Abstract
Green algae are a monophyletic lineage of Archaeplastida, a supergroup of photosynthetic eukaryotes exclusively originated from primary endosymbiosis. The green algae are divided into two clades, the Chlorophyta and Streptophyta. The Chlorophyta comprises the vast majority of green algae, with three major lineages (Trebouxiophyceae, Ulvophyceae, and Chlorophyceae) forming the “crown group” of Chlorophyta and, as their early offspring, an assemblage of various monophyletic lineages of unicellular prasinophyte algae. Chloroplast phylogenomics and multigene phylogenetic analyses not only largely confirmed previous assumptions based on single gene (18S or rbcL) analyses, but also revealed novel robust groupings. While a bifurcation of the Chlorophyceae and a clear distinction of four major lineages within the Ulvophyceae have been supported, the internal phylogenetic structure and circumscription of Trebouxiophyceae still remain ambiguous. The Streptophyta comprises relatively few algal lineages and embryophytes. The embryophyte land plants are a sister group of streptophyte green algae (Zygnematophyceae or a lineage comprising Zygnematophyceae and Coleochaetophyceae), but transitions of the green algae to the land have taken place several times, even outside the Streptophyta. Progress in green algal systematics at the species level revealed ITS2 rDNA as an appropriate candidate for DNA barcoding and species distinction. Approaches to integrate sequence analyses of several genes with morphology are now state of the art for species delimitations. An improved taxon sampling with consideration of traditional gathered expert knowledge combined with multigene phylogenetic analyses, and improved phylogeny inference methods will be required to clarify areas of ambiguity in the green algal phylogenies. A revisit of morphology is essential to establish synapomorphies for the novel clades in molecular groupings.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Green algae constitute a morphologically and ecologically diverse monophyletic lineage of Archaeplastida (Adl et al. 2005) which also includes embryophytes. Archaeplastida (also referred to as Plantae; Keeling 2004, 2010) is the only supergroup of photosynthetic eukaryotes exclusively originated from primary endosymbiosis. The plastid origin through primary endosymbiosis has been reviewed by a large number of articles, e.g., Bhattacharya et al. (2004), Reyes-Prieto et al. (2007), Baldauf (2008), Gould et al. (2008), Archibald (2009b), and Keeling (2010).
The Viridiplantae, to which all green algae and the embryophytes belong, the Rhodophyta, and the Glaucophyta form the Archaeplastida. The phylum Rhodophyta is often regarded as a sister group to Viridiplantae. The monophyletic origin of Archaeplastida has been supported by nuclear and chloroplast multigene analyses (Rodríguez-Ezpeleta et al. 2005; Archibald 2009b). Green algae share most of their features with embryophytes, including chloroplast structures, pigmentation, cell wall composition, and a characteristic stellate structure in the transition zone between flagellum and basal body (for review, see Lewis and McCourt 2004; Leliaert et al. 2011). Apart from being mostly not of true multicellular organization (see Coleochaetophyceae and Charophyceae; Leliaert et al. 2011) and often confined to aqueous habitats, it is often difficult to delimit the green algae from embryophytes without consideration of their very diverse gross morphologies. Morphological variation of green algae spans from the smallest known eukaryote (the prasinophyte Ostreococcus) and tiny flagellates to giant unicells with multiple nuclei or multicellular forms reminiscent of bryophyte gametophytes (Coloechaete). The majority of green algae thrive in freshwater or terrestrial habitats, but some microscopic forms (prasinophytes) are abundant in marine phytoplankton. Macroscopic marine forms are only known from the Ulvophyceae. It is typical for green algae to live in various terrestrial habitats (Holzinger 2009). Among green algae, there are many “land plants,” i.e., transitions to the land happened many times in the evolution of Chlorophyta and Streptophyta (Lewis and Lewis 2005; Lewis 2007). Examples for terrestrial green algal life are the biofilms of building facades (Barberousse et al. 2006, 2007), biological soil crusts (Smith et al. 2004; Büdel et al. 2009), or algal crusts on tree bark (Lüttge and Büdel 2010). Excellent reviews on the peculiarities of terrestrial green algae have been presented by López-Bautista et al. (2007) and Rindi (2011). Green algae, especially a variety of members of Trebouxiophyceae, are frequently involved in symbioses with ciliates and metazoa (for review, see Pröschold et al. (2011) and references therein) as well as lichenized fungi, where they are the dominating photobionts (review: Friedl and Büdel 2008).
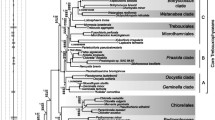
The Viridiplantae are divided into two major clades, the phyla Chlorophyta and Streptophyta (Lewis and McCourt 2004; Petersen et al. 2006; Pröschold and Leliaert 2007). The vast majority of green algae belongs to the Chlorophyta, whereas the Streptophta comprise relatively few algal lineages and embryophytes. The embryophytes are a sister group to certain assemblage of streptophyte green algae (recent reviews: Turmel et al. 2007; Becker and Marin 2009; Wodniok et al. 2011). With respect to the phylogeny of the Chlorophyta, recent work confirmed that an array of distinct lineages of prasinophyte green algae (formerly assigned to a single class “Prasinophyceae”) forms the most basal divergences and that four classes, the Chlorodendrophyceae (prasinophyte green algae), Trebouxiophyceae, Chlorophyceae, and Ulvophyceae form the core of Chlorophyta (Turmel et al. 2007; Marin and Melkonian 2010). The current view, which appears to become more and more substantiated particularly from chloroplast phylogenomics (Turmel et al. 2009a, b), supports the assumption that the Trebouxiophyceae emerged before Chlorophyceae and Ulvophyceae (Fig. 1). Therefore, this particular subdivision into three major chlorophytean classes may be called the “TUC crown group” of Chlorophyta (Pombert et al. 2005; Fig. 1). While the single origin of Chlorophyceae is not disputed, the monophyly of Ulvophyceae has only recently been robustly assessed (Cocquyt et al. 2010), but a sufficient evidence for the monophyly of Trebouxiophyceae is still lacking, and its circumscription not fully understood.
Simplified schematic consensus phylogeny of the Green Algae. Details of the various clades are discussed in the text. Several lineages of prasinophyte green algae have been omitted for clarity. For the Trebouxiophyceae only those clades that are robustly resolved in 18S rDNA phylogenies and which include several genera are shown
It should be noted that our current view of green algal systematics has been shaped by the intriguing results of molecular phylogenetic analyses which became feasible and widely employed in the 1990s following “traditional” algal systematics based on morphology and ultrastructure. Aspects of the taxonomic history of green algal systematics and evolution have recently been reviewed by Lewis and McCourt (2004) and Pröschold and Leliaert (2007), including a discussion of current knowledge. However, green algal systematics is progressing fast, particularly by analyses of novel data sets consisting of concatenated sequences from many loci of both nuclear and chloroplast genomes, as well as organelle-based phylogenomics. A comprehensive state-of-the-art review of green algal phylogenetics has been presented by Leliaert et al. (2011). Therefore, the intention of the present review is to give a brief introduction into current views of green algal systematics.
There is an important trend to focus on the systematics of green algal genera, i.e., their phylogenetic structures comprising the species. Species descriptions become more and more based on several markers and not only on a single gene, 18S, or rbcL. Most species-based works in green algae agree on a common molecular marker, i.e., ITS2 rDNA to unequivocally discriminate species (e.g., Müller et al. 2007; Coleman 2009). Certain nucleotide changes in ITS2 (CBCs), when mapped on the secondary structure model, which contains segments largely conserved over phylogenetic distances exceeding genera and families, help to guide decisions of what to include or exclude in a green algal species (Coleman 2003, 2007, 2009; Schultz et al. 2005). ITS2 rDNA is often discussed as the ideal candidate for a DNA barcode within green algae because there are many examples for its successful application in different major lineages of Chlorophyta (e.g., Pröschold et al. 2005, 2011; Luo et al. 2010) as well as the streptophyte green algae Klebsormidiophyceae (e.g. Rindi et al. 2011). ITS2 rDNA has even been suggested as a “double-edged tool” which is valuable from species to order levels or even higher (Coleman 2003). ITS2 sequence analyses have successfully been employed to assess the monophyly of Sphaeropleales within the Chlorophyceae, even more robustly than 18S rDNA. An automated data analysis approach based on a large (>1,500 sequences) ITS2 data set with a secondary structure-based alignment demonstrated the power to reconstruct evolutionary patterns for highly divergent lineages at the levels of orders and classes (Buchheim et al. 2011).
2 Prasinophyte Green Algae
Prasinophytes represent the earliest branches of the Chlorophyta and are also a morphologically heterogeneous assemblage of green algae (for reviews, see Nakayama et al. 1998, 2007; Turmel et al. 2009a; Marin and Melkonian 2010). Most prasinophycean algae are unicellular flagellates possessing a scaly covering and having an asymmetrical cell architecture. Some species lack flagella, others lack scales, and in some cases, both flagella and scales are absent, e.g., in Ostreococcus tauri, which is the smallest known eukaryote (Turmel et al. 2009a; Yamaguchi et al. 2011). Coccoid forms may have evolved in several lineages of prasinophytes (Guillou et al. 2004). Most prasinophytes occur in marine habitats where they can be important components of coastal phytoplankton communities (Yamaguchi et al. 2011). Earlier analyses based on 18S rDNA already revealed that there is no monophyletic class “Prasinophyceae” (e.g., Nakayama et al. 1998, 2007). Instead, a paraphyletic assemblage of seven monophyletic groups of prasinophytes have been identified at the base of the Chlorophyta (e.g., Guillou et al. 2004) of which only three have been formally described as independent classes until now, i.e., the Chlorodendrophyceae, Nephroselmidophyceae, and Mamiellophyceae (Marin and Melkonian 2010; Yamaguchi et al. 2011, and references therein). The position of the Chlorodendrophyceae appears to be basal to, or even within, the radiation of the TUC crown group (Guillou et al. 2004), whereas Nephroselmidophyceae and Mamiellophyceae occupy rather basal positions within the Chlorophyta (Marin and Melkonian 2010). Prasinophytes are regarded as central to understand evolutionary patterns that accompanied radiation within the Chlorophyta and reduction of cells in some lineages (Turmel et al. 2009a). The whole genomes of two species of Ostreococcus (Mamelliophyceae; Palenik et al. 2007; Derelle et al. 2008), the smallest eukaryote, and two strains of Micromonas (Mamelliophyceae; Worden et al. 2009) have been sequenced. Evidence for sexual reproduction has been found by genome analyses (Grimsley et al. 2010), despite not being observed in these algae (cyrptic sex), which may indicate that the ancestor of Chlorophyta already exhibited sexual reproduction (Archibald 2009a). Full genome analyses also showed that the observed reduction of gene content occured in conjunction with important changes in cell organization (Turmel et al. 2009a). To date, chloroplast genome data are available for Nephroselmis, Pyramimonas, Pycnococcus, Monomastix, and Ostreococcus (Turmel et al. 2009a and references therein), and revealed that the ancestor of euglenoid chloroplasts may have been a close relative of the Pyramimonadales lineage (Turmel et al. 2009a), which is probably one of the most basal chlorophytean lineages (Marin and Melkonian 2010). For the green algal origin of plastids of the Chlorarachniophyta, also a prasinophyte, Tetraselmis (Chlorodendrophyceae) seems to be a likely candidate (Takahashi et al. 2007). Consequently, prasinophytes may have been involved in two independent secondary endosymbioses (Rogers et al. 2007).
3 Trebouxiophyceae
The majority of presently known members of the class are coccoid unicells, in some lineages also colonial coccoids occur. A few lineages may also form filaments, which, however, easily disintegrate (e.g., Leptosira, Stichococcus). The only known trebouxiophyte genera with firm, often even multiseriate filaments are Prasiola and Rosenvingiella. Flagellated vegetative forms are not known for the class. Members of Trebouxiophyceae are mostly found in dryer habitats, e.g., in soil, or are aerophytic algae. Many lineages include minute freshwater phytoplankton (e.g., Chlorellales). There are numerous examples for symbioses in ciliates, metazoa (e.g., various species of Chlorella, Micractinium, Coccomyxa, and Elliptochloris), and lichens (e.g., Trebouxia and Asterochloris).
At present, there is consensus that 18S rDNA phylogenies resolve five well-supported clades which comprise multiple genera, i.e., Prasiola-clade, Trebouxiales, Watanabea-clade, Choricystis/Botryococcus-clade, and Chlorellales. There are several other lineages for which only a single species (e.g., Xylochloris; Neustupa et al. 2011) or genus (e.g., Leptosira, Lobosphaera) are known so far. The relationship between these lineages, however, still remains ambiguous.
The Prasiola-clade comprises numerous examples of morphological plasticity, which seems to be a characteristic feature of the whole class. Morphologically defined species (morphotypes, morphospecies) may be distributed over several phylogenetic lineages and morphologically rather distinct algae may appear closely related in phylogenetic analyses. For example, Stichococcus bacillaris, which forms short filaments that easily disintegrate into cylindrical cells, are distributed over multiple lineages within the clade (Neustupa et al. 2007 and references therein). In 18S rDNA as well as rbcL-based phylogenies, species and strains of the microscopic genus Stichococcus are the closest relatives of Prasiola and Rosenvingiella, which are macroscopic filamentous or blade-shaped marine algae, often with tissue-like cell layers (Rindi et al. 2004, 2007). Also for the uniseriate filamentous forms of Prasiola and Rosenvingiella, a higher genetic diversity has been found than their simple morphology may indicate (Rindi et al. 2004). Prasiola species are involved in a symbiotic relationship with the fungal genus Mastodia, which is regarded as a model to study interactions and processes in fungal–algal symbioses (Pérez-Ortega et al. 2010). Phylogenies based on rbcL gene sequences show that the algal partner is most closely related to Prasiola borealis. While most relationships within the Prasiola-clade remain ambiguous due to a relative poor resolution of the 18S rDNA sequences (Rindi et al. 2007), one lineage-including species of Koliella, Raphidonema, Pabia, and Pseudochlorella, is well supported as distinct from other members of the Prasiola-clade (Darienko et al. 2010; Neustupa et al. 2011).
The Trebouxiales (with three currently known genera, Trebouxia, Asterochloris, and Myrmecia) are common photobionts in lichen symbiosis (Friedl and Büdel 2008). Trebouxia is the most common lichen photobiont, and there is still a growing number of works which focus to reveal the algal species diversity within certain groups of lichens. Most often, algal-specific primers to directly amplify the Trebouxia ITS rDNA from lichen specimens without culturing are employed. Species diversity of Trebouxia appears larger than reflected by morphology (cryptic speciation) and has still not been fully explored. Intriguing examples have been presented by Blaha et al. (2006) and Skaloud and Peksa (2010). The 18S rDNA phylogenies showed that Trebouxia as previously defined was paraphyletic with Myrmecia and therefore, in a polyphasic approach using ITS and actin intron sequences as well as morphology, the genus Asterochloris has recently been segregated from Trebouxia (Skaloud and Peksa 2010).
Within the Choricystis/Botryococcus-clade, several genera thrive in symbioses as well as free living. Elliptochloris has been identified as symbiotic in lichens (Friedl and Büdel 2008) and marine invertebrates (Letsch et al. 2009), but species of Elliptochloris were also found free living on tree bark (Eliáš et al. 2008). Coccomyxa, the sister-group to Elliptochloris in the 18S rDNA phylogenies, includes symbionts of lichens and ciliates (previously summarized under “Zoochlorella”) (Pröschold et al. 2011) as well as free-living species (Zoller and Lutzoni 2003; Friedl and Büdel 2008). Species of Choricystis species are known as phytoplankton members (Fawley et al. 2005) and as symbionts in sponges (Handa et al. 2006; Pröschold et al. 2011). The oil-producing Botryococcus is a prominent member of this clade (Senousy et al. 2004; Weiss et al. 2010).
Most of the currently known members of the Watanabea-clade were traditionally assigned to Chlorella, but appeared distinct from the type of Chlorella, C. vulgaris, in molecular phylogenies as well as morphological features (Darienko et al. 2010). A new genus, Chloroidium, has been established to encompass the previous species C. ellipsoidea, C. trebouxioides, and C. saccharophila. The Watanabea-clade also includes Heterochlorella, a newly established genus to encompass previous “Chlorella” luteoviridis (Neustupa et al. 2009). Two more lineages of Chlorella-like algae which, however, are distinct from the two aforementioned lineages, are Kalinella (Neustupa et al. 2009) and Heveochlorella (Zhang et al. 2008). So far, only a single symbiotic member of the Watanabea-clade has been reported, i.e., as a still unidentified strain from the lichen Psoroglaena epiphylla (Nyati et al. 2007).
The Chlorellales have been the focus of intensive taxonomic research during recent years. This group unites the Chlorellaceae, which is subdivided into the Chlorella- and Parachlorella clades (Krienitz et al. 2004), with the “Auxenochlorella-clade” (Pröschold et al. 2011). While the latter comprises peculiar coccoid green algae without chlorophylls (Aslam et al. 2007), the order Chlorellales comprises mostly unicellular coccoids of minute size which thrive in an extremely broad range of habitats. For example, species of Chlorella (close relatives of the type species, C. vulgaris) have been described from terrestrial habitats (e.g., rock surfaces in Antarctica; Hu et al. 2008), symbioses with ciliates and sponges (C. vulgaris, C. variabilis, C. heliozoae; Hoshina et al. 2010; Pröschold et al. 2011), as well as freshwater phytoplankton (Henley et al. 2004; Krienitz et al. 2004). An overview of the “true” Chlorella species and genera most closely related with Chlorella, i.e., members of the Chlorella-clade of Chlorellales, has been presented by Luo et al. 2010. Several new genera which belong to the Chlorella-clade have been described from freshwater phytoplankton, e.g., Meyerella (Fawley et al. 2005), Hegewaldia (Pröschold et al. 2010), Heynigia, and Hindakia (Bock et al. 2010). Members of the same genus, Micractinium, are known as common components of freshwater phytoplankton (with cell wall appendices, bristles, inducible by grazers, Luo et al. 2006) as well as symbionts in the ciliate Paramecium bursaria (Pröschold et al. 2011). Many genera of the Chlorellales are common picoplanktonic green algae in freshwater, e.g., Picochlorum, Nannochloris, and Marvania (Henley et al. 2004). A new genus of marine phytoplankton, Marinichlorella, which is a member of the Parachlorella-clade, has recently been described on the basis of ultrastructural characters and 18S rDNA phylogenies (Aslam et al. 2007). A new genus of Chlorella-like phytoplankton, Chloroparva, a close relative of “Chlorella” minutissima within the “Nannochloris-like” clade of Chlorellales (Fig. 1), has also recently been described on the basis of 18S rDNA phylogenetic analyses, ultrastructure, and oleic fatty acids (Somogyi et al. 2011). Extensive ultrastructural studies in Trebouxiophyceae have become rare. However, an excellent ultrastructural study that focuses on the life cycle and reproduction by autospores in Chlorella and Parachlorella revealed ultrastructural features which support the separation of Parachlorella from Chlorella, independent of molecular phylogenetic analyses (Yamamoto et al. 2005). For the Auxenochlorella-clade, three genera of heterotrophic algae are currently known. Auxenochlorella has been described as a free-living aerophytic alga (Kalina and Punčochárová 1987), reported from symbioses with Hydra (Pröschold et al. 2011) as well as from a lichen symbiosis (Psoroglaena stigonemoides, Nyati et al. 2007). Prototheca species are well known as pathogens in vertebrates, e.g., cattle and humans, and the genus Helicosporidium in insects (Tartar et al. 2002; Tartar and Boucias 2004; de Koning and Keeling 2006). The close phylogenetic relationships of both parasitic genera have been resolved by 18S rDNA (Tartar et al. 2002) and phylogenomic analyses of mitochondrial (Pombert and Keeling 2010) and chloroplast DNA (de Cambiaire et al. 2007). As expected for parasitic algae not performing photosynthesis, the chloroplast genomes were rather reduced compared to other green algae (de Cambiaire et al. 2007). In 18S rDNA phylogenies, the three genera form a rather long branch, which almost certainly is due to systematic errors in phylogenetic inferences (long branch attraction, Verbruggen and Theriot 2008). Therefore, deciphering whether or not they form a single monophyletic lineage, and their position within the Chlorellales is still a challenging task.
The circumscription of Trebouxiophyceae is not fully understood yet. The Oocystaceae, a monophyletic group of coccoid green algae, may form another multigenera group within the Trebouxiophyceae. First, Hepperle et al. (2000) showed affinities of the Oocystaceae to the Trebouxiophyceae, despite it exhibiting rather long branches, but its relationship within the class could not be resolved. Later, an affiliation of the Oocystaceae with Chlorellales had been found (Krienitz et al. 2003; Pažoutová et al. 2010; Neustupa et al. 2011). Oocystaceae was even used as outgroup taxa in 18S rDNA phylogenies to address relationships within the Trebouxiophyceae (Pröschold et al. 2011). Species of Geminella were also resolved, forming a clade of the Trebouxiophyceae (Mikhailyuk et al. 2008), but appeared rather distant from other members of the class. The phylogenetic positions of Oocystaceae and the Geminella-clade are in urgent need of reevaluation; both clades may even represent two additional independent clades diverging within the TUC crown group.
Sexual reproduction in Trebouxiophyceae has so far been observed only in two genera, Micractinium (references in Pröschold et al. 2010) and Prasiola (references in Rindi et al. 2004). Analysis of the whole genome of Chlorella variabilis, symbiont of the ciliate P. bursaria, however, revealed evidence for the presence of sexual reproduction in which flagellated stages may have been involved or at least genes involved in these processes may have been retained (Blanc et al. 2010). Therefore, it is tempting to conclude that sexual reproduction is more widespread in Trebouxiophyceae than assumed from observations (cryptic sexuality). Absence of flagellated stages is very widespread in lineages of the Trebouxiophyceae (e.g. Watanabea- and Choricystis/Botryococcus-clades), but genomic evidence for flagellar motion in C. variabilis, a species for which only reproduction by nonmotile autospores is known, has been found (Blanc et al. 2010). This may indicate that the many autosporic lineages in Trebouxiophyceae may have ancestors with flagellated reproductive stages as well. Chloroplast phylogenomics could not resolve the monophyletic origin of the Trebouxiophyceae, but revealed an affiliation of the green flagellate Pedinomonas (whose phylogenetic position has been unclear so far) with the Trebouxiophyceae, particularly with the Chlorellales (Turmel et al. 2009b). Phylogenomic analyses based on mitochondrial DNA, however, did not support a relationship of Pedinomonas with Trebouxiophyceae, but both phylogenomic analyses certainly suffer from poor taxon sampling, which is a problem inherent to all phylogenomic analyses so far (Rodríguez-Ezpeleta et al. 2007).
4 Chlorophyceae
In contrast to the Trebouxiophyceae, the Chlorophyceae includes a variety of unicellular or colonial flagellates (e.g., Chlamydomonas, Volvox; Nakada et al. 2008) and many members with firm unbranched (e.g., Oedogonium; Alberghina et al. 2006) or branched filaments (e.g., Stigeoclonium, Michetti et al. 2010). Members of Chlorophyceae thrive mostly in freshwater or terrestrial habitats, and there are only very few symbiotic genera, e.g., Scenedesmus in ciliates (Pröschold et al. 2011). Chlorophyceae are not known yet from lichen symbioses (Friedl and Büdel 2008). Recent reviews of the Chlorophyceae are based on 18S rDNA (e.g., Müller et al. 2004) or chloroplast phylogenomics (e.g., Turmel et al. 2008). Consensus has been reached that the Chlorophyceae may be divided into five clades, Volvocales, Sphaeropleales, Oedogoniales, Chaetopeltidales, and Chaetophorales, which are robustly resolved in most phylogenetic analyses (Fig. 1). Monophyletic origin and distinction of Sphaeropleales has been substantiated by ITS2 secondary structure-based analyses (Keller et al. 2008). The actual conception of the group and its subdivision into clades has recently been reviewed by Krienitz et al. (2011). A recent overview of Volvocales [sometimes also called Chlamydomonadales, e.g., Müller et al. (2004)] can be found in Gerloff-Elias et al. (2005). Using a comprehensive large-scale data set of more than 400 18S rDNA sequences, the overall phylogenetic structure of the group has been revealed and phylogenetically-defined names have been applied to the monophyletic clades that constitute the Volvocales using PhyloCode (Nakada et al. 2008). Recent studies with expanded taxon samplings that would allow testing generic concepts or species delineations are still lacking for the Chaetopeltidales and Oedogoniales. A preliminary 18S rDNA analysis already indicated a polyphyletic origin for Oedogonium within the Oedogoniales (Alberghina et al. 2006). A most recent 18S rDNA-based overview of the Chaetophorales with its subdivisions can be found in Caisová et al. (2011).
Chloroplast phylogenomic analyses support a dichotomy of Chlorophyceae, i.e., a CS clade uniting the orders Chlamydomonadales (= Volvocales) and Sphaeropleales versus the OCC clade which comprises the orders Oedogoniales, Chaetopeltidales, and Chaetophorales (Turmel et al. 2008; Fig. 1). Until now, a total of eight chloroplast genomes has been sequenced for the six orders of both clades together (Brouard et al. 2011 and references therein). Recent analyses of 18S rDNAs alone have favored the dichotomy as seen in cpDNA phylogenetic analyses (Caisová et al. 2011), whereas analyses of concatenated 18S and LSU (28S) rDNA sequences (Buchheim et al. 2001) have supported only the CS clade (Shoup and Lewis 2003; Müller et al. 2004). As outlined by Lewis and McCourt (2004) and Leliaert et al. (2011), each of the two clades is defined by a unique absolute basal body configuration. Members of the CS clade produce biflagellated motile cells, while members of the OCC clade exhibit quadriflagellated stages or, in Oedogoniales, have multiple flagella (stephanokont zoids) (Alberghina et al. 2006). From chloroplast genomic data, it has been assumed that the ancestral state in Chlorophyceae may have been quadriflagellate motile cells with the DO + DO orientation, and the CW condition of flagellar basal bodies developed later convergently in the CS and OCC clades (Brouard et al. 2008; Turmel et al. 2008). Important conclusions for the evolution of green algae and the Chlorophyceae became possible by comparisons of genomes, i.e., that of the model organism Chlamydomonas reinhardtii with those of the multicellular Volvox (Prochnik et al. 2010) and the more ancestral prasinophytes Ostreococcus spp. (Peers and Niyogi 2008).
Many studies over the last 5 years focused more on taxa of the CS clade, less on members of the OCC clade. For both clades, phylogenetic analyses concluded that traditional species delineation based on morphology is not congruent with the molecular distinction. This general finding holds true from minute phytoplankton species (Krienitz et al. 2011), prominent examples of colonial coccoids [e.g., Pediastrum and Hydrodictyon; McManus and Lewis (2005, 2011)], to more complex branched filamentous forms [e.g., Chaetophora and Stigeoclonium; Caisová et al. (2011)]. In minute common phytoplankton species of the Sphaeropleales, the morphological difference in solitary versus colonial forms was found in disagreement with phylogenetic groupings and therefore the species were assigned to a single genus Mychonastes with the species distinguished primarily by differences in their ITS2 rDNAs (Krienitz et al. 2011). Sequence analyses of ITS2 rDNA became almost a standard marker to delineate species within Scenedesmus, Acutodesmus, and Desmodesmus and differences in ITS2 were found congruent with morphological features as seen by scanning-electron-microscopy (Hegewald et al. 2010). ITS2 sequence comparisons have also been successful in the unambiguous identification of Scenedesmus and Desmodesmus isolates from lake phytoplankton (Johnson et al. 2007). Chloroplast genes have frequently been used as molecular markers to test species and genus assignments in the Volvocales, but not in the Sphaeropleales. For example, a data set of three concatenated chloroplast genes revealed a new genus, Parallela, within the Volvocales of the CS clade in combination with light and electron microscopy (Novis et al. 2010). New species of the coccoid Asterococcus and the colonial flagellate Gonium have been recovered using plastid-encoded rbcL and nuclear-encoded ITS rDNA, combined with ultrastructure (Nakazawa et al. 2004; Hayama et al. 2010).
5 Ulvophyceae
The Ulvophyceae is probably morphologically the most diverse group of the Chlorophyta (Cocquyt et al. 2010; Leliaert et al. 2011). Morphologies range from microscopic unicells to multicellular thalli and giant cells with unique types of cytomorphology. The class may be regarded as predominantly marine; it includes all macroscopic marine representatives of the Chlorophyta. Only in recent years, monophyly of the class as well as its internal phylogenetic structure have been assessed using multigene analyses, i.e., concatenated 18S rDNA and a number of other nuclear and plastid genes. The Oltmannsiellopsidales, consisting of quadriflagellate unicells or coccoids arranged in disks or packets, may be the most basal lineage of Ulvophyceae (Friedl and O’Kelly 2002; Watanabe and Nakayama 2007; Fig. 1). Its basal position has been confirmed by mitochondrial genome sequence comparisons (Pombert et al. 2006). The Ignatius clade, comprising unicellular coccoids, is another more basal lineage of Ulvophyceae, which is independent of any other groups of the class (Watanabe and Nakayama 2007; Fig. 1). The Ulvales–Ulotrichales is a morphologically rather diverse clade, comprising most microscopic forms of Ulvophyceae. It includes two cytomorphological types: unicellular algae with a single nucleus and chloroplast, e.g., the unicellular or sarcinoid Pseudendoclonium, Planophila, and Desmochloris (Friedl and O’Kelly 2002; Darienko et al. 2009), and those with multicellular bodies composed of uninucleate cells, forming branched or unbranched filaments (e.g., Acrosiphonia, Ulothrix, Uronema; O’Kelly et al. 2004), blades or tubular forms (e.g., Ulva which is known to form “green tides,” e.g., the Qindao nuisance alga, Leliaert et al. 2009; O’Kelly et al. 2010). So far, there has been no support from phylogenetic analyses to separate the Ulvales–Ulotrichales clade into orders (Leliaert et al. 2011). The single origin for a rather large assemblage of lineages within the Ulvophyceae, termed the “Trentepohliales–Bryopsidales–Cladophorales–Dasycladales (TBCD)” clade (Fig. 1), has been revealed by multigene analyses (Cocquyt et al. 2010). The TBCD clade unites lineages of various cytomorphological organization (see Cocquyt et al. (2010) and references therein). The Trentepohliales is a unique lineage of exclusively terrestrial members forming filaments of uninucleate cells. The Trentepohliales includes a variety of important photobionts in lichen symbiosis (for review see Nelsen et al. 2011). For a comprehensive treatment of genus boundaries and a phylogenetic assessment of the Trentepohliales based on 18S rDNA and rbcL genes, see López-Bautista et al. (2006) and Rindi et al. (2009). The Bryopsidales (also referred to as Caulerpales) comprise mainly marine macroscopic lineages with siphonous forms consisting of a single giant cell with millions of nuclei or a single macronucleus. The order Cladophorales consists of mostly branched filaments of the siphonocladous type (Cocquyt et al. (2010) and references therein) and the marine tropical Dasycladales of siphonous forms. A large number of works have contributed to this present-day view of the phylogenetic structure of Ulvophyceae; they have been reviewed and summarized by Cocquyt et al. (2010) and Leliaert et al. (2011).
6 Streptophyte Green Algae
The streptophyte green algae comprise not more than four or five morphologically rather simple lineages and two advanced lineages with true multicellular organization (McCourt et al. 2004; Becker and Marin 2009): the scaly flagellate Mesostigmatophyceae (Marin and Melkonian 1999), the sarcinoid Chlorokybophyceae (McCourt et al. 2004), the unbranched filamentous Klebsormidiophyceae (for overview see Mikhailyuk et al. 2008; Sluiman et al. 2008), and the Zygnematophyceae which are characterized by conjugation as the method of sexual reproduction and the absence of flagellated cells (Fig. 1). The unicellular genus Spirotaenia, traditionally classified as a member of the Zygnematophyceae, likely represents another independent lineage of streptophyte green algae (Gontcharov and Melkonian 2004), confirming the earlier notion of Mollenhauer (1986). Multicellular organization and oogamous sexual reproduction exhibit the Coleochaetophyceae which form parenchymatous tissue (Coleochaete and Chaetosphaeridium; for review see Delwiche et al. 2002), and the Charophyceae (stoneworts) which form branched filaments with apical growth (McCourt et al. 2004; Becker and Marin 2009). A lineage uniting the flagellate Mesostigma and the sarcinoid Chlorokybus may represent the earliest diverging lineage of the Streptophyta as it was indicated by plastid phylogenomic analyses (Lemieux et al. 2007). Earlier studies suggested Mesostigma emerged even before the divergence of the Streptophyta and Chlorophyta (Lewis and McCourt 2004). Its basal position within the Streptophyta, however, is not debated anymore since a multigene family (BIP) and a GapA/GapB gene duplication shared with Mesostigma and other Streptophyta to the exclusion of Chlorophyta have been found (Nedelcu et al. 2006; Petersen et al. 2006). The Klebsormidiophyceae diverged after the Mesostigma/Chlorokybus lineage and is sister to a clade uniting the Zygnematophyceae, Charophyceae, and Coleochaetophyceae (Becker and Marin 2009; Wodniok et al. 2011). Which streptophyte algal group may be the closest living relatives of embryophytes has been the subject of a hot debate (see Chapman and Waters 2002 for comments), which began soon after an early multigene analysis suggested the Charophyceae as sister group to embryophytes (Karol et al. 2001). Most recent analyses of nuclear genes as well as chloroplast phylogenomic analyses, however, revealed either the Zygnematophyceae or a group uniting the Zygnematophyceae with Coleochaetophyceae is the sister group to embryophytes (Turmel et al. 2007; Wodniok et al. 2011). The Charophyceae is now seen as an earlier divergence and not as the closest relatives of land plants.
The Zygnematophyceae, in particular, the family Desmidiaceae, are the most species rich and taxonomically complex streptophyte green algae (Gontcharov and Melkonian 2011). However, due to a problematic species concept, the real taxonomic diversity of the Desmidiaceae family is not known with certainty (Kouwets 2008; Gontcharov and Melkonian 2011). The polyphyletic nature of almost all genera in Desmidiaceae has been recovered using nuclear and chloroplast-encoded molecular markers (e.g., Gontcharov and Melkonian 2008, 2011; Hall et al. 2008, and references therein), and there is still more work required to identify structural synapomorphies for the newly identified clades (Hall et al. 2008).
Recently, an increased interest in the common terrestrial Klebsormidiophyceae led to the discovery of a new member of the class, Interfilum (Mikhailyuk et al. 2008). Phylogenetic structures as well as species delineations within Klebsormidium have been analyzed using 18S, ITS, and rbcL sequences (Rindi et al. 2011), and new species have been described (Novis 2006). Concurrently, with the progress in Klebsormidium systematics, there have been further ecophysiological studies on the genus (Karsten et al. 2010; Karsten and Rindi 2010; Holzinger et al. 2011).
7 Conclusions
Phylogenies based on a single marker gene (mostly 18S rDNA) have already revealed most phylogenetic structures within Chlorophyta and Streptophyta; not much has changed since an already decent taxon sampling was achieved for most green algal groups in the early 2000s. The 18S rDNA is still the “universal marker” to uncover green algal diversity, e.g., in phytoplankton (Piganeau et al. 2011). Multigene sequencing and/or chloroplast phylogenomics not only confirmed previously resolved major lineages in the Chlorophyta, but revealed several novel robust groupings, e.g., in the Ulvophyceae or the assessment of the closest living relative of the embryophytes. There are still many “weak” areas in the phylogeny of Chlorophyta which continue to be ambiguous or unresolved. In particular, the circumscription and internal phylogenetic structure of the Trebouxiophyceae are still challenging. More analyses based on sequences from many single gene loci of at least the nuclear and chloroplast genomes are required, but systematic errors need to be overcome by increased taxon sampling and improved phylogeny inferences methods (Rodríguez-Ezpeleta et al. 2007; Verbruggen and Theriot 2008; Cocquyt et al. 2010). Transcriptome analyses using next generation sequencing (e.g., Timme and Delwiche 2010) is another promising and powerful tool for testing phylogenetic hypotheses in the green algae. Systematics, in general, attempts to recover the actual phylogenetic relationships as close as possible by using as many characters as possible, and, therefore, more than just phylogenies are needed. In many groups we now are left with a molecular grouping that cannot be explained satisfactorily by morphology or other phenotypical characters. The situation may be best exemplified in the Desmidiaceae (Zygnematophyceae) where the traditional generic concepts almost constantly fail. Morphology needs to be revisited and much work is needed to find structural or other phenotypical synapomorphies to define the molecular groupings. There are several good examples (e.g., in the Chlorellales, Trebouxiophyceae) where a reevaluation of morphology resulted in congruence with the molecular groupings and features whose phylogenetic significance has not been understood so far. Taxon sampling will become even more important for future studies in green algal systematics, not only in terms of numbers of taxa to be included, but also which taxa to select. Previous sequence analyses may guide us well to find the right critical taxa, but to consider traditional gathered expert knowledge from morphology, ultrastructure and ecology of a certain group of green algae will be pivotal.
References
Adl SM, Simpson AGB, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor MFJR (2005) The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol 52:399–451
Alberghina J, Vigna M, Confalonieri V (2006) Phylogenetic position of the Oedogoniales within the green algae (Chlorophyta) and the evolution of the absolute orientation of the flagellar apparatus. Plant Syst Evol 261:151–163
Archibald JM (2009a) Green evolution, green revolution. Science 324:191–192
Archibald JM (2009b) The puzzle of plastid evolution. Curr Biol 19:R81–R88
Aslam Z, Shin W, Kim MK, Im W-T, Lee S-T (2007) Marinichlorella kaistiae gen. et sp. nov. (Trebouxiophyceae, Chlorophyta) based on polyphasic taxonomy. J Phycol 43:576–584
Baldauf SL (2008) An overview of the phylogeny and diversity of eukaryotes. J Syst Evol 46:263–273
Barberousse H, Tell G, Yéprémian C, Couté A (2006) Diversity of algae and cyanobacteria growing on building facades in France. Algol Stud 120:81–105
Barberousse H, Brayner R, Rego AMBD, Castaing J-C, Beurdeley-Saudou P, Colombet J-F (2007) Adhesion of façade coating colonisers, as mediated by physico-chemical properties. Biofouling 23:15–24
Becker B, Marin B (2009) Streptophyte algae and the origin of embryophytes. Ann Bot 103:999–1004
Bhattacharya D, Yoon HS, Hackett JD (2004) Photosynthetic eukaryotes unite: endosymbiosis connects the dots. Bioessays 26:50–60
Blaha J, Baloch E, Grube M (2006) High photobiont diversity associated with the euryoecious lichen-forming ascomycete Lecanora rupicola (Lecanoraceae, Ascomycota). Biol J Linn Soc 88:283–293
Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, Salamov A, Terry A, Yamada T, Dunigan DD, Grigoriev IV, Claverie J-M, Van Etten JL (2010) The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell Online 22:2943–2955
Bock C, Pröschold T, Krienitz L (2010) Two new Dictyosphaerium-morphotype lineages of the Chlorellaceae (Trebouxiophyceae): Heynigia gen. nov. and Hindakia gen. nov. Eur J Phycol 45:267–277
Brouard J-S, Otis C, Lemieux C, Turmel M (2008) Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer. BMC Genomics 9:290
Brouard J-S, Otis C, Lemieux C, Turmel M (2011) The chloroplast genome of the green alga Schizomeris leibleinii (Chlorophyceae) provides evidence for bidirectional DNA replication from a single origin in the Chaetophorales. Genome Biol Evol 3:505–15
Buchheim M, Michalopulos E, Buchheim J (2001) Phylogeny of the Chlorophyceae with special reference to the Sphaeropleales: a study of 18 S and 26 S rDNA data. J Phycol 37:819–835
Buchheim MA, Keller A, Koetschan C, Förster F, Merget B, Wolf M (2011) Internal transcribed spacer 2 (nu ITS2 rRNA) sequence-structure phylogenetics: towards an automated reconstruction of the green algal tree of life. Plos One 6:e16931
Büdel B, Darienko T, Deutschewitz K, Dojani S, Friedl T, Mohr K, Salisch M, Reisser W, Weber B (2009) Southern African biological soil crusts are ubiquitous and highly diverse in drylands, being restricted by rainfall frequency. Microb Ecol 57:229–247
Caisová L, Marin B, Sausen N, Pröschold T, Melkonian M (2011) Polyphyly of Chaetophora and Stigeoclonium within the Chaetophorales (Chlorophyceae), revealed by sequence comparisons of nuclear-encoded SSU rRNA genes. J Phycol 47:164–177
Chapman RL, Waters DA (2002) Green algae and land plants – an answer at last? J Phycol 38:237–240
Cocquyt E, Verbruggen H, Leliaert F, De Clerck O (2010) Evolution and cytological diversification of the green seaweeds (Ulvophyceae). Mol Biol Evol 27:2052–2061
Coleman AW (2003) ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet 19:370–375
Coleman AW (2007) Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res 35:3322–3329
Coleman AW (2009) Is there a molecular key to the level of “biological species” in eukaryotes? A DNA guide. Mol Phylogenet Evol 50:197–203
Darienko T, Friedl T, Pröschold T (2009) Desmochloris mollenhaueri – a new terrestrial ulvophycean alga from south-west African soils. (Molecular phylogeny and systematics of terrestrial Ulvophyceae I.). Algol Stud 129:25–40
Darienko T, Gustavs L, Mudimu O, Menendez CR, Schumann R, Karsten U, Friedl T, Pröschold T (2010) Chloroidium, a common terrestrial coccoid green alga previously assigned to Chlorella (Trebouxiophyceae, Chlorophyta). Eur J Phycol 45:79–95
de Cambiaire J, Otis C, Lemieux C, Turmel M (2007) The chloroplast genome sequence of the green alga Leptosira terrestris: multiple losses of the inverted repeat and extensive genome rearrangements within the Trebouxiophyceae. BMC Genomics 8:213
de Koning A, Keeling P (2006) The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol 4:12
Delwiche CF, Karol KG, Cimino MT, Sytsma KJ (2002) Phylogeny of the genus Coleochaete (Coleochaetales, Charophyta) and related taxa inferred by analysis of the chloroplast gene rbcL. J Phycol 38:394–403
Derelle E, Ferraz C, Escande M-L, Eychenié S, Cooke R, Piganeau G, Desdevises Y, Bellec L, Moreau H, Grimsley N (2008) Life-cycle and genome of OtV5, a large DNA virus of the pelagic marine unicellular green alga Ostreococcus tauri. Plos One 3:e2250
Eliáš M, Neustupa J, Škaloud P (2008) Elliptochloris bilobata var. corticola var. nov. (Trebouxiophyceae, Chlorophyta), a novel subaerial coccal green alga. Biologia 63:791–798
Fawley MW, Fawley KP, Owen HA (2005) Diversity and ecology of small coccoid green algae from Lake Itasca, Minnesota, USA, including Meyerella planktonica, gen. et sp. nov. Phycologia 44:35–48
Friedl T, Büdel B (2008) Photobionts. In: Nash TI (ed) Lichen biology, 2nd edn. Cambridge University Press, Cambridge, pp 9–26
Friedl T, O’Kelly C (2002) Phylogenetic relationships of green algae assigned to the genus Planophila (Chlorophyta): evidence from 18 S rDNA sequence data and ultrastructure. Eur J Phycol 37:373–384
Gerloff-Elias A, Spijkerman E, Pröschold T (2005) Effect of external pH on the growth, photosynthesis and photosynthetic electron transport of Chlamydomonas acidophila Negoro, isolated from an extremely acidic lake (pH 2.6). Plant. Cell Environ 28:1218–1229
Gontcharov AA, Melkonian M (2004) Unusual position of the genus Spirotaenia (Zygnematophyceae) among streptophytes revealed by SSU rDNA and rbcL sequence comparisons. Phycologia 43:105–113
Gontcharov AA, Melkonian M (2008) In search of monophyletic taxa in the family Desmidiaceae (Zygnematophyceae, Viridiplantae): the genus Cosmarium. Am J Bot 95:1079–1095
Gontcharov AA, Melkonian M (2011) A study of conflict between molecular phylogeny and taxonomy in the Desmidiaceae (Streptophyta, Viridiplantae): analyses of 291 rbcL sequences. Protist 162:253–267
Gould SB, Waller RF, McFadden GI (2008) Plastid evolution. Annu Rev Plant Biol 59:491–517
Grimsley N, Péquin B, Bachy C, Moreau H, Piganeau G (2010) Cryptic sex in the smallest eukaryotic marine green alga. Mol Biol Evol 27:47–54
Guillou L, Eikrem W, Chrétiennot-Dinet M-J, Le Gall F, Massana R, Romari K, Pedrós-Alió C, Vaulot D (2004) Diversity of picoplanktonic prasinophytes assessed by direct nuclear SSU rDNA sequencing of environmental samples and novel isolates retrieved from oceanic and coastal marine ecosystems. Protist 155:193–214
Hall JD, Karol KG, McCourt RM, Delwiche CF (2008) Phylogeny of the conjugating green algae based on chloroplast and mitochondrial nucleotide sequence data. J Phycol 44:467–477
Handa S, Nakahara M, Tsubota H, Deguchi H, Masuda Y, Nakano T (2006) Choricystis minor (Trebouxiophyceae, Chlorophyta) as a symbiont of several species of freshwater sponge. Hikobia 14:365–373
Hayama M, Nakada T, Hamaji T, Nozaki H (2010) Morphology, molecular phylogeny and taxonomy of Gonium maiaprilis sp. nov. (Goniaceae, Chlorophyta) from Japan. Phycologia 49:221–234
Hegewald E, Wolf M, Keller A, Friedl T, Krienitz L (2010) ITS2 sequence-structure phylogeny in the Scenedesmaceae with special reference to Coelastrum (Chlorophyta, Chlorophyceae), including the new genera Comasiella and Pectinodesmus. Phycologia 49:325–335
Henley WJ, Hironaka JL, Guillou L, Buchheim MA, Buchheim JA, Fawley MW, Fawley KP (2004) Phylogenetic analysis of the ‘Nannochloris-like’ algae and diagnoses of Picochlorum oklahomensis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta). Phycologia 43:641–652
Hepperle D, Hegewald E, Krienitz L (2000) Phylogenetic position of the Oocystaceae (Chlorophyta). J Phycol 36:590–595
Holzinger A (2009) Desiccation tolerant green algae: implications of physiological adaptation and structural requirements. In: Hagen KN (ed) Algae: nutrition, pollution control and engergy sources. Nova Science, New York, pp 41–56
Holzinger A, Lütz C, Karsten U (2011) Desiccation stress causes structural and ultrastructural alterations in the aeroterrestrial green alga Klebsormidium crenulatum (Klebsormidiophyceae, Streptophyta) isolated from an alpine soil crust. J Phycol 47(3):591–602
Hoshina R, Iwataki M, Imamura N (2010) Chlorella variabilis and Micractinium reisseri sp. nov. (Chlorellaceae, Trebouxiophyceae): Redescription of the endosymbiotic green algae of Paramecium bursaria (Peniculia, Oligohymenophorea) in the 120th year. Phycol Res 58:188–201
Hu H, Li H, Xu X (2008) Alternative cold response modes in Chlorella (Chlorophyta, Trebouxiophyceae) from Antarctica. Phycologia 47(1):28–34
Johnson JL, Fawley MW, Fawley KP (2007) The diversity of Scenedesmus and Desmodesmus (Chlorophyceae) in Itasca State Park, Minnesota, USA. Phycologia 46:214–229
Kalina T, Punčochárová M (1987) Taxonomy of the subfamily Scotiellocystoideae Fott 1976 (Chlorellaceae, Chlorophyceae). Arch Hydrobiol Suppl 73(4) (Algol Stud 45):473–451
Karol KG, McCourt RM, Cimino MT, Delwiche CF (2001) The closest living relatives of land plants. Science 294:2351–2353
Karsten U, Rindi F (2010) Ecophysiological performance of an urban strain of the aeroterrestrial green alga Klebsormidium sp. (Klebsormidiales, Klebsormidiophyceae). Eur J Phycol 45:426–435
Karsten U, Lütz C, Holzinger A (2010) Ecophysiological performance of the aeroterrestrial green alga Klebsormidium crenulatum (Charophyceae, Streptophyta) isolated from an alpine soil crust with an emphasis on desiccation stress. J Phycol 46:1187–1197
Keeling PJ (2004) Diversity and evolutionary history of plastids and their hosts. Am J Bot 91:1481–1493
Keeling PJ (2010) The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc B Biol Sci 365:729–748
Keller A, Schleicher T, Forster F, Ruderisch B, Dandekar T, Muller T, Wolf M (2008) ITS2 data corroborate a monophyletic chlorophycean DO-group (Sphaeropleales). BMC Evol Biol 8:218
Kouwets F (2008) The species concept in desmids: the problem of variability, infraspecific taxa and the monothetic species definition. Biologia 63:881–887
Krienitz L, Hegewald E, Hepperle D, Wolf M (2003) The systematics of coccoid green algae: 18 S rRNA gene sequence data versus morphology. Biologia 58:437–446
Krienitz L, Hegewald E, Hepperle D, Huss VAH, Rohr T, Wolf M (2004) Phylogenetic relationship of Chlorella and Parachlorella gen. nov. (Chlorophyta, Trebouxiophyceae). Phycologia 43:529–542
Krienitz L, Bock C, Dadheech PK, Pröschold T (2011) Taxonomic reassessment of the genus Mychonastes (Chlorophyceae, Chlorophyta) including the description of eight new species. Phycologia 50:89–106
Leliaert F, Zhang X, Ye N, E-j M, Engelen AH, Mineur F, Verbruggen H, De Clerck O (2009) Research note: identity of the Qingdao algal bloom. Phycol Res 57:147–151
Leliaert F, Smith DR, Moreau H, Herron M, Verbruggen H, Delwiche CF, De Clerck O (2011) Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci (in press)
Lemieux C, Otis C, Turmel M (2007) A clade uniting the green algae Mesostigma viride and Chlorokybus atmophyticus represents the deepest branch of the Streptophyta in chloroplast genome-based phylogenies. BMC Biol 5:2
Letsch MR, Muller-Parker G, Friedl T, Lewis LA (2009) Elliptochloris marina sp. nov. (Trebouxiophyceae, Chlorophyta), symbiotic green alga of the temperate pacific sea anemones Anthopleura xanthogrammica and A. elegantissima (Anthozoa, Cnidaria). J Phycol 45:1127–1135
Lewis LA (2007) Chlorophyta on land. In: Seckbach J (ed) Algae and cyanobacteria in extreme environments. Springer, Dordrecht, pp 569–582
Lewis LA, Lewis PA (2005) Unearthing the molecular phylodiversity of desert soil green algae (Chlorophyta). Syst Biol 54:936–947
Lewis LA, McCourt RM (2004) Green algae and the origin of land plants. Am J Bot 91:1535–1556
López-Bautista JM, Rindi F, Guiry MD (2006) Molecular systematics of the subaerial green algal order Trentepohliales: an assessment based on morphological and molecular data. Int J Syst Evol Microbiol 56:1709–1715
López-Bautista JM, Rindi F, Casamatta D (2007) The systematics of subaerial algae. In: Seckbach J (ed) Algae and cyanobacteria in extreme environments. Springer, Dordrecht, pp 601–617
Luo W, Pflugmacher S, Pröschold T, Walz N, Krienitz L (2006) Genotype versus phenotype variability in Chlorella and Micractinium (Chlorophyta, Trebouxiophyceae). Protist 157:315–333
Luo W, Pröschold T, Bock C, Krienitz L (2010) Generic concept in Chlorella-related coccoid green algae (Chlorophyta, Trebouxiophyceae). Plant Biol 12:545–553
Lüttge U, Büdel B (2010) Resurrection kinetics of photosynthesis in desiccation-tolerant terrestrial green algae (Chlorophyta) on tree bark. Plant Biol 12:437–444
Marin B, Melkonian M (1999) Mesostigmatophyceae, a new class of streptophyte green algae revealed by SSU rRNA sequence comparisons. Protist 150:399–417
Marin B, Melkonian M (2010) Molecular phylogeny and classification of the Mamiellophyceae class. nov. (Chlorophyta) based on sequence comparisons of the nuclear- and plastid-encoded rRNA operons. Protist 161:304–336
McCourt RM, Delwiche CF, Karol KG (2004) Charophyte algae and land plant origins. Trends Ecol Evol 19:661–666
McManus HA, Lewis LA (2005) Molecular phylogenetics, morphological variation and colony-form evolution in the family Hydrodictyaceae (Sphaeropleales, Chlorophyta). Phycologia 44:582–595
McManus HA, Lewis LA (2011) Molecular phylogenetic relationships in the freshwater family Hydrodictyaceae (Sphaeropleales, Chlorophyceae), with an emphasis on Pediastrum duplex. J Phycol 47:152–163
Michetti KM, Leonardi PI, Caceres EJ (2010) Morphology, cytology and taxonomic remarks of four species of Stigeoclonium (Chaetophorales, Chlorophyceae) from Argentina. Phycol Res 58:35–43
Mikhailyuk TI, Sluiman HJ, Massalski A, Mudimu O, Demchenko EM, Kondratyuk SY, Friedl T (2008) New streptophyte green algae from terrestrial habitats and an assessment of the genus Interfilum (Klebsormidiophyceae, Streptophyta). J Phycol 44:1586–1603
Mollenhauer D (1986) Contribution towards a revision of the genus Spirotaenia (Mesotaeniaceae). Nova Hedwigia Beiheft 56:61–90
Müller T, Rahmann S, Dandekar T, Wolf M (2004) Accurate and robust phylogeny estimation based on profile distances: a study of the Chlorophyceae (Chlorophyta). BMC Evol Biol 4:20–27
Müller T, Philippi N, Dandekar T, Schultz J, Wolf M (2007) Distinguishing species. RNA 13:1469–1472
Nakada T, Misawa K, Nozaki H (2008) Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18 S rRNA phylogenetic analyses. Mol Phylogenet Evol 48:281–291
Nakayama T, Marin B, Kranz HD, Surek B, Huss VAR, Inouye I, Melkonian M (1998) The basal position of scaly green flagellates among the green algae (Chlorophyta) is revealed by analyses of nuclear-encoded SSU rRNA sequences. Protist 149:367–380
Nakayama T, Suda S, Kawachi M, Inouye I (2007) Phylogeny and ultrastructure of Nephroselmis and Pseudoscourfieldia (Chlorophyta), including the description of Nephroselmis anterostigmatica sp nov and a proposal for the Nephroselmidales ord. nov. Phycologia 46:680–697
Nakazawa A, Yamada T, Nozaki H (2004) Taxonomic study of Asterococcus (Chlorophyceae) based on comparative morphology and rbcL gene sequences. Phycologia 43:711–721
Nedelcu AM, Borza T, Lee RW (2006) A land plant–specific multigene family in the unicellular Mesostigma argues for its close relationship to Streptophyta. Mol Biol Evol 23:1011–1015
Nelsen MP, Plata ER, Andrew CJ, Lücking R, Lumbsch HT (2011) Phylogenetic diversity of trentepohlialean algae associated with lichen-forming fungi. J Phycol 47:282–290
Neustupa J, Eliáš M, Šejnohová L (2007) A taxonomic study of two Stichococcus species (Trebouxiophyceae, Chlorophyta) with a starch-enveloped pyrenoid. Nova Hedwigia 84:51–63
Neustupa J, Němcová Y, Eliáš M, Škaloud P (2009) Kalinella bambusicola gen. et sp. nov. (Trebouxiophyceae, Chlorophyta), a novel coccoid Chlorella-like subaerial alga from Southeast Asia. Phycol Res 57:159–169
Neustupa J, Eliáš M, Škaloud P, Němcová Y, Šejnohová L (2011) Xylochloris irregularis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta), a novel subaerial coccoid green alga. Phycologia 50:57–66
Novis PM (2006) Taxonomy of Klebsormidium (Klebsormidiales, Charophyceae) in New Zealand streams and the significance of low-pH habitats. Phycologia 45:293–301
Novis PM, Lorenz M, Broady PA, Flint EA (2010) Parallela Flint: its phylogenetic position in the Chlorophyceae and the polyphyly of Radiofilum Schmidle. Phycologia 49:373–383
Nyati S, Beck A, Honegger R (2007) Fine structure and phylogeny of green algal photobionts in the microfilamentous genus Psoroglaena (Verrucariaceae, Lichen-Forming Ascomycetes). Plant Biol 9:390–399
O’Kelly CJ, Kurihara A, Shipley TC, Sherwood AR (2010) Molecular assessment of Ulva spp. (Ulvophyceae, Chlorophyta) in the Hawaiian Islands. J Phycol 46:728–735
O’Kelly CJ, Wysor B, Bellows WK (2004) Collinsiella (Ulvophyceae, Chlorophyta) and other ulotrichalean taxa with shell-boring sporophytes form a monophyletic clade. Phycologia 43:41–49
Palenik B, Grimwood J, Aerts A, Rouzé P, Salamov A, Putnam N, Dupont C, Jorgensen R, Derelle E, Rombauts S, Zhou K, Otillar R, Merchant SS, Podell S, Gaasterland T, Napoli C, Gendler K, Manuell A, Tai V, Vallon O, Piganeau G, Jancek S, Heijde M, Jabbari K, Bowler C, Lohr M, Robbens S, Werner G, Dubchak I, Pazour GJ, Ren Q, Paulsen I, Delwiche C, Schmutz J, Rokhsar D, Van de Peer Y, Moreau H, Grigoriev IV (2007) The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci 104:7705–7710
Pažoutová M, Škaloud P, Nemjová K (2010) Phylogenetic position of Ooplanctella planoconvexa, gen. et comb. nova and Echinocoleum elegans (Oocystaceae, Trebouxiophyceae, Chlorophyta). Fottea 10:75–82
Peers G, Niyogi KK (2008) Pond scum genomics: The genomes of Chlamydomonas and Ostreococcus. Plant Cell Online 20:502–507
Pérez-Ortega S, Adl R, Crespo A, Sancho LG (2010) Symbiotic lifestyle and phylogenetic relationships of the bionts of Mastodia tessellata (Ascomycota, incertae sedis). Am J Bot 97:738–752
Petersen J, Teich R, Becker B, Cerff R, Brinkmann H (2006) The GapA/B gene duplication marks the origin of Streptophyta (Charophytes and Land Plants). Mol Biol Evol 23:1109–1118
Piganeau G, Eyre-Walker A, Grimsley N, Moreau H (2011) How and why DNA barcodes underestimate the diversity of microbial eukaryotes. PLoS ONE 6:e16342
Pombert J-F, Keeling PJ (2010) The mitochondrial genome of the entomoparasitic green alga Helicosporidium. PLoS ONE 5:e8954
Pombert J-F, Otis C, Lemieux C, Turmel M (2005) The chloroplast genome sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) reveals unusual structural features and new insights into the branching order of Chlorophyte lineages. Mol Biol Evol 22:1903–1918
Pombert J-F, Beauchamp P, Otis C, Lemieux C, Turmel M (2006) The complete mitochondrial DNA sequence of the green alga Oltmannsiellopsis viridis evolutionary trends of the mitochondrial genome in the Ulvophyceae. Curr Genet 50:137–147
Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, Hellsten U, Chapman J, Simakov O, Rensing SA, Terry A, Pangilinan J, Kapitonov V, Jurka J, Salamov A, Shapiro H, Schmutz J, Grimwood J, Lindquist E, Lucas S, Grigoriev IV, Schmitt R, Kirk D, Rokhsar DS (2010) Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329:223–226
Pröschold T, Leliaert F (2007) Systematics of the green algae: conflict of classic and modern approaches. In: Brodie J, Lewis J (eds) Unravelling the algae: the past, present, and future of algal systematics. CRC, Boca Raton, pp 123–153
Pröschold T, Harris EH, Coleman AW (2005) Portrait of a species. Genetics 170:1601–1610
Pröschold T, Bock C, Luo W, Krienitz L (2010) Polyphyletic distribution of bristle formation in Chlorellaceae: Micractinium, Diacanthos, Didymogenes and Hegewaldia gen. nov. (Trebouxiophyceae, Chlorophyta). Phycol Res 58:1–8
Pröschold T, Darienko T, Silva PC, Reisser W, Krienitz L (2011) The systematics of Zoochlorella revisited employing an integrative approach. Environ Microbiol 13:350–364
Reyes-Prieto A, Weber APM, Bhattacharya D (2007) The origin and establishment of the plastid in algae and plants. Annu Rev Genet 41:147–168
Rindi F (2011) Terrestrial green algae: systematics, biogeography and expected responses to climate change. In: Hodkinson TR, Jones MB, Waldren S, Parnell JAN (eds) Climate change, ecology and systematics. Cambridge University Press, Cambridge, pp 201–230
Rindi F, McIvor L, Guiry MD (2004) The Prasiolales (Chlorophyta) of atlantic Europe: an assessment based on morphological, molecular, and ecological data, including the characterization of Rosenvingiella radicans (Kützing) comb. nov. J Phycol 40:977–997
Rindi F, McIvor L, Sherwood AR, Friedl T, Guiry MD, Sheath RG (2007) Molecular phylogeny of the green algal order Prasiolales (Trebouxiophyceae, Chlorophyta). J Phycol 43:811–822
Rindi F, Lam DW, López-Bautista JM (2009) Phylogenetic relationships and species circumscription in Trentepohlia and Printzina (Trentepohliales, Chlorophyta). Mol Phylogenet Evol 52:329–339
Rindi F, Mikhailyuk TI, Sluiman HJ, Friedl T, Lopez-Bautista JM (2011) Phylogenetic relationships in Interfilum and Klebsormidium (Klebsormidiophyceae, Streptophyta). Mol Phylogenet Evol 58:218–231
Rodríguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Löffelhardt W, Bohnert HJ, Philippe H, Lang BF (2005) Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol 15:1325–1330
Rodríguez-Ezpeleta N, Brinkmann H, Roure B, Lartillot N, Lang BF, Philippe H (2007) Detecting and overcoming systematic errors in genome-scale phylogenies. Syst Biol 56:389–399
Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ (2007) The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol 24:54–62
Schultz J, Maisel S, Gerlach D, Müller T, Wolf M (2005) A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 11:361–364
Senousy HH, Beakes GW, Hack E (2004) Phylogenetic placement of Botryococcus braunii (Trebouxiophyceae) and Botryococcus sudeticus isolate UTEX 2629 (Chlorophyceae). J Phycol 40:412–423
Shoup S, Lewis LA (2003) Polyphyletic origin of parallel basal bodies in swimming cells of chlorophycean green algae (Chlorophyta). J Phycol 39:789–796
Skaloud P, Peksa O (2010) Evolutionary inferences based on ITS rDNA and actin sequences reveal extensive diversity of the common lichen alga Asterochloris (Trebouxiophyceae, Chlorophyta). Mol Phylogenet Evol 54:36–46
Sluiman HJ, Guihal C, Mudimu O (2008) Assessing phylogenetic affinities and species delimitations in Klebsormidiales (Streptophyta): nuclear-encoded rDNA phylogenies and ITS secondary structure models in Klebsormidium, Hormidiella, and Entransia. J Phycol 44:183–195
Smith SM, Abed RMM, Gercia-Pichel F (2004) Biological soil crusts of sand dunes in Cape Cod National Seashore, Massachusetts, USA. Microb Ecol 48:200–208
Somogyi B, Felföldi T, Solymosi K, Makk J, Homonnay ZG, Horváth G, Turcsi E, Böddi B, Márialigeti K, Vörös L (2011) Chloroparva pannonica gen. et sp. nov. (Trebouxiophyceae, Chlorophyta) – a new picoplanktonic green alga from a turbid, shallow soda pan. Phycologia 50:1–10
Takahashi F, Okabe Y, Nakada T, Sekimoto H, Ito M, Kataoka H, Nozaki H (2007) Origins of the secondary plastids of Euglenophyta and Chlorarachniophyta as revealed by an analysis of the plastid-targeting, nuclear-encoded gene psbO1. J Phycol 43:1302–1309
Tartar A, Boucias DG (2004) The non-photosynthetic, pathogenic green alga Helicosporidium sp. has retained a modified, functional plastid genome. FEMS Microbiol Lett 233:153–157
Tartar A, Boucias DG, Adams BJ, Becnel JJ (2002) Phylogenetic analysis identifies the invertebrate pathogen Helicosporidium sp. as a green alga (Chlorophyta). Int J Syst Evol Microbiol 52:273–279
Timme R, Delwiche C (2010) Uncovering the evolutionary origin of plant molecular processes: comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol 10:96
Turmel M, Pombert JF, Charlebois P, Otis C, Lemieux C (2007) The green algal ancestry of land plants as revealed by the chloroplast genome. Int J Plant Sci 168:679–689
Turmel M, Brouard J, Gagnon C, Otis C, Lemieux C (2008) Deep division in the Chlorophyceae (Chlorophyta) revealed by chloroplast phylogenomic analyses. J Phycol 44:739–750
Turmel M, Gagnon M-C, O’Kelly CJ, Otis C, Lemieux C (2009a) The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of Prasinophytes and the origin of the secondary chloroplasts of Euglenids. Mol Biol Evol 26:631–648
Turmel M, Otis C, Lemieux C (2009b) The chloroplast genomes of the green algae Pedinomonas minor, Parachlorella kessleri, and Oocystis solitaria reveal a shared ancestry between the Pedinomonadales and Chlorellales. Mol Biol Evol 26:2317–2331
Verbruggen H, Theriot EC (2008) Building trees of algae: some advances in phylogenetic and evolutionary analysis. Eur J Phycol 43:229–252
Watanabe S, Nakayama T (2007) Ultrastructure and phylogenetic relationships of the unicellular green algae Ignatius tetrasporus and Pseudocharacium americanum (Chlorophyta). Phycol Res 55:1–16
Weiss TL, Spencer Johnston J, Fujisawa K, Sumimoto K, Okada S, Chappell J, Devarenne TP (2010) Phylogenetic placement, genome size, and GC content of the liquid-hydrocarbon-producing green microalga Botryococcus braunii strain Berkeley (Showa) (Chlorophyta)1. J Phycol 46:534–540
Wodniok S, Brinkmann H, Glockner G, Heidel A, Philippe H, Melkonian M, Becker B (2011) Origin of land plants: do conjugating green algae hold the key? BMC Evol Biol 11:104
Worden AZ, Lee J-H, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, Foulon E, Grimwood J, Gundlach H, Henrissat B, Napoli C, McDonald SM, Parker MS, Rombauts S, Salamov A, Von Dassow P, Badger JH, Coutinho PM, Demir E, Dubchak I, Gentemann C, Eikrem W, Gready JE, John U, Lanier W, Lindquist EA, Lucas S, Mayer KFX, Moreau H, Not F, Otillar R, Panaud O, Pangilinan J, Paulsen I, Piegu B, Poliakov A, Robbens S, Schmutz J, Toulza E, Wyss T, Zelensky A, Zhou K, Armbrust EV, Bhattacharya D, Goodenough UW, Van de Peer Y, Grigoriev IV (2009) Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324:268–272
Yamaguchi H, Suda S, Nakayama T, Pienaar R, Chihara M, Inouye I (2011) Taxonomy of Nephroselmis viridis sp. nov. (Nephroselmidophyceae, Chlorophyta), a sister marine species to freshwater N. olivacea. J Plant Res 124:49–62
Yamamoto M, Kurihara I, Kawano S (2005) Late type of daughter cell wall synthesis in one of the Chlorellaceae, Parachlorella kessleri (Chlorophyta, Trebouxiophyceae). Planta 221:766–775
Zhang J, Huss VAR, Sun X, Chang K, Pang D (2008) Morphology and phylogenetic position of a trebouxiophycean green alga (Chlorophyta) growing on the rubber tree, Hevea brasiliensis, with the description of a new genus and species. Eur J Phycol 43:185–193
Zoller S, Lutzoni F (2003) Slow algae, fast fungi: exceptionally high nucleotide substitution rate differences between lichenized fungi Omphalina and their symbiotic green algae Coccomyxa. Mol Phylogenet Evol 29:629–640
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Friedl, T., Rybalka, N. (2012). Systematics of the Green Algae: A Brief Introduction to the Current Status. In: Lüttge, U., Beyschlag, W., Büdel, B., Francis, D. (eds) Progress in Botany 73. Progress in Botany, vol 73. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-22746-2_10
Download citation
DOI: https://doi.org/10.1007/978-3-642-22746-2_10
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-22745-5
Online ISBN: 978-3-642-22746-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)