Vibrio cholerae is a Gram negative, rod-shaped bacterium. It thrives naturally in fresh and salty waters and when invading a human host colonizes the gastrointestinal tract causing cholera. The capacity of V. cholerae to form biofilms is crucial for both its survival in aquatic environments and its ability to colonize human hosts (Hall-Stoodley and Stoodley, 2005). As with numerous other bacteria, discussed above, it seems that iron is an important player in biofilm formation by V. cholerae. Biofilm formation by the E1 Tor V. cholerae strain N16961 has been examined under different conditions. In the presence of iron chelators (low iron conditions) N16961 produces little biofilm and represses rugose switching, a sort of colony morphology switch that relates to the expression level of exopolysaccharide. This phenotype accords with earlier data as typically rugose switching indicates increased exopolysaccharide expression and is associated with more biofilm formation (Wai et al., 1998). The effect of iron on biofilm formation appears to be mediated by RyhB, a small regulatory RNA responsible for down-regulating expression of sodB, TCA cycle enzymes, and energy metabolism proteins. ryhB transcription is controlled by a Fur and iron-responsive promoter, induced >tenfold in low iron conditions and repressed by Fur in replete iron conditions. To validate that iron regulates biofilm formation by V. cholerae, a N16961 strain mutated in the rhyB locus was generated and examined. The ryhB mutant is unable to form normal biofilms in low iron conditions, but addition of excess iron restores normal biofilm formation, suggesting that the rhyB mutant is iron stressed. Notably, the rhyB mutant was observed to exhibit decreased chemotaxis in low iron conditions implying that RhyB is required also for normal cellular iron metabolism. Although these data seemingly support iron as a player in biofilm formation, RhyB could influence biofilm formation in low iron conditions via mechanisms independent of iron metabolism. Since RyhB expression is induced in limiting iron conditions and RyhB represses energy metabolism, it is possible that in the ryhB mutant certain energy metabolism enzymes are inappropriately expressed. Indeed, it has been shown that in the rhyB mutant under low iron conditions the succinate dehydrogenase operon exhibits up-regulated expression, which presumably lowers substantially the cellular levels of succinate, a component of the exopolysaccharide produced by V. cholerae during biofilm growth. Therefore, RyhB involvement in biofilm regulation under limiting iron conditions may be mediated by repression of succinate dehydrogenase (Mey et al., 2005). Furthermore, the role of RyhB in biofilm regulation in low iron conditions may be via its effect on motility, which is considered important for biofilm formation. Microarray analyses reveal decreased expression of several flagellar genes in the ryhB mutant. In conclusion, the iron-regulated repressor small RNA RyhB influences biofilm formation in low iron conditions most likely via several different mechanisms, including iron metabolism, energy metabolism and motility (Mey et al., 2005).
4.6 Oral Pathogens
The relationship between iron and biofilm formation has been studied in three oral pathogens: Streptococcus mutans, Actinomyces naeslundii, and Haemophilus actinomycetemcomitans. S. mutans is a Gram positive bacterium and the major cause of dental cavities. Colonization of S. mutans on the tooth surface promotes adhesion of other oral pathogens, thus initiating formation of a mixed species biofilm that is dental plaque (Rolerson et al., 2006). A study of this pathogen concluded that aggregation, which eventually leads to biofilm formation, is induced in iron-restricted saliva and in normal saliva when the iron concentrations are kept within the range 0.1–1 µM. Increasing the iron concentration in saliva was found to cause a decrease in cell aggregation and biofilm formation (Francesca et al., 2004).
Similarly, it has been observed that low environmental iron conditions promote biofilm formation by the oral pathogen A. naeslundii, a Gram positive bacterium that is an early colonizer of the oral cavity. Accordingly, excess iron inhibits biofilm formation by this pathogen. An iron-dependent repressor thought to regulate biofilm formation by A. naeslundii in high iron concentrations is AmdR. For an amdR mutant produces biofilms even in a medium with increased metal ion concentration (Moelling et al., 2007).
The third oral pathogen for which iron regulation of biofilm development has been studied is H. actinomycetemcomitans, a Gram negative bacterium associated with periodontitis, an inflammatory disease that affects dental tissue. H. actinomycetemcomitans does not use siderophores to acquire iron from the environment. Rather, it uses mostly host haemin as an iron source. In agreement with the phenotypes of other Gram negative bacteria, it has been reported that the presence of DIP, an iron chelator, reduces biofilm formation by certain strains of H. actinomycetemocomitans (Rhodes et al., 2007).
4.7 Mycobacterium Smegmatis
Mycobacterium smegmatis, a saprophytic, motile, Gram positive bacterium is found mostly in the environment near large bodies of water (Brown-Elliott and Wallace, 2002). M. smegmatis serves as a model organism for the more pathogenic species belonging to its genus, like Mycobacterium leprae and Mycobacterium tuberculosis. Biofilm development by M. smegmatis requires the presence of at least 1 µM iron in the growth medium (Ojha et al., 2005). This observation and the finding that iron acquisition genes are upregulated in biofilms relative to planktonic cells, suggest that iron is an important player in biofilm formation by M. smegmatis. Accordingly, mutations affecting synthesis of exochelin, one of the siderophores synthesized by M. smegmatis, or affecting its uptake, are associated with impaired biofilm formation. However, mutations in mycobactin, another siderophore synthesized by M. smegmatis, or in iron-ABC transporters, do not cause impaired biofilm formation. Thus, it appears that specifically the exochelin iron-uptake system is required for M. smegmatis biofilm formation. Notably, iron regulates also production of C56–C68 fatty acids, which are building blocks for some biofilm matrix components. M. smegmatis grown in iron concentrations below 1 µM produce very low amounts of C56–C68 fatty acids, which could explain the poor biofilm formation in these conditions (Ojha and Hatfull, 2007).
4.8 Iron and the Biofilm Matrix
As alluded above, changes in iron concentration can influence synthesis of the biofilm matrix. The building blocks of the biofilm matrix are polysaccharides, proteins, and DNA (Chen and Stewart, 2002; Whitchurch et al., 2002). For example, Yang et al. showed that the amount of ecDNA, an important matrix component of P. aeruginosa biofilms, is reduced in response to elevated iron concentrations (Yang et al., 2007). One of the forces thought to be involved in cohesion of the biofilm matrix is electrostatic interactions. In agreement with this, Chen and Stewart demonstrated that the viscosity of a biofilm suspension can be affected by changes in ionic strength and composition (Chen and Stewart, 2002). Also, treatment of P. aeruginosa–Klebsiella pneumoniae mixed biofilm suspensions with FeCl2 or Fe(NO3)3 has been shown to increase biofilm viscosity, by 56% and 44% respectively (Chen and Stewart, 2002). Similarly, Banin et al. have found that chelation of iron, mediated by the addition of EDTA (50 mM), initiates detachment of cells from mature P. aeruginosa biofilms (Banin et al., 2006). Taken together, these results indicate that iron may not only be an important signal for biofilm development but also a cross-linking agent, which promotes covalent bonding between polymers and stabilizes the biofilm matrix.
5 Concluding Remarks
A few themes emerge from the data accumulated to date. First, there seems to be a general difference between Gram positive and Gram negative bacteria with regards to biofilm formation and iron. For the most part, biofilm formation by Gram positive bacteria increases in response to low iron conditions. This is true for both S. aureus and Staphylococcus epidermidis and two of the three oral pathogens discussed here, S. mutans and A. naeslundii, all of which are Gram positive bacteria. In contrast, typically Gram negative bacteria, including P. aeruginosa, E. coli, and V. cholerae, repress biofilm formation in low iron conditions. However, this dichotomy is not universally accurate. The Gram positive bacterium Mycobacterium smegmatis demonstrates impaired biofilm formation in medium lacking a minimal iron concentration (Ojha et al., 2005) and the nonmotile Gram negative bacterium Acinetobacter baumannii exhibits increased biofilm formation in the presence of iron chelators (Tomaras et al., 2003).
Another theme concerns the role of motility in iron-regulated biofilm development. All nonmotile bacteria including S. aureus, S. epidermidis, S. mutans, A. naeslundii, and A. baumannii demonstrate increased biofilm formation in low iron conditions. In contrast, all motile bacteria including P. aeruginosa, E. coli, V. cholerae, and M. smegmatis display decreased biofilm formation in low iron conditions. Notably, in the case of P. aeruginosa, induction of surface motility has been suggested to explain the impaired biofilm formation observed under iron limiting condition (Singh et al., 2002; Singh, 2004). The only exception to this “motility classification” is the nonmotile, oral pathogen H. actinomycetemcomitans, which displays reduced biofilm formation in iron-chelated medium, but this microbe has not been studied extensively. An important question arising from this classification is whether iron regulates directly surface motility or controls cell–cell adhesion, which in turn affects motility and biofilm formation. Future work should characterize in more detail the role of motility in iron-regulated biofilm development in various bacterial species.
The biofilm mode of growth is an important feature of bacterial lifestyle that affects our daily life. These are exciting times in biofilm research as newly improved molecular and microscopy tools should allow us to begin to dissect the physiology of bacterial biofilms in the environment. A better understanding of biofilm growth is important not only for our intellectual appreciation of the living world, but also required urgently in order to control bacterial biofilm formation in industrial and medical settings. Specifically, the influence of iron on biofilm formation, the topic discussed in this review, represents a potential new approach to controlling biofilm formation.
References
Andrews, S.C., Robinson, A.K. and Rodriguez-Quinones, F. (2003) Bacterial iron homeostasis. FEMS Microbiol. Rev. 27: 215–237.
Banin, E., Vasil, M.L. and Greenberg, E.P. (2005) Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. USA 102: 11076–11081.
Banin, E., Brady, K.M. and Greenberg, E.P. (2006) Chelator-induced dispersal and killing of Pseudomonas aruginoa. Appl. Environ. Microbiol. 72: 2064–2069.
Beech, I.B. and Sunner, J. (2004) Biocorrosion: towards understanding interactions between biofilms and metals. Curr. Opin. Biotechnol. 15: 181–186.
Beenken, K.E., Dunman, P.M., McAleese, F., Macapagal, D., Murphy, E., Projan, S.J., Blevins, J.S. and Smeltzer, M.S. (2004) Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186: 4665–4684.
Bollinger, N., Hassett, D.J., Iglewski, B.H., Costerton, J.W. and McDermott, T.R. (2001) Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J. Bacteriol. 183: 1990–1996.
Braun, V. and Herrmann, C. (2007) Docking of the periplasmic FecB binding protein to the FecCD transmembrane proteins in the ferric citrate transport system of Escherichia coli. J. Bacteriol. 189: 6913–6918.
Brooks, B.E. and Buchanan, S.K. (2008) Signaling mechanisms for activation of extracytoplasmic function (ECF) sigma factors. Biochim. Biophys. Acta 1778: 1930–1945.
Brown-Elliott, B.A. and Wallace, R.J. Jr. (2002) Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15: 716–746.
Cescau, S., Cwerman, H., Letoffe, S., Delepelaire, P., Wandersman, C. and Biville, F. (2007) Heme acquisition by hemophores. Biometals 20: 603–613.
Chan, C.S., De Stasio, G., Welch, S.A., Girasole, M., Frazer, B.H., Nesterova, M.V., Fakra, S. and Banfield, J.F. (2004) Microbial polysaccharides template assembly of nanocrystal fibers. Science 303: 1656–1658.
Chen, X. and Stewart, P.S. (2002) Role of electrostatic interactions in cohesion of bacterial biofilms. Appl. Microbiol. Biotechnol. 59: 718–720.
Claverys, J.P. (2001) A new family of high-affinity ABC manganese and zinc permeases. Res. Microbiol. 152: 231–243.
Cornelis, P. and Aendekerk, S. (2004) A new regulator linking quorum sensing and iron uptake in Pseudomonas aeruginosa. Microbiology 150: 752–756.
Cramton, S.E., Gerke, C., Schnell, N.F., Nichols, W.W. and Gotz, F. (1999) The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67: 5427–5433.
Davies, D.G., Parsek, M.R., Pearson, J.P., Iglewski, B.H., Costerton, J.W. and Greenberg, E.P. (1998) The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280: 295–298.
Deighton, M. and Borland, R. (1993) Regulation of slime production in Staphylococcus epidermidis by iron limitation. Infect. Immun. 61: 4473–4479.
Delany, I., Rappuoli, R. and Scarlato, V. (2004) Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 52: 1081–1090.
Dryla. A., Gelbmann. D., von Gabain, A. and Nagy, E. (2003) Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol. Microbiol. 49: 37–53.
Duan. K. and Surette, M.G. (2007) Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J. Bacteriol. 189: 4827–4836.
Dubiel, M., Hsu, C.H., Chien, C.C., Mansfeld, F. and Newman, D.K. (2002) Microbial iron respiration can protect steel from corrosion. Appl. Environ. Microbiol. 68: 1440–1445.
Ekins, A., Khan, A.G., Shouldice, S.R. and Schryvers, A.B. (2004) Lactoferrin receptors in Gram-negative bacteria: insights into the iron acquisition process. Biometals 17: 235–243.
Francesca, B., Ajello, M., Bosso, P., Morea, C., Andrea, P., Giovanni, A. and Piera, V. (2004) Both lactoferrin and iron influence aggregation and biofilm formation in Streptococcus mutans. Biometals 17: 271–278.
Götz, F. (2002) Staphylococcus and biofilms. Mol. Microbiol. 43: 1367–1378.
Hall-Stoodley, L. and Stoodley, P. (2005) Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 13: 7–10.
Hamilton, W.A. (1985) Sulphate-reducing bacteria and anaerobic corrosion. Annu. Rev. Microbiol. 39: 195–217.
Hamilton, W.A. (1998) Bioenergetics of sulphate-reducing bacteria in relation to their environmental impact. Biodegradation 9: 201–212.
Hamilton, W.A. (2003) Microbially influenced corrosion as a model system for the study of metal microbe interactions: a unifying electron transfer hypothesis. Biofouling 19: 65–76.
Hancock, V., Ferrieres, L. and Klemm, P. (2007) Biofilm formation by asymptomatic and virulent urinary tract infectious Escherichia coli strains. FEMS Microbiol. Lett. 267: 30–37.
Hancock, V., Ferrieres, L. and Klemm, P. (2008) The ferric yersiniabactin uptake receptor FyuA is required for efficient biofilm formation by urinary tract infectious Escherichia coli in human urine. Microbiology 154: 167–175.
Hunt, T.A., Peng, W.T., Loubens, I. and Storey, D.G. (2002) The Pseudomonas aeruginosa alternative sigma factor PvdS controls exotoxin A expression and is expressed in lung infections associated with cystic fibrosis. Microbiology 148: 3183–3193.
Hussain, M., Becker, K., von Eiff, C., Schrenzel, J., Peters, G. and Herrmann, M. (2001) Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J. Bacteriol. 183: 6778–6786.
Jacques, J.F., Jang, S., Prevost, K., Desnoyers, G., Desmarais, M., Imlay, J. and Masse, E. (2006) RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol. Microbiol. 62: 1181–1190.
Jayaraman, A., Sun, A.K. and Wood, T.K. (1998) Characterization of axenic Pseudomonas fragi and Escherichia coli biofilms that inhibit corrosion of SAE 1018 steel. J. Appl. Microbiol. 84: 485–492.
Jayaraman, A., Hallock, P.J., Carson, R.M., Lee, C.C., Mansfeld, F.B. and Wood, T.K. (1999a) Inhibiting sulfate-reducing bacteria in biofilms on steel with antimicrobial peptides generated in situ. Appl. Microbiol. Biotechnol. 52: 267–275.
Jayaraman, A., Ornek, D., Duarte, D.A., Lee, C.C., Mansfeld, F.B. and Wood, T.K. (1999b) Axenic aerobic biofilms inhibit corrosion of copper and aluminum. Appl. Microbiol. Biotechnol. 52: 787–790.
Johnson, M., Cockayne, A., Williams, P.H. and Morrissey, J.A. (2005) Iron-responsive regulation of biofilm formation in Staphylococcus aureus involves fur-dependent and fur-independent mechanisms. J. Bacteriol. 187: 8211–8215.
Johnson, M., Cockayne, A. and Morrissey, J.A. (2008) Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect. Immun. 76: 1756–1765.
Jones. D.A. and Amy, P.S. (2000) Related electrochemical characteristics of microbial metabolism and iron corrosion. Ind. Eng. Chem. Res. 39: 575–582.
Kim, S.J., Park, R.Y., Kang, S.M., Choi, M.H., Kim, C.M. and Shin, S.H. (2006) Pseudomonas aeruginosa alkaline protease can facilitate siderophore-mediated iron-uptake via the proteolytic cleavage of transferrins. Biol. Pharm. Bull. 29: 2295–2300.
Klausen, M., Aaes-Jorgensen, A., Molin, S. and Tolker-Nielsen, T. (2003) Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50: 61–68.
Koch, G.H., Brongers, M.P.H., Thompson, N.G., Virmani, Y.P. and Payer, J.H. (2001) Corrosion Costs and Preventative Strategies in the United States. Report by CC Technologies Laboratories to the Federal Highway Administration. Report FHWA-RD-01-156. Office of Infrastructure Research and Development, Washington, DC.
Larsen, N.A., Lin, H., Wei, R., Fischbach, M.A. and Walsh, C.T. (2006) Structural characterization of enterobactin hydrolase IroE. Biochemistry 45: 10184–10190.
Lee, A.K. and Newman, D.K. (2003) Microbial iron respiration: impacts on corrosion processes. Appl. Microbiol. Biotechnol. 62: 134–139.
Lee, N.S., Kim, B.T., Kim, D.H. and Kobashi, K. (1995) Purification and reaction mechanism of arylsulfate sulfotransferase from Haemophilus K-12, a mouse intestinal bacterium. J. Biochem. 118: 796–801.
Lequette, Y., Lee, J.H., Ledgham, F., Lazdunski, A. and Greenberg, E.P. (2006) A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J. Bacteriol. 188: 3365–3370.
Maresso, A.W. and Schneewind, O. (2006) Iron acquisition and transport in Staphylococcus aureus. Biometals 19: 193–203.
Maresso, A.W., Garufi, G. and Schneewind, O. (2008) Bacillus anthracis secretes proteins that mediate heme acquisition from hemoglobin. PLoS. Pathog. 4 :e1000132.
Masse, E., Vanderpool, C.K. and Gottesman, S. (2005) Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187: 6962–6971.
Matsukawa. M. and Greenberg, E.P. (2004) Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186: 4449–4456.
Mey, A.R., Craig, S.A. and Payne, S.M. (2005) Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 73: 5706–5719.
Miethke, M. and Marahiel, M.A. (2007) Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71: 413–451.
Miethke, M., Klotz, O., Linne, U., May, J.J., Beckering, C.L. and Marahiel, M.A. (2006) Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol. Microbiol. 61: 1413–1427.
Miller, C.E., Rock, J.D., Ridley, K.A., Williams, P.H. and Ketley, J.M. (2008) Utilization of lactoferrin-bound and transferrin-bound iron by Campylobacter jejuni. J. Bacteriol. 190: 1900–1911.
Moelling, C., Oberschlacke, R., Ward, P., Karijolich, J., Borisova, K., Bjelos, N. and Bergeron, L. (2007) Metal-dependent repression of siderophore and biofilm formation in Actinomyces naeslundii. FEMS Microbiol. Lett. 275: 214–220.
Musk, D.J., Banko, D.A. and Hergenrother, P.J. (2005) Iron salts perturb biofilm formation and disrupt existing biofilms of Pseudomonas aeruginosa. Chem. Biol. 12: 789–796.
O’Toole, G.A. and Kolter, R. (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30: 295–304.
O’Toole, G., Kaplan, H.B. and Kolter, R. (2000) Biofilm formation as microbial development. Annu. Rev. Microbiol. 54: 49–79.
Oglesby, A.G., Farrow, J.M., 3rd, Lee, J.H., Tomaras, A.P., Greenberg, E.P., Pesci, E.C. and Vasil, M.L. (2008) The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J. Biol. Chem. 283: 15558–5567.
Ojha, A. and Hatfull, G.F. (2007) The role of iron in Mycobacterium smegmatis biofilm formation: the exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth. Mol. Microbiol. 66: 468–483.
Ojha, A., Anand, M., Bhatt, A., Kremer, L., Jacobs, W.R., Jr. and Hatfull, G.F. (2005) GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123: 861–873.
Ornek, D., Jayaraman, A., Syrett, B.C., Hsu, C.H., Mansfeld, F.B. and Wood, T.K. (2002) Pitting corrosion inhibition of aluminum 2024 by Bacillus biofilms secreting polyaspartate or γ-polyglutamate. Appl. Microbiol. Biotechnol. 58: 651–657.
Orsi, N. (2004) The antimicrobial activity of lactoferrin: current status and perspectives. Biometals 17: 189–196.
Park, R.Y., Sun, H.Y., Choi, M.H., Bai, Y.H., Chung, Y.Y. and Shin, S.H. (2006) Proteases of a Bacillus subtilis clinical isolate facilitate swarming and siderophore-mediated iron uptake via proteolytic cleavage of transferrin. Biol. Pharm. Bull. 29: 850–853.
Patriquin, G.M., Banin, E., Gilmour, C., Tuchman, R., Greenberg, E.P. and Poole, K. (2008) Influence of quorum sensing and iron on twitching motility and biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 190: 662–671.
Pesci, E.C., Milbank, J.B., Pearson, J.P., McKnight, S., Kende, A.S., Greenberg, E.P. and Iglewski, B.H. (1999) Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96: 11229–11234.
Pichon, C. and Felden, B. (2007) Proteins that interact with bacterial small RNA regulators. FEMS Microbiol. Rev. 31: 614–625.
Pohlmann, V. and Marahiel, M.A. (2008) Delta-amino group hydroxylation of L-ornithine during coelichelin biosynthesis. Org. Biomol. Chem. 6: 1843–1848.
Poole, K. and McKay, G.A. (2003) Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front. Biosci. 8: 661–686.
Potekhina, J.S., Shericheva, N.G., Povetkina, L.B., Pospelov, A.P., Rakitina, T.A., Warnecke, F. and Gottschalk, G. (1999) Role of microorganisms in corrosion inhibition of metals in aquatic habitats. Appl. Microbiol. Biotechnol. 52: 639–646.
Raymond, K.N., Dertz, E.A. and Kim, S.S. (2003) Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 100: 3584–3588.
Rhodes, E.R., Shoemaker, C.J., Menke, S.M., Edelmann, R.E. and Actis, L.A. (2007) Evaluation of different iron sources and their influence in biofilm formation by the dental pathogen Actinobacillus actinomycetemcomitans. J. Med. Microbiol. 56: 119–128.
Rohwerder, T., Gehrke, T., Kinzler, K. and Sand, W. (2003) Bioleaching review Part A: progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation. Appl. Microbiol. Biotechnol. 63: 239–248.
Rolerson, E., Swick, A., Newlon, L., Palmer, C., Pan, Y., Keeshan, B. and Spatafora, G. (2006) The SloR/Dlg metalloregulator modulates Streptococcus mutans virulence gene expression. J. Bacteriol. 188: 5033–5044.
Roos, V., Nielsen, E.M. and Klemm, P. (2006) Asymptomatic bacteriuria Escherichia coli strains: adhesins, growth and competition. FEMS Microbiol. Lett. 262: 22–30.
Rudolph, G., Hennecke, H. and Fischer, H.M. (2006) Beyond the Fur paradigm: iron-controlled gene expression in rhizobia. FEMS Microbiol. Rev. 30: 631–648.
Schröder, I., Johnson, E. and de Vries, S. (2003) Microbial ferric iron reductases. FEMS Microbiol. Rev. 27: 427–447.
Sebulsky, M.T. and Heinrichs, D.E. (2001) Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J. Bacteriol. 183: 4994–5000.
Shigematsu, T., Fukushima, J., Oyama, M., Tsuda, M., Kawamoto, S. and Okuda, K. (2001) Iron-mediated regulation of alkaline proteinase production in Pseudomonas aeruginosa. Microbiol. Immunol. 45: 579–590.
Singh, P.K. (2004) Iron sequestration by human lactoferrin stimulates P. aeruginosa surface motility and blocks biofilm formation. Biometals 17: 267–270.
Singh, P.K., Schaefer, A.L., Parsek, M.R., Moninger, T.O., Welsh, M.J. and Greenberg, E.P. (2000) Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407: 762–764.
Singh, P.K., Parsek, M.R., Greenberg, E.P. and Welsh, M.J. (2002) A component of innate immunity prevents bacterial biofilm development. Nature 417: 552–555.
Sprencel, C., Cao, Z., Qi, Z., Scott, D.C., Montague, M.A., Ivanoff, N., Xu, J., Raymond, K.M., Newton, S.M. and Klebba, P.E. (2000) Binding of ferric enterobactin by the Escherichia coli periplasmic protein FepB. J. Bacteriol. 182: 5359–5364.
Tam, R. and Saier, M.H., Jr. (1993) Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57: 320–346.
Tomaras, A.P., Dorsey, C.W., Edelmann, R.E. and Actis, L.A. (2003) Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149: 3473–3484.
Touati, D. (2000) Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373: 1–6.
Vasil, M.L. (2007) How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events. Biometals 20: 587–601.
Visca, P., Leoni, L., Wilson, M.J. and Lamont, I.L. (2002) Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 45: 1177–1190.
Vuong, C., Kidder, J.B., Jacobson, E.R., Otto, M., Proctor, R.A. and Somerville, G.A. (2005) Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J. Bacteriol. 187: 2967–2973.
Wai, S.N., Mizunoe, Y., Takade, A., Kawabata, S.I. and Yoshida, S.I. (1998) Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64: 3648–3655.
Wandersman, C. and Delepelaire, P. (2004) Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58: 611–647.
Whitchurch, C.B., Tolker-Nielsen, T., Ragas, P.C. and Mattick, J.S. (2002) Extracellular DNA required for bacterial biofilm formation. Science 295: 1487.
Whiteley, M., Bangera, M.G., Bumgarner, R.E., Parsek, M.R., Teitzel, G.M., Lory, S. and Greenberg, E.P. (2001) Gene expression in Pseudomonas aeruginosa biofilms. Nature 413: 860–864.
Wilderman, P.J., Vasil, A.I., Johnson, Z., Wilson, M.J., Cunliffe, H.E., Lamont, I.L. and Vasil, M.L. (2001) Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 69: 5385–5394.
Wilderman, P.J., Sowa, N.A., FitzGerald, D.J., FitzGerald, P.C., Gottesman, S., Ochsner, U.A. and Vasil, M.L. (2004) Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA 101: 9792–9797.
Williams, H.D., Zlosnik, J.E. and Ryall, B. (2007) Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 52: 1–71.
Wolff, N., Izadi-Pruneyre, N., Couprie, J., Habeck, M., Linge, J., Rieping, W., Wandersman, C., Nilges, M., Delepierre, M. and Lecroisey, A. (2008) Comparative analysis of structural and dynamic properties of the loaded and unloaded hemophore HasA: functional implications. J. Mol. Biol. 376: 517–525.
Yang, L., Barken, K.B., Skindersoe, M.E., Christensen, A.B., Givskov, M. and Tolker-Nielsen, T. (2007) Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 153: 1318–1328.
Zuo, R. (2007) Biofilms: strategies for metal corrosion inhibition employing microorganisms. Appl. Microbiol. Biotechnol. 76: 1245–1253.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer Science+Business Media B.V.
About this chapter
Cite this chapter
Avidan, O., Satanower, S., Banin, E. (2010). Iron and Bacterial Biofilm Development. In: Seckbach, J., Oren, A. (eds) Microbial Mats. Cellular Origin, Life in Extreme Habitats and Astrobiology, vol 14. Springer, Dordrecht. https://doi.org/10.1007/978-90-481-3799-2_19
Download citation
DOI: https://doi.org/10.1007/978-90-481-3799-2_19
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-90-481-3798-5
Online ISBN: 978-90-481-3799-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)