Abstract
Evolution is a stochastic process, resulting from a combination of deterministic and random factors. We present results from a general theory of directional evolution that reveals how random variation in fitness, heritability, and migration influence directional evolution. First, we show how random variation in fitness produces a directional trend toward phenotypes with minimal variation in fitness. Furthermore, we demonstrate that stochastic variation in population growth rate amplifies the expected change due to directional selection in small populations. Second, we show that the evolutionary impacts of migration depend on the entire distribution of migration rates such that increasing the variance in migration rates reduces the impact of migration relative to selection. This means that changing the variance in migration rates, holding the mean constant, can substantially change the potential for local adaptation. Finally, we show that covariation between stochastic selection and stochastic heritability can drive directional evolutionary change, and that this can substantially alter the outcome of evolution in variable environments.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Evolutionary biologists have long recognized the importance of stochastic processes in the mechanics of evolution. The best studied stochastic evolutionary process is genetic drift – change in allele frequency resulting from random variation in fitness and segregation – which plays a critical role in the modern theory of molecular evolution. Drift is nondirectional, meaning that the expected change in allele frequency due to drift alone is zero, and it is often assumed that this will be true of any stochastic evolutionary process.
In fact, the potential of stochastic variation in fitness to contribute to directional evolution was noted by a number of authors in the 1970s (Hartl and Cook 1973; Karlin and Liberman 1974; Gillespie 1974). These authors recognized that differences in the variances of individual fitness distributions could contribute to directional change, just as differences in the mean values can.
Differential fitness is not the only factor influencing evolution. Both migration between populations and the process of inheritance itself can drive directional evolution – and, like selection, both of these are inherently stochastic processes (this is most obvious in the case of inheritance, since both mutation and recombination are chemical processes subject to quantum uncertainty). In this chapter, we will discuss some of the ways in which random variation in fitness, migration, or inheritance can lead to directional evolutionary change.
2 Modeling Stochastic Evolution
Introducing stochasticity into our models of evolution requires that we treat values like fitness and migration rate as random variables. For our purposes, a random variable differs from an ordinary variable in a mathematical equation in that a random variable has a distribution of possible values, rather than a single value.
It is important to note that saying that fitness, migration, or anything else, is stochastic is not the same as just saying that it varies over time. If we specify that fitness values will alternate, across generations, between specific values, we are defining a deterministic (not stochastic) process in which the value varies in a predictable manner over time. By contrast, making fitness stochastic means that we cannot say what value it will have at any particular time, only that it has a distribution of possible values at that time.
This distinction is important; treating a variable as deterministic but temporally variable can yield very different results than does treating it as a stochastic random variable. This is illustrated, for the case of fitness, in Fig. 2.1. In Fig. 2.1a, fitness is treated as an ordinary variable that fluctuates over time – alternating between 1 and 2. In Fig. 2.1b, fitness is a random variable that, in any particular generation, has a 50% chance of being 1 and a 50% chance of being 2.
Though we might be tempted to treat the deterministic case (Fig. 2.1a) as an “average” instance of the stochastic case (Fig. 2.1b), that would be misleading. After four generations, a population following the deterministic case will have increased in size by a factor of 4, corresponding to a per generation change of \( \sqrt {2} \approx 1.414 \), which is the geometric mean of 1 and 2. By contrast, the expected size of a population following the stochastic case for four generations is just over 5 (specifically, 5.0625), corresponding to a per generation change of 1.5 – the arithmetic mean of 1 and 2. The different outcomes illustrated in Fig. 2.1 are not results of the short time interval considered; the same effective fitness values arise if we consider an arbitrary number of generations. This should be kept in mind when evaluating arguments about the utility of geometric mean fitness.
Treating fitness, migration, and other values as random variables requires that we consider the variances and covariances of their distributions. This can lead to notational confusion, since we are also concerned with means, variances, and covariances of the same values within a population. For example, we will be concerned with both the mean fitness of an individual (the mean of its fitness distribution) and the mean fitness in the entire population.
We thus will distinguish between two different sets of statistical operators: frequency and probability. Frequency operators, denoted by straight symbols (ā for mean, \( [{[^2}a]] \) for variance, and \( [[a,b]] \) for covariance), describe operations over some collection of things. For instance, \( \overline w \) is the average fitness across individuals in a population, and \( [[\phi, w]] \) is the covariance, across all individuals in the population, between phenotype and fitness.
Probability operators, denoted by angled symbols (â for mean, \( \left\langle {\left\langle {^2a} \right\rangle } \right\rangle \) for variance, and \( \left\langle {\left\langle {a,b} \right\rangle } \right\rangle \) for covariance), describe operations over distributions of random variables. For example, \( \widehat{w} \) is the expected fitness of an individual – the mean of its fitness distribution – while \( \left\langle {\left\langle {^2w} \right\rangle } \right\rangle \) is the variance of the same distribution (the variance in fitness values that the individual might have). A detailed discussion of these two kinds of operators, and the rules for manipulating them, is given in Rice and Papadopoulos (2009). Table 2.1 lists the main symbols that we will use in this chapter.
3 Stochastic Fitness
An individual’s fitness is the number of descendants that it has after some chosen time interval. We often choose the time interval to be a single generation and think of fitness as simply the number of offspring, but in the general case, we need to consider all descendants, including grand offspring and the individual itself at the future time.
Because we cannot know with certainty how many descendants each individual in a population will have, we need to treat fitness as a random variable – having a distribution of possible values. The vast majority of evolutionary models consider only the mean of this distribution – the expected number of descendants – and in fact fitness is often defined as this expected value. We will see, though, that accurately describing evolution requires consideration of the entire distribution.
Using the notation in Table 2.1, the general equation for evolution in a closed population (no migration in or out) with stochastic fitness and inheritance is (Rice 2008):
Fitness enters into Eq. (2.1) through the term Ω, which is the ratio of an individual’s fitness to mean population fitness. We will refer to this as “relative fitness” (note, though, that the term “relative fitness” is sometimes used in other ways). Strictly, Ω is defined under the condition that \( \overline w \ne 0, \) for both mathematical and biological reasons. Mathematically, the ratio is undefined if \( \overline w = 0 \). Biologically, mean population fitness being zero corresponds to extinction, and change in mean phenotype is undefinable when the population ceases to exist.
It is common to treat mean population fitness (\( \overline w \)) as a constant, even when individual fitness (w) is a random variable. This is done, for example, in both the Wright-Fisher and Moran models of genetic drift. This assumption, though, is made purely for the sake of simplifying the mathematics – nobody expects \( \overline w \) to be constant in most real populations. We will thus treat mean population fitness as another random variable. As we show below, relaxing this seemingly inoffensive assumption exposes an entire class of evolutionary processes that are otherwise invisible.
Acknowledging that both w and \( \overline w \) are random variables complicates our interpretation of \( \widehat{\Omega } \). The expected value of a ratio of random variables can behave in surprising ways. For instance, given random variables a and b, it can be the case that the expected value of \( \tfrac{a}{b} \) and that of \( \tfrac{b}{a} \) are both greater than 1. Rice (2008) showed that expected relative fitness can be written as an infinite series, the terms of which contain moments of the individual fitness distributions. When the fitness values of different individuals are independent, then this series can be written as:
Substituting this series into the first term on the right-hand side of Eq. (2.1) yields:
The \( \left\langle {\left\langle {^iw} \right\rangle } \right\rangle \) terms in Eqs. (2.2) and (2.3) are the central moments of an individual’s fitness distribution (\( \langle {\langle {^2w} \rangle } \rangle \) being the variance, and \( \langle {\langle {^3w} \rangle } \rangle \) being the third central moment, etc.). Eq. (2.3), thus, shows how different aspects of the shapes of individual fitness distributions contribute to directional evolution.
The standard interpretation of selection is captured by the first term on the right-hand side of Eq. (2.3), which contains the covariance between phenotype and expected fitness (\( \left[ {\left[ {\phi, \hat{w}} \right]} \right] \)). All of the subsequent terms, involving the various moments of the fitness distribution, represent directional stochastic effects in evolution. Note that these disappear if we treat fitness as a fixed value, since, in that case, all of the \( \left\langle {\left\langle {^iw} \right\rangle } \right\rangle \) terms are zero.
To illustrate how these directional stochastic effects influence evolution, we consider the second term, containing the covariance between phenotype and variance in fitness (\( \left[ {\left[ {\phi, \left\langle {\left\langle {^2w} \right\rangle } \right\rangle } \right]} \right] \)). This term is negative (as are all terms involving even-valued moments) – telling us that there is a force pulling the population toward phenotypes with minimum variance in fitness.
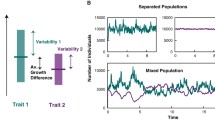
Figure 2.2 illustrates schematically how a population can be pulled toward phenotypes that minimize variance in fitness. The figure shows a case in which two different phenotypic values have the same expected fitness (i.e., the same \( \widehat{w} \)), but different variances in their fitness distributions (i.e., different values of \( \langle {\langle {^2w} \rangle } \rangle \)). The key is to note that the magnitude of change in mean phenotype is inversely proportional to mean population fitness (\( \bar{w} \)). Thus, in case (B), when individuals with φ = 0 have low fitness (and thus they decrease in frequency), the change in mean phenotype is relatively large, because \( \bar{w} \) is low in that case. By contrast, when φ = 0, individuals are doing well (and thus increasing in frequency), their increase is relatively small because \( \bar{w} \) is larger in this case (A). The result is that, even though mean phenotype increases half of the time and decreases half of the time, the step sizes are different – large when \( \overline \phi \) increases and small when it decreases – leading to a net positive expected change.
A simple case of directional stochastic evolution. Individuals with phenotype (φ) of 0 leave two descendants or none, each with probability 0.5. Individuals with phenotypic value 1 always leave one descendant. Initial mean phenotype (\( \overline \phi \)) is 0.5. If individuals with phenotype 0 leave 2 offspring each, then the mean phenotype changes to \( \overline \phi = \tfrac{1}{3} \), so \( \Delta \overline \phi = - \tfrac{1}{6} \). If these individuals leave no offspring, then \( \Delta \overline \phi = \tfrac{1}{2} \). Since these two outcomes occur with equal probability, the expected change in mean phenotype is \( \widehat{{\Delta \overline \phi }} = \tfrac{1}{2}\left( { - \tfrac{1}{6}} \right) + \tfrac{1}{2}\ \tfrac{1}{2} = \tfrac{1}{6} \)
This example illustrates the basic principle underlying directional stochastic evolution. When population growth rate (here captured by \( \bar{w} \)) is large, the step size in evolutionary change tends to be smaller than when growth rate is low. Strategies that contribute disproportionately to variation in population growth rate (such as the strategy φ = 0 in Fig. 2.2) thus tend to take smaller steps when they increase than when they decrease in frequency. In Fig. 2.2, the expected fitness values (\( \widehat{w} \)) are the same for the two strategies, so there is no directional selection acting. If this is not the case, then the expected change is a function of both selection and the directional stochastic effects.
Note that, in Eq. (2.3), the terms on the right-hand side are each divided by increasing powers of population size (N). This is because Eqs. (2.2) and (2.3) assume that each individual’s realized fitness is independent of that of other individuals. This is what we expect when variation in fitness is due to pure demographic stochasticity. In such cases, the strength of directional stochastic evolutionary effects declines with increasing population size. By contrast, when variation in fitness is due to stochastic environmental variation, such that all individuals with a particular phenotype either do well or poorly together, then directional stochastic effects remain strong even in large populations (Rice 2008).
Though we have discussed only the effects of variance, it is clear from Eq. (2.1) that all of the moments of an individual’s fitness distribution can influence evolution. To see the general pattern, note that the terms containing even moments (2nd, 4th, etc.) are all negative, while those containing odd moments are positive. Since even moments measure symmetrical spread about the mean, and odd moments measure asymmetry, we can say that directional stochastic evolution tends to shift populations toward phenotypes with minimum symmetrical variation in fitness and maximum positive skewness in fitness.
Finally, we note that even the selection term in Eq. (2.1) is influenced by stochasticity. The \( {\hbox{H(}}\overline {\hbox{w}} {)} \) in the denominator of the first term on the right-hand side represents the harmonic mean of \( \bar{w} \). Because the harmonic mean is strongly influenced by small values, this term will get smaller as the variance in \( \bar{w} \) increases – thus amplifying the selection differential (Rice 2008). Since \( \bar{w} \) is the mean of a finite set of individuals, its variance is expected to go up as population size declines (corresponding to taking the mean of a smaller sample). Thus, the expected change due to selection will tend to increase as population size gets very small. Note, though, that the variance in \( \Delta \overline \phi \) will also increase in small populations, so it will be necessary to examine a large number of cases to see the amplifying effect on the mean.
4 Stochastic Migration
Migration, like fitness, influences population growth. We thus might expect that stochastic variation in migration rates will generate the same kinds of directional evolutionary effects that we saw with stochastic fitness. The general equation for change in mean phenotype in an open population (one subject to immigration and emigration) is (Rice and Papadopoulos 2009):
Here, the left superscript ds indicate that the values are measured within a deme – a subpopulation subject to migration.
The various terms in Eq. (2.4) capture all of the ways that selection, transmission, and migration can influence directional change. We will focus here only on the last term on the right-hand side, \( \hat{\Xi }\left( {\widehat{\gamma } - \widehat{{\overline \delta }}} \right) \) (Rice and Papadopoulos (2009) present the full derivation, and discuss the meaning of each of the terms). \( \left( {\widehat{\gamma } - \widehat{{\overline \delta }}} \right) \) is simply the difference between the expected phenotype of immigrants and that of native offspring who stay in the deme. The expected relative immigration rate is captured by \( \widehat{\Xi } \), which is the expected value of the number of immigrants divided by the total deme growth rate.
Note that the number of immigrants directly influences the deme growth rate; so \( \widehat{\Xi } \), like \( \widehat{\Omega } \), is the expectation of the ratio of correlated random variables. Defining ξ and \( \varepsilon \) as the numbers of immigrants and emigrants divided by deme size, we can expand \( \widehat{\Xi } \) to yield:
Equation 2.5 shows only the first- and second-order terms in the expansion, but already we can see that directional evolution will be influenced not only by the expected immigration rate (\( \widehat{\xi } \)), but also by the variance in immigration (\( \langle {\langle {^2\xi } \rangle } \rangle \)), the covariance between immigration and fitness within the deme (\( \langle {\langle {\bar{w},\xi } \rangle } \rangle \)), and the covariance between immigration and emigration (\( \langle {\langle {\varepsilon, \,\xi } \rangle } \rangle \)).
Focusing on the variance in immigration rate (\( \langle {\langle {^2\xi } \rangle } \rangle \)), the fact that the second term on the right-hand side of Eq. (2.5) is negative shows that increasing the variance in migration reduces the impact of migration on directional change. This effect is illustrated in Fig. 2.3 (the numbers are from Fig. 2.3a in Rice and Papadopoulos (2009)).
The effect of changing the variance in immigration rates in a continent–island model. The bar graphs show the distribution of immigration rates to the island, ranging from low variance (case 1) to very high variance (case 6). In each case, the mean immigration rate is two individuals per generation. Selection on the continent favors a phenotypic value of 0, while selection on the island favors a phenotypic value of 1. In the lower figures, mean phenotype is indicated by shading, with 0 being white and 1 being black
Figure 2.3 shows the consequences of changing the variance in immigration rate in a continent–island model, where evolution on the island is a consequence of both local selection and migration from the continent. Here, selection on the continent favors phenotypic value zero (white) while selection on the island favors phenotypic value 1 (black). In all cases, the expected number of migrants from the continent to the island is two per generation. The distribution of migration rates varies though, from a case with relatively low variance (case 1, in which either one or three immigrants arrive, each with probability 0.5) to a case of very high variance (case 5, in which 200 immigrants may arrive at once, but none arrive in most generations).
In the example shown, selection on the island favors a phenotypic value of 1 (black), but with a mean immigration rate of two individuals per generation and low variance, the equilibrium mean phenotype on the island is only 0.29. Increasing the variance in migration rate (while holding the mean rate constant) significantly increases the degree to which the island population can diverge from the continental population.
We thus see that the potential for local adaptation is a function not only of expected migration rates, but of the entire distribution of rates at which individuals arrive or depart. The reason that high variance in immigration rate reduces the impact of migration relative to selection is that when many immigrants arrive together, deme growth rate (here defined as R, which combines reproduction within the deme with immigration and emigration) is large. Large R (just like large \( \bar{w} \) in the example in Fig. 2.2) reduces the magnitude of change in mean phenotype.
This result also has consequences for speciation. A number of authors (Schluter 2001; Rundle and Nosil 2005; Fitzpatrick et al. 2009) have noted that pure allopatric or pure sympatric speciation are extreme cases, and that many actual cases of speciation will involve alternation of allopatry and sympatry. The result presented above illustrates that even if the average migration rate is held constant, lengthening the time between immigration pulses (even when those pulses involve more individuals) greatly increases the opportunity for the fixation of traits that facilitate reproductive isolation.
Finally, these results have consequences for our interpretation of traditional migration models. In nearly all natural populations, there will be variation in migration rates. The fact that such variation reduces the impact of migration relative to selection means that models that treat migration as a single parameter (and thus assume no variance in migration rates) will always tend to overestimate the relative importance of migration as an evolutionary force.
5 Stochastic Inheritance
Unlike reproduction and migration, the degree to which offspring resemble their parents does not directly influence population size. Stochastic variation in inheritance, thus, does not lead to the kind of directional stochastic evolution that we see in the cases of fitness and migration. In fact, so long as inheritance is independent of fitness or migration rates, we need not know anything more than the expected phenotype of offspring in order to calculate \( \widehat{{\Delta \overline \phi }} \). The situation changes, though, if inheritance covaries with selection.
Though it is generally treated as a fixed parameter in quantitative genetic models, heritability often changes as a function of the environment in which organisms develop (Merila and Sheldon 2001; Charmantier and Garant 2005; Wilson et al. 2006). We thus expect that, to the extent that the environment is stochastic, so will be heritability. More significantly, if any of the environmental factors that influence heritability also influence fitness, then we expect heritability and fitness to covary. That this happens in many natural populations is suggested by the observation that environments that confer low fitness also tend to confer low heritability (Charmantier and Garant 2005; Wilson et al. 2006).
To illustrate how covariation between heritability and selection can influence evolution, we consider a simple case. Following the standard assumption of quantitative genetics, we assume that the mean phenotype of offspring is a linear function of parental phenotype (or, properly, midparent phenotype). This assumption allows us to capture inheritance with a single term, heritability (h 2), defined as the slope of the linear regression of offspring phenotype on midparent phenotype. We could just as well use the covariance between offspring and midparent phenotype – which is the “additive genetic variance”. (Note that, for sexually reproducing organisms, an “individual” parent with respect to Eq. (2.1) is really a mated pair, with φ being the mean phenotype of the pair. We were thus tacitly already using “midparent” phenotype.)
Inheritance enters into Eq. (2.1) through the term δ, the difference between the mean phenotype of an individual’s offspring and that individual’s own phenotype. Under the standard quantitative genetics assumptions, δ can be derived from the parent’s phenotype and population wide heritability, as shown in Fig. 2.4.
(a) The relationship between δ and heritability (h 2) under the assumptions of quantitative genetics. Heritability is the slope of the regression of offspring phenotype on midparent phenotype. For a given midparent phenotype (φ), δ φ is the vertical distance between the 45° line (defined by φ° = φ) and the regression line. (b) When heritability is stochastic, the regression of offspring on parents becomes a random variable – having a distribution of possible slopes, rather than a single slope
The key results, derivable from Fig. 2.4, are that, under the quantitative genetics assumptions, δ = (h 2 − 1)(\( \phi - \overline \phi \)) and \( \widehat{{\overline \delta }} = 0 \). Substituting these results into Eq. (2.1) and simplifying yields:
The first term on the right-hand side of Eq. (2.6), \( \widehat{{{h^2}}}[ {[ {\phi, \widehat{\Omega }} ]} ] \), is just the expected heritability multiplied by the selection differential. This is written “h 2 S” in quantitative genetics (S is the selection differential), so the first term is just the standard “breeder’s equation”.
The second term on the right of Eq. (2.6), \( \left[ {\left[ {\phi, \langle \!{\langle {{h^2},\Omega } \rangle } \! \rangle } \right]} \right] \), captures the evolutionary consequences of covariation between heritability and selection. This term is read as: the covariance across the population (frequency covariance) between an individual’s phenotype (φ) and the covariance (probability covariance) between that individual’s relative fitness and heritability of φ. (Note that the distinction between frequency and probability covariance is critical here).
Figure 2.5 shows one way that covariation between heritability and selection can influence the outcome of evolution. In this example, the environment that a population experiences varies unpredictably across generations such that, in any one generation, there is a 50% chance of experiencing Environment 1 and a 50% chance of experiencing Environment 2. (e.g., these might correspond to wet and dry years experienced by an annual plant). The solid lines show the fitness in each environment as a function of phenotype, and the dashed line shows the expected fitness across both environments.
Illustration of one consequence of covariation between selection and heritability. Heritabilities in Environments 1 and 2 are represented by \( h_1^2 \) and \( h_2^2 \), respectively. The arrows and vertical dotted lines indicate the equilibrium phenotype for the case of constant heritability (\( h_1^2 = h_2^2 = 0.5 \)) and the case in which heritability covaries with the environment (\( h_1^2 = 0.6,\quad h_2^2 = 0.4 \)). The equilibria were calculated from (2.6) under the assumption that the population variance is low enough that the regressions of fitness on phenotype are approximated by the slopes of the fitness curves
If heritability is equal in both environments, then the evolutionary equilibrium is (in this case) the strategy that maximizes mean fitness. This outcome changes when heritability covaries with the selective regime. If heritability is higher in Environment 1 than in Environment 2, then the equilibrium shifts to a point where the population is much better adapted to Environment 1.
Biologically, this is because selection is more efficient at driving evolution in the environment in which heritability is higher. Mathematically, we can see how this result follows from Eq. (2.6) by noting how \( \langle \!{\langle {{h^2},\widehat{\Omega }} \rangle }\! \rangle \, \) varies with phenotype. For individuals with large values of the trait, near the optimum for Environment 1, high fitness (Environment 1) co-occurs with high heritability (also Environment 1), so \( \langle\! {\langle {{h^2},\widehat{\Omega }} \rangle }\! \rangle \, >\, 0 \). By contrast, for individuals with lower values of the trait, near the optimum for Environment 2, high fitness co-occurs with low heritability, so \( \langle\! {\langle {{h^2},\widehat{\Omega }} \rangle } \!\rangle \, <\, 0 \). Thus \( \left[ {\left[ {\phi, \langle \!{\langle {{h^2},\Omega } \rangle } \!\rangle } \right]} \right] \) is positive, shifting the population toward higher phenotypic values.
Two points should be noted from this example: First, covariance between heritability and selection shifts the equilibrium substantially toward the optimal phenotype for Environment 1, and substantially away from the optimum for environment 2. Thus, even though the population encounters Environment 2 roughly half of the time, and has significant heritability in that environment, it ends up rather poorly adapted to Environment 2. Second, the equilibrium does not maximize \( \bar{w} \). This example thus illustrates that, even with frequency-independent selection, mean population fitness is not necessarily maximized when fitness and heritability are stochastic.
6 Conclusions
Many of the factors that influence evolution, including individual fitness, migration, and genetic transmission, are inherently stochastic. For the sake of mathematical simplicity, many evolutionary models treat some or all of these factors as deterministic, on the assumption that any stochasticity in real systems will simply add noise to the outcome, without changing the expected value.
We have demonstrated that, contrary to the common assumption, stochastic variation in fitness or migration, even when that variation is completely symmetrical, imposes a directionality on evolution that is not apparent in deterministic models. Stochastic heritability, while not directional itself, can significantly influence adaptation when heritability covaries with selection.
In this chapter, we have chosen only a few examples for the sake of illustration. However, the number of terms in Eqs. (2.1) and (2.4), and the fact that each of these terms can be expanded as in Eqs. (2.2) and (2.5), suggest that we have only scratched the surface in the study of directional stochastic effects in evolution.
References
Charmantier A, Garant D (2005) Environmental quality and evolutionary potential: lessons from wild populations. Proc Biol Sci 272(1571):1415–1425
Fitzpatrick BM, Fordyce JA, Gavrilets S (2009) Pattern, process and geographic modes of speciation. J Evol Biol 22:2342–2347
Gillespie JH (1974) Natural selection for within-generation variance in offspring number. Genetics 76(3):601–606
Hartl DL, Cook RD (1973) Balanced polymorphisms of quasineutral alleles. Theor Popul Biol 4(2):163–172
Karlin S, Liberman U (1974) Random temporal variation in selection intensities: case of large population size. Theor Popul Biol 6(3):355–382
Merila J, Sheldon BC (2001) Avian quantitative genetics. In: Nolan V Jr (ed) Current ornithology, Chapter 4, vol 16. Kluwer/Plenum, New York, p 179
Rice SH (2008) A stochastic version of the Price equation reveals the interplay of deterministic and stochastic processes in evolution. BMC Evol Biol 8:262
Rice SH, Papadopoulos A (2009) Evolution with stochastic fitness and stochastic migration. PLoS ONE 4(10):e7130
Rundle HD, Nosil P (2005) Ecological speciation. Ecol Lett 8(17):336–352
Schluter D (2001) Ecology and the origin of species. Trends Ecol Evol 16(7):372–380
Wilson AJ, Pemberton JM, Pilkington JG, Coltman DW, Mifsud DV, Clutton-Brock TH, Kruuk LEB (2006) Environmental coupling of selection and heritability limits evolution. PLoS Biol 4(7):e216
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Rice, S.H., Papadopoulos, A., Harting, J. (2011). Stochastic Processes Driving Directional Evolution. In: Pontarotti, P. (eds) Evolutionary Biology – Concepts, Biodiversity, Macroevolution and Genome Evolution. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-20763-1_2
Download citation
DOI: https://doi.org/10.1007/978-3-642-20763-1_2
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-20762-4
Online ISBN: 978-3-642-20763-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)