Abstract
The wild-relative species of spinach comprise Spinacia turkestanica iljin and S. tetrandra Stev., with closely related genera Beta and Chenopodium. The wild species of Spinacia have not been collected for conservation in gene banks to a large extent, but they do not seem to be threatened seriously in their growing areas of Western Asia. They may be an important source of genes for disease resistance for cultivated spinach in the future, and they have contributed to the elucidation of the development of the heteromorphic sex expression in the genera. In situ collection of the two wild Spinacia species should be encouraged to generate good sources of disease-resistance genes for future breeding of vegetable spinach.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
13.1 Basic Botany of the Species
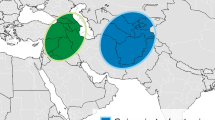
The cultivated spinach, Spinacia oleracea L., belongs to the family Chenopodiaceae with two known wild relative species S. turkestanica iljin and S. tetrandra Stev. A study of taxonomic position of Spinacia with its two closely related genera Beta and Chenopodium and their taxonomic position in Caryophyllales based on chloroplast restriction polymorphism can be found in Downie and Palmer (1994). The two wild species are found distributed over western parts of Asia, S. turkistanica in Turkmenistan, Uzbekistan, and Kazakstan and S. tetrandra in the Caucasus area, in Armenia and Kurdistan between Iran, Iraq, and Turkey. The two wild species have some tendency to become weedy, but remain geographically restricted (Astley and Ford-Lloyd 1981; Gabrielian and Fragman-Sapir 2008). Cultivated spinach is diploid with 2n = 2x = 12 chromosomes (Bemis and Wilson 1953). It is widely cultivated as a leaf vegetable all over the world with a worldwide production in 2007 of 14 MT fresh weight, most of these (12 MT) produced in China (FAO 2007).
13.2 Conservation Initiatives
There are quite large collections worldwide of cultivated spinach, while activities concerning the wild species seem sporadic. The largest organized conservation initiative for the wild species seems to be the Crop Wild Relative initiative with reference to 48 accessions of S. tetrandra registered in the Armenian database in vivo (Crop Wild Relatives 2009). Most gene banks such as the International Spinach Data Base of The Netherlands (12 S. turkestanica and 14 S. tetrandra) (International Spinach Database 2009), the National Plant Germplasm System USDA/ARS, GRIN (4 S. turkestanica and 2 S. tetrandra) (National Plant Germplasm System 2009), and the Plant Genetics Resources of Germany (2 S. turkestanica and 3 S. tetrandra) (Plant Genetic Resources 2009) have few accessions of the wild species. It has been argued that for reasons of species conservation at least in the Russian area the wild Spinacia does not need conservation in gene banks because they are not threatened (Ul'janova 1977); however, for use in the improvement of cultivated spinach, larger seed bank collections would be highly desirable.
13.3 Role in Elucidation of Origin and Evolution of Spinach
The exact origin of the cultivated spinach is not known, and the species is not found as wild types. The geographical distribution of the close relatives S. turkestanica and S. tetrandra is the main reason for the general assumption that cultivated spinach originated from the same geographical area of West Asia. The general high sexual compatibility between cultivated S. oleracea and the two wild species supports the idea that cultivated spinach originated through domestication of one or both of the wild species. The cultivated spinach seems to have spread to China during the seventh century and to Europe during the twelfth century (Sneep 1982; Sauer 1993; Rubatzky and Yamaguchi 1997).
13.5 Role in Classical and Molecular Genetic Studies
Spinacia turkestanica has been used as a parent for construction of genetically broad segregating offspring populations for construction of a genetic map and for mapping of the genetic factor determining dioecious sex expression in spinach (Khattak et al. 2006). S. tetrandra has been used in a genetic diversity study of spinach using target region amplification markers (TRAP), and the single accession of S. tetrandra did not cluster much distant from the 47 S. oleracea accessions of the study. This indicates that the genetic differences between cultivated and wild species are small and that a rather large genetic variation is still maintained in the cultivated material (Hu et al. 2007). Large genetic variation maintained within cultivated S. oleracea is also concluded from a diversity study using genic microsatellite markers (Khattak et al. 2007).
13.6 Role in Spinach Improvement Through Traditional and Advanced Tools
A major problem in production of cultivated spinach is the frequent attack by specialized races of downy mildew (Peronospora farinose f.sp. spinaciae). These fungal epidemics pose a threat to spinach production without widespread chemical sprays. Specific highly efficient genes for resistance against at least seven different races of downy mildew have been identified and used in breeding, but the duration of effective resistance of such genes become increasingly shorter as spinach production intensifies. At least one such race-specific gene against the race D3 is known to be transferred to cultivated spinach from a line of S. turkestanica, and another specific gene conferring resistance against Peronospora race LN4 has also been transferred from S. turkestanica (Hanke et al. 2000). Other diseases such as leaf spot caused by the fungus Stemphylium botryosum (Mou et al. 2008) as well as several pests including leaf miners Liriomzyza spp. (Mou 2008a) cause problems in spinach production, and more resistant plant types may be found in future in the wild material.
13.7 Genomics Resources Developed
The complete annotated chloroplast genome is available for spinach (Schmitz-Linneweber et al. 2001), but further published sequence information in Spinacia is still limited. The NCBI database (NCBI 2009) contained only 363 genomic sequences, 284 mRNA sequences, and 16 rRNA sequences. The sequences in the database was used by Groben and Wricke (1998) and Khattak et al. (2007) to develop a set of 35 primer pairs for amplification of microsatellites also known as simple sequence repeats (SSRs) of spinach, 13 of which were used for a diversity study of the cultivated spinach.
13.8 Scope for Domestication and Commercialization
There seems to be little reason to domesticate or commercialize the two wild relatives of spinach, since they do not seem very different from the already cultivated material. The two wild species, however, may be a valuable source of genes for resistance to common diseases of cultivated spinach. If such variation is conserved in the wild material, it may be used to introgress the resistance genes into the cultivated spinach and, thus, to maintain a sustainable low chemical input for vegetable production worldwide. Apparently, so far, the wide use of genes from the wild relatives has not been necessary, since enough resistance has been available within the cultivated material; however, in the future the resource of the wild gene pool may be valuable. An undesirable feature of cultivated spinach in addition to disease and pest susceptibility is the tendency to accumulate high amounts of oxalates, which may induce nutritional problems. Screening for low oxalate genotypes have been performed with mainly the cultivated types of spinach, with some progress (Mou 2008b). Screening of larger collections of wild types of spinach may provide further useful genes for this type of breeding.
13.9 Some Dark Sides and Their Addressing
The wild and cultivated types of spinach are reported to be only slightly weedy and apparently not very invasive, since they are still only found wild in limited geographic areas. They are, however, strong wind-pollinated outbreeders whose pollen can spread over long distances. Thus, they will be expected to be difficult to contain in case of transgenic cultivated types being marketed.
13.10 Recommendations for Future Actions
The wild species and their genetic variation have already been shown to be valuable as sources of disease resistance genes for cultivated types of spinach. Further international support for local conservation both in situ and ex situ collections should be encouraged. With new generations of sequencing technology, it has become efficient to establish much better maps and synteny studies, which should be used to ease future introgression of important traits from wild to cultivated spinach. Furthermore, the new marker techniques are efficient tools to characterize diversity of wild species for efficient stratification of the material during conservation initiatives.
References
Astley D, Ford-Lloyd BV (1981) The evolutionary significance of multigermicity in the genus Spinacia (Chenopodiaceae). Plant Syst Evol 137:57–61
Bemis WP, Wilson GB (1953) A new hypothesis explaining the genetics of sex determination in Spinacea oleracea L. J Hered 44:91–95
Crop wild Relatives (2009) Bioversity International. Rome, Italy. http://www.cropwildrelatives.org/. Accessed 20 May 2009
Downie SR, Palmer JD (1994) A chloroplast DNA phylogeny of the Caryophyllales based on structure and inverted repeat restriction site variation. Syst Bot 19:236–252
FAO (2007) Food and Agriculture Organisation of the United Nations. FAOSTAT database, Rome, Italy
Gabrielian E, Fragman-Sapir O (2008) Flowers of the Transcaucasus and adjacent areas. Koeltz Scientific Books, Koenigstein, Germany
Groben R, Wricke G (1998) Occurrence of microsatellites in spinach sequences from computer databases and development of polymorphic SSR markers. Plant Breed 117:271–274
Hanke S, Seehous C, Radies M (2000) Detection of a linkage of the four dominant mildew resistance genes “M1M2M3M4” in spinach from the wild type Spinacia turkestanica. Gartenbauwissenschaft 65:73–78
Hu J, Mou B, Bick BA (2007) Genetic diversity of 38 spinach (Spinacia oleracea L.) germplasm accessions and 10 commercial hybrids assessed by TRAP markers. Genet Resour Crop Evol 54:1667–1674
Iizuka M, Janick J (1971) Sex chromosome variation in Spinacia oleracea L. J Hered 62:349–352
International Spinach DataBase (2009) Centre for Genetic Resources. Wageningen, Netherlands. http://www.cgn.wur.nl/NL. Accessed 20 May 2009
Khattak JZK, Torp AM, Andersen SB (2006) A genetic linkage map of Spinacia oleracea and localization of a sex determination locus. Euphytica 148:311–318
Khattak JZK, Christiansen JL, Torp AM, Andersen SB (2007) Genic microsatellite markers for discrimination of spinach cultivars. Plant Breed 126:454–456
Mou BQ (2008a) Leafminer resistance in spinach. HortScience 43:1716–1719
Mou BQ (2008b) Evaluation of oxalate concentration in the U.S. spinach germplasm collection. HortScience 43:1690–1693
Mou BQ, Koike ST, du Toit LJ (2008) Screening for resistance to leaf spot diseases of spinach. HortScience 43:1706–1710
National Plant Germplasm System (2009) Agricultural Research System, Washington, DC, USA. http://www.ars-grin.gov/npgs. Accessed 20 May 2009
NCBI (2009) National Institute of Health. Rockville, MA, USA. http://www.ncbi.nlm.nih.gov. Accessed 20 May 2009
Plant Genetic Resources (2009) Bundesanstalt für Landwirtschaft und Ernährung. Bonn, Germany. http://www.genres.de. Accessed 20 May 2009
Ramanna MS (1976) Are there heteromorphic sex chromosomes in spinach (Spinacia oleracea L.)? Euphytica 25:277–284
Rubatzky VE, Yamaguchi M (1997) World vegetables: principles, production and nutritive values, 2nd edn. Chapman & Hall, New York, USA
Sauer JD (1993) Historical geography of crop plants: a select roster. CRC, Boca Raton, FL, USA
Schmitz-Linneweber C, Maier RM, Alcaraz J-P, Cottet A, herrmann RG, Mache R (2001) The plastid chromosome of spinach (Spinacia oleracea): complete nucleotide sequence and gene organization. Plant Mol Biol 45:307–315
Sneep J (1982) The domestication of spinach and the breeding history of its varieties. Euphytica (Suppl 2):27
Ul'janova TN (1977) Segetal and ruderal relatives of cultivated plants and the problems of their conservation and use: a Russian perspective. Genet Resour Crop Evol 44:5–8
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Andersen, S.B., Torp, A.M. (2011). Spinacia. In: Kole, C. (eds) Wild Crop Relatives: Genomic and Breeding Resources. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-20450-0_13
Download citation
DOI: https://doi.org/10.1007/978-3-642-20450-0_13
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-20449-4
Online ISBN: 978-3-642-20450-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)