Abstract
Desiccation tolerance (DT) is defined as the equilibration of protoplasmic water potential with that of the surrounding air (generally dry) without loss of viability upon rehydration. Vegetative DT is widespread among mosses and lichens, but is relatively rare in vascular plants (0.15%). Recent studies of selected resurrection species indicate that while resurrection plants might have evolved unique adaptive proteins, enzymes, and antioxidants, the molecular genetic basis of DT lies in the orchestration of transcriptional and posttranscriptional regulatory programs that operate during drying and rehydration. DT requires signaling pathways and regulatory mechanisms, aspects of which resemble developmental programs present in orthodox seeds, which result in the accumulation of oligosaccharides, stress adaptive proteins, antioxidants, reactive oxygen scavenging enzymes, as well as alterations in the composition and structure of membrane lipids. Functional genomics studies using transcriptome, proteome, and metabolome analyses are just beginning to unravel the system complexity required to orchestrate the metabolic symphony that is DT. The status of current gene discovery efforts is summarized along with major transcriptome technologies available currently to conduct desiccation sensitive versus tolerant species comparisons. These strategies, integrated with large-scale proteomic and metabolomic investigations currently in progress, promise to revolutionize our understanding of the mechanistic basis of desiccation tolerance in resurrection plants.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Relative Water Content
- Subtractive Suppression Hybridization
- Late Embryogenesis Abundant Protein
- Massively Parallel Signature Sequencing
- Sucrose Accumulation
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Desiccation tolerance (DT), the ability of cells to survive the air-dried state (equilibration to the water potential of the air which is generally low), is not uncommon in the plant kingdom. The vast majority of plants develop tissues that can withstand desiccation and for the most part these tissues are propagules: spores, pollen, and seeds. Vegetative DT is relatively common in the less complex plants that make up the algae, lichens, bryophytes, and perhaps the hornworts (Alpert 2006; Wood 2007), but is rare in the more complex vascular plants (Porembski and Barthlott 2000; Oliver et al. 2000; Proctor and Pence 2002). Vegetative DT was likely lost during the first steps in the evolution of tracheophytes (Oliver et al. 2000) around 415 million years ago (Soltis et al. 2002). Early mechanisms of vegetative desiccation tolerance have been postulated to be energetically expensive and perhaps structurally demanding (requiring simplicity) and thus were discarded in lieu of increased growth rates, structural and morphological complexity, and mechanisms for increased water conservation and efficient carbon fixation. The distribution of desiccation-tolerant plants is described in detail in Chap. 8.
Mechanistically, one can see trends within the evolutionary pathway for vegetative DT. DT has been postulated to have first evolved in spores of the land plants, and probably their charophyte alga relatives, prior to expression in vegetative tissues. The mechanism for tolerance in the early land plants resembles that seen in the modern day desiccation-tolerant bryophytes: a mechanism based upon constitutive cellular protection coupled with a rehydration-induced recovery process that involves cellular repair (Oliver et al. 2005). The available data suggest that the early reappearance of vegetative DT in vascular plants, using modern day ferns as a model, involved an environmentally inducible cellular protection mechanism as well as a rehydration-induced recovery process, indicating, at least conceptually, a simple progression from the mechanisms employed by bryophytes (Bewley et al. 1993). Seed plants evolved around 340 million years ago (Soltis et al. 2002), and it is at this time that DT appeared as a developmentally controlled program of what appears to be almost exclusively a cellular protection process (Oliver et al. 2000; Berjak et al. 2007; LePrince and Buitink 2007). In the angiosperms, the predominant mechanism for vegetative DT is the environmental induction of a cellular protection process with apparently little need for a rehydration-induced recovery or repair mechanisms (Ingram and Bartels 1996; Oliver et al. 2000; Hoekstra et al. 2001; Moore et al. 2009).
Although we have DT has played in the evolution of the land plants, much is yet to be uncovered. Our understanding of the underlying mechanisms, their components, the genes, gene networks, and the regulatory processes that control them is still in its infancy. The attainment of an understanding of vegetative DT mechanisms has profound importance, for elucidating not only the intricacies of plant evolution, but also the more practical aspects of the plant sciences that lie within the agricultural arena. Unraveling the genetic components that control and establish tolerance to severe dehydration will generate novel strategies for the improvement of drought tolerance in crops for which the toleration of dehydration is a major component. This is proving to be the case as discussed in a recent review by Moore et al. (2009) and as we allude to in this chapter.
The recent advances in systems biology and the tools that have been developed to look at biological processes on a large scale, the “omics” technologies, have not yet been fully employed in the study of DT. These technologies offer the most exciting possibilities for expanding our knowledge of DT in plants. Here, we review the gene discovery and transcriptomic studies that have been performed in resurrection species and discuss how future studies and improvements in these are expected to revolutionize our understanding of DT. Although less commonly performed, we also summarize recent proteomic and metabolite profiling studies using various resurrection plant species.
2 Targeted Gene Discovery
Early targeted gene isolation and expression studies in resurrection plants have been limited to a relatively few species including the moss Tortula ruralis (Scott and Oliver 1994; O’Mahony and Oliver 1999a, b; Wood et al. 2000; Zeng and Wood 2000; Chen et al. 2002; Zeng et al. 2002), the clubmosses Selaginella lepidophylla (Zentella et al. 1999) and Selaginella tamariscina (Liu et al. 2008), the dicotyledonous species Craterostigma plantagineum, which has received the most attention (Bartels et al. 1990, 1992; Iturriaga et al. 1992, 1996; Michel et al. 1994; Velasco et al. 1994, 1998; Bernacchia et al. 1995, 1996; Chandler and Bartels 1997; Furini et al. 1997; Ingram et al. 1997; Frank et al. 1998; Mariaux et al. 1998; Kleines et al. 1999; Kirch et al. 2001; Deng et al. 2002, 2006; Hilbricht et al. 2002; Phillips et al. 2002a, b; Rodrigo et al. 2004; Ditzer and Bartels 2006), and the monocotyledonous species Sporobulus stapfianus (Gaff et al. 1997; Clugston et al. 1998; Blomstedt et al. 1998a, b; O’Mahony and Oliver 1999a, b; Neale et al. 2000; Le et al. 2007), Xerophyta viscosa (Mundree et al. 2000; Mowla et al. 2002; Garwe et al. 2003, 2006; Lehner et al. 2008), Xerophyta humilis (Collett et al. 2003; Illing et al. 2005; Mulako et al. 2008), and Xerophyta villosa (Collett et al. 2004).
Other resurrection plant species have been characterized, but no gene sequence information has yet been reported for them including the moss Polytrichum formosum (Proctor et al. 2007a, b), the lichen Cladonia convoluta (Tuba et al. 1998), the clubmoss Selaginella bryopteris (Deeba et al. 2009), the fern Polypodium virginianum (Reynolds and Bewley 1993), the homoiochlorophyllous angiosperms Haberlea rhodopensis (Georgieva et al. 2007, 2009), Boea hygrometrica (Jiang et al. 2007), and Boea hygroscopica (Bochicchio et al. 1998), the angiosperms Ramonda serbica (Živković et al. 2005; Degl’Innocenti et al. 2008; Veljovic-Jovanovic et al. 2008), Craterostigma wilmsii (Cooper and Farrant 2002; Vicré et al. 2004), and Lindernia brevidens, a close relative of C. plantagineum (Smith-Espinoza et al. 2007; Phillips et al. 2008), the resurrection grass Eragrostis nindensis (Vander Willigen et al. 2003; Illing et al. 2005), and the dicotyledonous, woody, medicinal shrub, Myrothamnus flabellifolia (Moore et al. 2006, 2007).
3 Gene Discovery Using Expressed Sequence Tags
The most basic approach toward transcriptome analysis has been to collect expressed sequence tags (ESTs) using traditional Sanger sequencing. ESTs are typically automatically curated, single-read sequences of cDNA (complementary DNA molecules derived from reverse-transcribed cellular mRNA populations) (Rudd 2003). The relatively inexpensive nature of random sampling of cDNA libraries has made EST sequencing a very attractive and popular route for sampling transcriptomes (Rudd 2003) and also provides the raw material for the fabrication of cDNA or oligonucleotide microarrays (Alba et al. 2004). In the absence of complete genome sequence data, EST data can provide a low-cost, accessible way to efficiently sample the actively transcribed portions of a genome (Rudd 2003). Comparison of available EST data among diverse taxa of resurrection species or closely related “sister groups” also provides a means to provide novel insights into shared or unique molecular processes required for DT. Once whole-genome sequence information becomes available for a resurrection species, EST data will provide an invaluable resource for genome annotation.
Despite its vast utility, EST sequencing has a number of weaknesses. First, the sampling depth of most traditional EST projects is often limited to only a few hundred or thousand sequenced cDNAs. Although the cDNA library might accurately reflect the relative abundance of a particular transcript, some transcripts, such as low abundance, will be represented poorly in the cDNA library and genes, which are not expressed in the particular organ or tissue from which the library was prepared, will be absent (Rudd 2003). Enrichment strategies, such as normalization and subtraction, can partially overcome inadequate sampling (Bonaldo et al. 1996) and can dramatically improve the sequence diversity within a particular cDNA library by equalizing the relative occurrence of abundant versus rare transcripts. However, such strategies can never fully compensate for inadequate sampling leading to unreliable estimates of relative transcript abundance, particularly of low to moderately expressed mRNAs. Fortunately, more high-throughput sampling technologies have been developed that allow for more cost-effective, in-depth quantitative mRNA expression profiling (see Sect. 15.9).
Second, the sequence quality of cDNA-derived ESTs derived from Sanger sequencing is typically about 97% accurate when compared with genomic reference sequences considering all types of errors including insertions, deletions, and substitutions (Hillier et al. 1996). These errors can be the result of the poor fidelity of the reverse transcriptase and sequencing polymerase (Arezi and Hogrefe 2007) and base-calling accuracy (Li et al. 2004; Prosdocimi et al. 2007). Furthermore, if EST sequences are not carefully cleansed, they can contain xenocontaminants (e.g., vector, polylinker, and primer-adaptor sequences) or sequences from foreign organisms (e.g., E. coli and fungi) and abundant structural or regulatory RNAs (e.g., rRNAs and organellar transcripts). Typically, such contaminants can represent 1–3% of all ESTs (Lee and Shin 2009). In general, high-throughput sequencing strategies that provide highly redundant sampling technologies will result in far more accurate transcriptome sequencing data (Sect. 15.9).
Third, EST sequencing will often not provide a complete representation of gene models or full-length cDNAs from which they were derived. To overcome this limitation, full-length cDNA collections have been developed for many important model desiccation-sensitive species including Arabidopsis (Seki et al. 2002, 2004), soybean (Umezawa et al. 2008), and the model halophyte, Thellungiella halophila (Taji et al. 2008). Such full-length cDNA collections should also be developed as a key component for whole-genome annotation for selected resurrection species. In addition, full-length cDNA collections provide for the efficient exploration of plant gene function in heterologous hosts and plant improvement by using heterologous gene resources (Ichikawa et al. 2006; Kondou et al. 2009).
4 Transcriptome Analysis of Nonvascular Resurrection Plants
Targeted gene characterization studies have expanded to become large-scale EST sequencing efforts within a handful of resurrection model species (Table 15.1). Early small-scale EST sequencing (152 ESTs), from a cDNA library from polysomal mRNP fractions of desiccated leaves of the desiccation-tolerant bryophyte, T. ruralis, showed that 71% of the ESTs in the library represented novel sequences (Wood et al. 1999). More extensive EST sequencing of cDNA library from rehydrated, rapid-dried T. ruralis resulted in the characterization of ~10,368 ESTs representing 5,563 genes of which 2,242 (40.3%) were classified as unknowns, indicating the possibility that this species serves as a genetic reservoir for novel genes involved in stress tolerance (Oliver et al. 2004). Some of the most abundant transcripts in this cDNA library encode late embryogenesis abundant (LEA) proteins, suggesting that these proteins might play a role in the recovery from desiccation upon rehydration (Oliver et al. 2004). Comparison of these T. ruralis ESTs to available ESTs from the desiccation-sensitive moss, Physcomitrella patens, revealed that while T. ruralis is closely related, as both species are bryophytes, there is substantial phylogenetic distance between these two species (Oliver et al. 2004). These studies also led to the identification of desiccation- and rehydration-specific ubiquitin genes (O’Mahony and Oliver 1999a, b).
5 Transcriptome Analysis in Vascular Resurrection Plants
Large-scale gene discovery efforts in vascular resurrection plants have been limited to all but a few model species. One of the earliest reports of cloning and sequencing large numbers of cDNA clones from a resurrection species used differential, subtractive, or cold-plaque screening to isolate and characterize 200 cDNA clones from C. plantagineum leaves dried for 1 h or to complete dryness (Bockel et al. 1998). Recently, a very comprehensive transcriptome analysis has become available for cDNAs generated from different physiological stages of C. plantagineum (Rodriguez et al. 2010). Genes encoding abundant drought-induced genes correlated with DT or low abundance transcripts encoding gene products not previously associated with drought stress in S. stapfianus were isolated by differential screening (Blomstedt et al. 1998a) or by “cold-plaque” hybridization procedures (Neale et al. 2000a, b), respectively, suggesting that resurrection plants may possess unique genes and/or regulatory processes that confer DT.
In the new world lycophyte, S. lepidophylla, native to Mexico and the southwestern United States, 1,046 ESTs were obtained from a cDNA library constructed from plants undergoing desiccation representing 873 unique transcripts (Iturriaga et al. 2006). Comparison of the S. lepidophylla ESTs with 1,301 unigenes from Selaginella moellendorffii revealed that 63% of genes were unique to S. lepidophylla. In contrast to the 2,181 ESTs from S. moellendorffii, the desiccation-tolerant S. lepidophylla EST collection (Weng et al. 2005) contained a much greater relative percentage of stress response (i.e., LEA proteins), chaperones, and heat shock protein (HSP) ESTs. More importantly, analysis of the most abundant transcripts sampled by EST sequencing revealed that S. lepidophylla preferentially expressed genes whose primary assignable function is in stress response pathways (Iturriaga et al. 2006). Currently, a total of 8,355 ESTs have been sequenced using traditional Sanger sequencing from a cDNA library constructed from plants undergoing dehydration or rehydration at intervals of approximately 10% relative water content (RWC) loss or gain (Table 15.1).
Large-scale EST collections have also been generated from the poikilochlorophyllous, monocotyledonous, Xerophyta humilis, a resurrection species native of southern Africa (Collett et al. 2004). Four individually normalized cDNA libraries were generated from dehydrating leaf, dehydrating root, rehydrating leaf, and rehydrating root, respectively, at a range of seven different RWCs. Approximately 100 cDNA clones from each library were sequenced, resulting in an annotated set of 424 cDNAs of which 94% of clones were unique. On the basis of this limited evaluation, the cDNA libraries were judged to be normalized successfully. The libraries obtained from dehydrating root and leaf tissues were also found to be enriched for genes known to be associated with water-deficit, osmotic, cold, and pathogen stress responses, relative to the rehydration libraries (Collett et al. 2004). These same 424 cDNAs were then used to fabricate a printed, cDNA microarray, the results of which were also validated using reverse northern slot blot analysis. Between the two hybridization approaches, a total of 55 cDNA clones (13%) exhibited increased relative transcript abundance upon dehydration at either 26 or 9% RWC. Notably, cDNAs encoding LEA proteins, metallothioneins, an oleosin, and biosynthetic enzymes for the biosynthesis of polyols and raffinose family oligosaccharides were included in this group. A total of 79 cDNA clones (18.6%) showed decreased relative transcript abundance upon dehydration. Of these, 25% encoded cDNAs with functions related to photosynthesis and metabolism consistent with the notion that this poikilochlorophyllous species deactivates photosynthetic functions during dehydration. A follow-on study focused on the analysis of 16 cDNAs representing seven different LEA protein groups derived from this X. humilis EST collection (Illing et al. 2005). The relative mRNA expression of 13 of these cDNAs was validated by northern blot analysis and all exhibited similar expression profiles with peak expression occurring during desiccation (<65% RWC) and stably stored in dry (<6% RWC) leaves. In this same study, a comparison of LEA protein mRNA expression, antioxidant enzyme mRNA expression and activity patterns, and sucrose accumulation patterns among desiccation-sensitive and desiccation-tolerant species revealed discrete commonalities with desiccation-tolerant plants and the acquisition of DT in orthodox seeds. Namely, both DT vegetative tissues and seeds shared sucrose accumulation and the expression of a LEA-6 gene and a 1-cys-peroxiredoxin gene as common DT mechanisms (Illing et al. 2005).
6 Subtractive Suppression Hybridization
Most EST sequencing projects are conducted from randomly selected clones from cDNA libraries from a specific tissue or developmental stage. Such a random sampling approach is relatively inefficient because it does not selectively target a subpopulation of transcripts within a particular cDNA library. In contrast, the Subtractive Suppression Hybridization (SSH) approach allows cDNA libraries to be constructed so that they are specifically enriched for a subpopulation of transcripts (Diatchenko et al. 1996). SSH combines normalization and subtraction into a single process that involves the use of two hybridizations that normalize and enrich, respectively, for differentially expressed target genes in “tester” versus “driver” cDNA populations. Differentially expressed target cDNAs become preferentially amplified, whereas amplification of nontarget cDNA is suppressed during the PCR stages of the process. The SSH procedure reportedly achieves a greater than 1,000-fold enrichment of differentially expressed cDNAs (Diatchenko et al. 1996). This enrichment reduces the cloning of abundantly expressed genes common to control (driver) and treatment (tester) samples, a process that increases the probability of cloning differentially expressed genes of interest. Many genes of interest are expressed at relatively low abundance or in specific cell or tissue types or under a particular environmental condition, making selective enrichment by SSH an important and useful strategy.
The SSH approach was first used in the DT lycopod S. tamariscina to enrich for and identify genes whose mRNA expression is increased following 2–4 h dehydration of fronds (Liu et al. 2008). Out of more than 300 cDNA clones obtained, reverse northern blot analysis revealed that 96 were differentially expressed with 4 and 92 clones had reduced or increased relative mRNA abundance, respectively, following dehydration stress. The most abundantly expressed cDNA encoded an early light-inducible protein B (ELIPB) closely related to the homologous gene from T. ruralis (Zeng et al. 2002). A subset (11) of the increased abundance clones were also verified to be abscisic acid (ABA)-responsive genes adding evidence to the notion that ABA mediates dehydration stress responses in S. tamariscina (Liu et al. 2008).
The SSH approach was also used in the DT moss T. ruralis to enrich for and identify genes with low relative mRNA abundance that might be involved in slow drying or rehydration (Oliver et al. 2009). A total of 768 cDNAs were sequenced. Of these, 614 (80%) were unique demonstrating the effectiveness of the normalization strategy. Half of these cDNAs (298) were not previously obtained from an earlier EST sequencing effort that generated over 10,000 ESTs (Oliver et al. 2004). Furthermore, 59% of the SSH-derived EST contigs could not be annotated by similarity matches to public sequence databases, which might be expected of less well-expressed transcripts. Interestingly, the dehydration SSH EST collection only contained a single gene encoding a protein with a photosynthetic function. In contrast, functional categorization of EST collections from S. lepidophylla and X. humilis revealed that up to 17% of ESTs had functions related to photosynthesis. This bias against photosynthetic genes in the T. ruralis SSH EST collection might be an indication that the photosynthetic apparatus of this DT moss is relatively undamaged by desiccation and rehydration (Oliver et al. 2009) and that its protection constitutes a major component of DT in bryophytes (Oliver et al. 2005; Proctor et al. 2007a, b) (see Chap. 7). In contrast, the rehydration SSH EST collection was enriched for cDNAs that encode components of the protein synthetic apparatus and the translation process, consistent with the important role played by protein synthesis in the recovery of T. ruralis from desiccation following rehydration (Oliver et al. 2005, 2009).
A major motivation for the generation of EST collections is also to provide physical probe collections for cDNA microarrays. cDNA collections derived from two different SSH libraries enriched for sequestered transcripts within slow-dried T. ruralis gametophytes or transcripts translated after rehydration were used to develop a printed cDNA microarray containing 768 cDNAs (Oliver et al. 2009). Expression profiles within total RNA derived from hydrated gametophytes compared with rapid-dried rehydrated (RDR) and slow-dried (SD), respectively, or within polyA RNA from the polysomal fraction of hydrated gametophytes compared with the polysomal fractions of SD or RDR gametophytes revealed existence of several novel components of the DT mechanism including jasmonic acid signaling, proteosomal activation, and alternative splicing (Oliver et al. 2009).
7 cDNA-Amplified Fragment Length Polymorphism
Quantitative cDNA-Amplified Fragment Length Polymorphism (AFLP) is a powerful gene discovery technique for the identification of differentially expressed genes (Bachem et al. 1996; Breyne et al. 2003). The basic approach involves combinatorial restriction enzyme digestion of cDNA followed by selective amplifications of the resulting fragments to produce less complex subsets of transcript tags, which are then separated by electrophoresis on high-resolution polyacrylamide gels and visualized by autoradiography or an automated LI-COR system (Vuylsteke et al. 2007). The original cDNA-AFLP method has undergone several refinements to screen and visualize a majority of both abundant and weak differentially expressed genes on a genome-wide basis (Breyne et al. 2003). Despite the accessibility of cDNA-AFLP analysis for the typical laboratory, to our knowledge there have been no reports of its use on any resurrection species. However, the related technique of mRNA differential display visualized by silver staining was reported to examine mRNA expression patterns in the dicotyledonous, homoiochlorophyllous resurrection plant B. hygrometrica (Deng et al. 1999) or by radioactive labeling to isolate a desiccation-rehydration-responsive small GTP-binding protein gene (O’Mahony and Oliver 1999b).
8 Comparative Transcriptome Analysis in Resurrection Plants
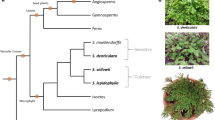
The identification of differentially expressed genes is only the beginning of the gene discovery process, especially in relation to understanding the underlying genetic components of DT. Desiccation and subsequent rehydration are extreme environmental insults to plant cells whether or not they are tolerant to them. A large proportion of the differential gene expression responses are likely to result from cellular injury. Injury might trigger the up- or downregulation of specific genes that are not involved in promoting adaptation to dehydration per se and are thus misleading when it comes to unraveling mechanisms of tolerance and perhaps of less use in drought improvement strategies. The only means by which the adaptive nature of a gene (and its expression) can be inferred is from a phylogenetic perspective, which entails species-to-species comparisons. There are two basic types of comparisons that one can employ to analyze changes in gene expression across species (Fig. 15.1): (1) ancestor–descendant comparisons where one attempts to reconstruct the evolutionary history of a gene and its association with a trait such as DT, and (2) sister-group contrasts where one compares the transcriptomes and gene expression profiles for two closely related species that differ in a critical phenotype, in this case DT. Direct comparisons between species-specific alterations in gene expression associated with DT have not been made using either strategy. The necessary transcriptomic resources for an ancestor–descendant comparison for desiccation-tolerant plants are not yet available, but their development is in progress in several labs. There have been some limited efforts to compare tolerant and nontolerant species, but generally they are of limited value because of the evolutionary distance between the two species within the contrast. Illing et al. (2005) compared expression profiles for a number of X. humilis genes with published profiles for Arabidopsis homologues and was able to infer that there are similarities between the response to desiccation in the resurrection plant to the acquisition of DT in Arabidopsis seeds (see below). This is an important observation and adds credence to some of the earlier hypotheses regarding the evolution of DT, but says little about the adaptive nature of the genes investigated. Oliver et al. (2004) compared the transcriptomes of rehydrating T. ruralis, a desiccation-tolerant bryophyte, to a publicly available transcriptome for P. patens, a desiccation-tolerant bryophyte, and could identify general categories of transcripts that might have indicated adaptive responses to desiccation, in particular those related to maintaining plastid integrity. However, this was a very limited comparison and the two mosses are not particularly closely related. A much closer species-specific transcriptome comparison was reported by Iturriaga et al. (2006), for S. lepidophylla and S. moellendorffii (see earlier discussion). This comparison requires more direct expression profiling data before any substantial hypotheses can be drawn.
9 High-Throughput Sequencing Approaches
Although EST sequencing and its variations described above have been used widely for gene discovery, the depth of coverage is typically limited for digital measurements of gene expression in which the relative abundance of each sequence tag is used to infer the relative abundance of its corresponding transcript present within a particular tissue or condition (Audic and Claverie 1997). Therefore, a number of methods have been developed to provide greater depth of sequencing of short tags, and thereby provide a more accurate, quantitative measure of the relative abundance of a transcript. High-throughput, sequence-based gene expression methods such as Serial Analysis of Gene Expression (SAGE) or Massively Parallel Signature Sequencing (MPSS) can be regarded as complementary to hybridization-based or “closed” transcriptome approaches provided that a sufficient number of biological replicates are performed with each platform (Liu et al. 2007; Vega-Sanchez et al. 2007) and generally provide similar results at high transcript abundance ranges (Nygaard et al. 2008). However, high-throughput sequencing-based methods exhibit increased sensitivity to the detection of low abundance transcripts, increased dynamic range, and have the inherent advantage of measuring new transcripts (Liu et al. 2007). Here, we provide a brief overview of the major transcriptome technologies available currently for genomic and transcriptomic analysis of resurrection plants.
9.1 Serial Analysis of Gene Expression
The SAGE method relies on Sanger sequencing of short (10–14 bp), concatenated cDNA tags derived from a cDNA region immediately 3′ to the 3′-most restriction site present in a double-stranded cDNA synthesized on magnetic beads (Velculescu et al. 1995). Although this original method represented an up to 30-fold improvement in tag coverage for the same number of Sanger sequencing reads, such short tags did not always permit unambiguous assignment of tags to corresponding transcripts. Therefore, improved versions of the original method called LongSAGE, which generates 17–19-mer tags (Saha et al. 2002), and SuperSAGE, which generates 26-mer tags (Matsumura et al. 2003, 2008), were developed subsequently. Despite the improved throughput and reduced cost compared to EST sequencing and its widespread use in diverse species (Anisimov 2008), no SAGE-related profiling studies using a resurrection or related species have been reported to date. Furthermore, newer, massively parallel, high-throughput sequencing technologies have recently eclipsed traditional SAGE approaches in both tag size and sampling depth.
9.2 Next-Generation Sequencing Technologies
Recent advances in high-throughput sequencing made possible by the commercial introduction of so-called second-generation sequencing instrumentation, which is capable of producing millions of DNA sequence reads in a single run, promise to rapidly transform the pace and scope of functional genomic analyses (Mardis 2008a, b; Simon et al. 2009). Such high-throughput sequencing systems promise to not only drive down the cost of gene discovery by transcriptome or genome sequencing, but also provide genome-wide sequence readouts as endpoints for a wide variety of applications including mutation or polymorphism discovery, comparative genomics, chromatin structural analysis, epigenetic regulation, and discovery of noncoding RNAs (Mardis 2008a, b). Application of these new “open” transcriptome technologies will have a profound influence on our future understanding of resurrection plant gene expression dynamics.
The first so-called next-generation Roche (454) Genome Sequencer (GS) FLX sequencer was commercialized in 2004 by 454 Life Sciences/Roche Applied Science (http://www/454/com) and is based on pyrosequencing (Ronaghi et al. 1998). This system is capable of producing 100–200 Mb during a typical run with an average read length of up to ~500 bp, making this platform well suited for de novo sequencing. (Margulies et al. 2005; Droege and Hill 2008). Given its attractiveness, Roche/454 Life Sciences pyrosequencing has recently been used to characterize a major fraction of the transcriptomes of S. lepidophylla and S. stapfianus using mixed cDNA libraries prepared from dehydrating and rehydrating tissues in an effort to capture the full repertoire of expressed genes in these species (Table 15.1).
Illumina’s SBS Genome Analyzer II system introduced in 2006 uses sequencing-by-synthesis (http://www.illumina.com) technology, which results in shorter (currently ~60 bases) but more abundant ESTs than the Roche/454 Life Sciences platform; however, longer read lengths (>100 bases) are expected to be possible in the near future (Mardis 2008a, b). A typical run will yield 30–50 million sequence tags, making this platform highly desirable for transcriptome profiling, SNP detection (genome resequencing), and genome-wide detection of protein–DNA interactions using ChIP sequencing in organisms with fully sequenced genomes (Bentley 2006; Smith et al. 2008; Simon et al. 2009).
The Applied Biosystems, Inc. Sequencing by Oligo Ligation and Detection (SOLiD) System (http://www.appliedbiosystems.com) generates an adapter-ligated fragment library using emulsion PCR to amplify the fragments on the surface of small magnetic beads similar to the Roche/454 Life Sciences system. However, after amplification, the beads are covalently attached to the surface of a specially treated glass slide, which is placed inside a fluidics cassette inside the sequencer (Mardis 2008a, b). A unique attribute of the SOLiD system is that it contains an inherent quality check feature, called “2 base encoding,” to identify miscalled bases from correct base calls during the data analysis step (Mardis 2008a, b; Simon et al. 2009). Each SOLiD run produces 2–3 Gb of sequence data. The major applications for SOLiD include very deep transcriptome profiling, SNP characterization, dissection of chromatin architecture (nucleosome positioning) using ChIP sequencing, and resequencing of bacterial genomes (Simon et al. 2009).
10 Protein Expression and Proteomics
Despite the enormous utility of transcriptome analyses, mRNA abundance does not always correlate well with the relative expression of the corresponding protein in eukaryotes (Gygi et al. 1999; Ideker et al. 2001). Posttranslational modifications such as phosphorylation, glycosylation, or N- or C-terminal processing events cannot be assessed using nucleic acid approaches, and small open reading frames are often difficult to confirm without direct sequence information from expressed proteins (Wasinger and Humphery-Smith 1998). Furthermore, proteomic methods are necessary to define the quantity, structure, and function of proteins within the systems biology framework (Phizicky et al. 2003).
The analysis of protein expression changes in resurrection plants has gained increasing attention recently, but also has a long history dating back to the 1980s. One of the earliest studies targeting S. lepidophylla investigated changes in the pattern of protein synthesis that occurred during rehydration (Eickmeier 1982). Cytosol-directed protein synthesis occurred within the first 12 h following hydration, whereas organelle-directed protein synthesis remained low until after 12 h of hydration and increased rapidly thereafter coincident with extensive thylakoid membrane proliferation and chloroplast polysome formation within this same time frame (Eickmeier 1982). Alterations in the pattern of protein synthesis were also characterized by comparisons between hydrated and rehydrated gametophytes (2 h of rehydration following rapid desiccation) in T. ruralis (Oliver 1991). This study was able to detect the termination or decrease in synthesis of 25 proteins (termed hydrins) and the initiation or substantial increase in the synthesis of 74 others (termed rehydrins). Using a timed labeling strategy, this study was also able to demonstrate that the controls over the reduction or increase in the synthesis of these two groups of proteins are not linked mechanistically. A certain amount of prior water loss was needed to fully activate the synthesis of rehydrins upon rehydration. Discrete protein expression changes have also been documented in the resurrection fern P. virginianum undergoing dehydration and rehydration (Reynolds and Bewley 1993). A 22 kDa early-light-induced protein (ELIP) was found to increase in abundance during desiccation and ABA treatment in the thylakoid membranes of C. plantegineum chloroplasts and was found to colocalize with the carotenoid zeaxanthin (Alamillo and Bartels 2001). This protein is thought to contribute to protection against photoinhibition caused by dehydration. Analysis of plastidic antenna protein complexes in the resurrection plant H. rhodopensis revealed movement of a portion of this complex from PSII to PSI during plant desiccation (Georgieva et al. 2009). This movement was also accompanied by a large increase in zeaxanthin accumulation. The expression of two small heat shock proteins was induced by dehydration and heat stress and exogenous ABA treatment and surprisingly in unstressed vegetative tissues (roots and lower parts of shoots) of C. plantegineum, which resembles the expression patterns typically found in desiccation-tolerant zygotic embryos and germinating seeds (Alamillo et al. 1995). Protein expression changes have also been compared in the desiccation-tolerant, resurrection grass S. stapfianus and the desiccation-sensitive grass S. pyramidalis (Kuang et al. 1995) and the desiccation-tolerant S. elongatus versus the desiccation-sensitive S. pyramidalis (Ghasempour and Kianian 2007). In vivo isotopic labeling of newly synthesized proteins was monitored following 6, 12, and 24 h rehydration in C. plantagineum by two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) analysis (Bernacchia et al. 1996). The expression pattern did not change appreciably after 12 h rehydration and none of the proteins were identified.
More recent and more comprehensive proteomic analyses have been performed in several resurrection species. 2D-PAGE analysis of the leaves of the small, dicotyledonous, homoiochlorophyllous resurrection plant B. hygrometrica reproducibly identified 223 proteins of which most (60%) were unchanged in abundance in dehydrated versus rehydrated leaves (Jiang et al. 2007). This species differs from many other resurrection species in that detached leaves retain the same ability of DT as that found in intact plants. Analysis of detached leaves avoids potential interference from developmental regulation and long-distance signaling events from other organs. In detached leaves, 35% of the proteins surveyed showed increased abundance upon dehydration, whereas only 5% showed increased abundance following rehydration (Jiang et al. 2007). Of the 14 dehydration-responsive proteins that were analyzed by mass spectrometry (MS), eight were identified as having functional roles in reactive oxygen scavenging, photosynthesis, and metabolism with the remainder being unknown or unidentified.
A more comprehensive proteomic analysis in the monocotyledonous, poikilochlorophyllous, resurrection plant X. viscosa surveyed approximately 430 protein spots (Ingle et al. 2007). During dehydration to 65 and 35% RWC, 20 proteins increased in abundance, 13 decreased in abundance, and 21 were specific to dehydration. Of these 54 proteins, 17 were identified by matrix-assisted laser desorption ionization (MALDI)–time of flight (TOF) MS. Proteins with increased abundance during leaf drying included an RNA-binding protein, chloroplast FtsH protease, desiccation-related proteins, and glycolytic and antioxidant enzymes. In contrast, proteins that declined in abundance included four components of PSII, indicating that desiccation involves the dismantling of the thylakoid membranes. In contrast, the abundance of these PSII proteins was largely maintained in the homoiochlorophyllous species C. plantagineum, which does not dismantle its thylakoid membranes upon drying (Ingle et al. 2007).
A recent study compared the protein expression profiles in dehydrated and rehydrated S. bryopteris fronds, a species in which its detached fronds have the ability to survive desiccation in the same way as the intact plant (Deeba et al. 2009). Analysis of detached fronds avoids potential interference from developmental regulation and long-distance signaling (Jiang et al. 2007). A total of about 250 spots were reproducibly detected by 2D-PAGE with 21 and 27 spots exhibiting increases or decreases in abundance, respectively. Among the 30 most differentially expressed proteins, 9 were identified by LC-MS/MS. Two of the proteins with increased relative abundance included a putative F-box/LRR repeat protein identified as an E3 ubiquitin ligase involved in proteasomal protein degradation and a putative DEAD-box ATP-dependent RNA helicase 5 protein whose expression is known to be increased by salt, dehydration, wounding, and low temperature stresses in Arabidopsis (Deeba et al. 2009).
In addition to protein analysis, changes in reversible protein phosphorylation patterns have also been studied in C. plantagineum undergoing dehydration stress (Röhrig et al. 2006). Desiccation-induced phosphoproteins were shown to accumulate in desiccated roots and leaves with two phosphoproteins, CDet11-24, a dehydration and ABA-responsive protein and CDeT6-19, a group 2 LEA protein, being especially abundant. Phosphorylation sites were mapped within predicted coiled-coil regions of CDet11-24, indicating that these phosphorylations might influence the stability of coiled-coil interactions with itself or other proteins (Röhrig et al. 2006). In a follow-up study, more than 20 desiccation-induced putative phosphoproteins were enriched using a modified metal oxide affinity chromatography approach, separated by 2D-PAGE, detected by Pro-Q Diamond staining, and identified by MALDI–TOF MS and MS/MS (Röhrig et al. 2008). Of the 20 putative phosphoproteins, 16 were suggested to be bona fide phosphoproteins from published evidence. More recently, protocols for the extraction and analysis of nuclear proteins from X. viscosa have also been optimized (Abdalla et al. 2009).
11 Metabolomics and Fluxomics
In addition to transcriptomic and proteomic analyses, the study of metabolite concentrations and how their abundance changes during the dehydration–rehydration process of resurrection plants is an absolutely essential part of understanding the biochemical and regulator aspects of DT. Quantitative knowledge of the complete set of metabolites and knowledge of which distinct metabolic processes are involved in the production and degradation or flux of discrete sets of metabolites can provide a better representation of the phenotype of an organism than other methods (Wiechert et al. 2007; Cascante and Marin 2008). Although large-scale metabolomics studies have been reported for Arabidopsis undergoing dehydration (Urano et al. 2009), similar studies have not yet been performed for any resurrection species; however, such studies are currently in progress for S. lepidophylla and S. stapfianus.
11.1 Sugar Metabolism
Despite the lack of large-scale metabolite studies, many targeted or small-scale investigations into the composition and abundance of key metabolites or changes in the activities of key enzymes have been reported. For example, desiccation-tolerant mosses such as T. ruralis maintain a high sucrose content (>100 mg g−1 DW) (Smirnoff 1992). Similarly, Selaginella species maintain high concentrations of both sucrose and the nonreducing, disaccharide, trehalose (White and Towers 1967; Adams et al. 1990; Iturriaga et al. 2000; Liu et al. 2008), whereas trehalose is only a minor component of accumulated sugars in many other DT species (Ghasempour et al. 1998). Either hydrated or desiccated fronds of S. lepidophylla or S. tamariscina maintain a high sugar content (>130 mg g−1 DW) (Adams et al. 1990; Iturriaga et al. 2000; Liu et al. 2008). In hydrated, growing fronds, trehalose content is high, but then declines 0.8-fold compared with desiccated fronds, whereas sucrose concentrations increase three-fold upon desiccation (Adams et al. 1990). The angiosperm, C. plantagineum, maintains a substantial sugar content (>400 mg g−1 DW) in both hydrated and desiccated leaves (Bianchi et al. 1991). The monosaccharide 2-octulose is the major soluble sugar in hydrated leaves (430 mg g−1 DW), whereas sucrose becomes the dominant sugar upon desiccation (374 mg g−1 DW) in C. plantagineum and C. wilmsii (Cooper and Farrant 2002). A similar, but less dynamic, interconversion of 2-octulose and sucrose has also been observed in L. brevidens (Phillips et al. 2008). In comparison, most orthodox seeds accumulate sucrose and large quantities of raffinose series oligosaccharides (RFOs) during the late maturation stages of seed development (Amuti and Pollard 1977). In the intermediate homoiochlorophyllous resurrection grass S. stapfianus, hexose sugars (glucose and fructose) increase along with sucrose during dehydration above 50% RWC. However, as RWC declines below 50% during the later stages of dehydration, both hexose sugars decline in abundance, whereas sucrose accumulation continues to increase (Ghasempour et al. 1998; Whittaker et al. 2001). The transient accumulation of hexose sugars might play an osmoregulatory role during the intermediate stages of dehydration (Ghasempour et al. 1998). A similar temporal accumulation pattern is observed in the monocotyledonous, poikilochlorophyllous X. viscosa, except that hexose sugars do not accumulate to large concentrations during the dehydration process (Whittaker et al. 2001). In addition to sucrose, RFOs, particularly raffinose, are prominent soluble carbohydrates that accumulate in dehydrated leaves of X. viscosa (Peters et al. 2007) as well as other resurrection species including C. plantagineum (Norwood et al. 2000), R. serbica (Živković et al. 2005), and various resurrection species where they accumulate to about 10% of sucrose content (Ghasempour et al. 1998). The sucrose-to-raffinose mass ratio was very low (1.3:1) in X. viscosa compared with other resurrection species where this ratio was much higher (5:1 to 10:1), suggesting that raffinose might also serve a dual role in stress protection and carbon storage (Peters et al. 2007). The myo-inositol 1-phosphate synthase (MIPS) gene encoding the enzyme catalyzing the committed step in the formation of myo-inositol, which is required for RFOs biosynthesis, has been cloned from X. viscosa (Lehner et al. 2008). The MIPS gene and protein increase in abundance following salinity stress and the loss of water to >65% RWC (Lehner et al. 2008). Sucrose accumulates in the drying leaves of the desiccation-tolerant E. nindensis, but not in those of the desiccation-sensitive Eragrostis species, providing compelling evidence for the importance of sucrose in conferring desiccation tolerance (Illing et al. 2005).
Soluble sugars, such as trehalose, sucrose, and other oligosaccharides, function as compatible solutes in water replacement, preferential exclusion of destabilizing molecules, and vitrification, an immobilized glassy state formed within the cytosol to prevent the occurrence of deleterious reactions in desiccated cells (Buitink et al. 1998; Hoekstra et al. 2001). Sucrose accumulates in all leaf cell types during dehydration (<56% RWC) in S. stapfianus as shown by in situ staining (Martinelli 2008). Sucrose has also been shown to lower membrane phase transition temperatures of dried membranes in vitro, which is beneficial during desiccation (Hoekstra and Golovina 1999). The membranes of desiccation-tolerant species also tend to contain higher unsaturated phospholipid concentrations, which help prevent phase transition of membranes (Hoekstra and Golovina 1999).
11.2 Enzyme Activities
Early studies examining the conservation of enzyme activities in dried tissues revealed a mean conservation of 94% in the bryophyte Acrocladium cuspidatum (Stewart and Lee 1972), with T. ruralis showing 70% conservation for Rubisco along with substantial conservation of several other enzymes (Sen Gupta 1977). Dried S. lepidophylla fronds retained an average of 75% activity of ten enzymes (Harten and Eickmeier 1986), whereas desiccation-tolerant angiosperm species, such as X. viscosa and M. flabellifolia, generally showed lower amounts of enzyme content. The conservation of these enzyme activities is thought to aid in the rapid resumption of metabolic activity in resurrection plants following rehydration.
During dehydration, hexokinase activities increase concomitantly with increased sucrose accumulation and decreased hexose sugar accumulation in both S. stapfianus and X. viscosa (Whittaker et al. 2001). Hexokinases catalyze the conversion of glucose, and with fructose at a lesser efficiency, to hexose monophosphates with the conversion of ATP to ADP for sucrose production. Leaf Glc-6-P, Fru-6-P, and ATP concentrations also increased in support of the observed increases in hexokinase activity. In contrast, fructokinase activity was unchanged during dehydration. The expression of genes encoding both sucrose-phosphate synthase (SPS) (Ingram et al. 1997) and sucrose synthase (SuSy) (Kleines et al. 1999) have been shown to increase during dehydration in C. plantagineum. SPS is considered the major enzyme of sucrose biosynthesis. The increase in SuSy has been proposed to be important for supplying carbon via sucrose catabolism to fuel glycolysis during dehydration and/or following rehydration (Kleines et al. 1999). SPS activity has also shown to increase during dehydration of S. stapfianus leaves coincident with sucrose accumulation (Whittaker et al. 2007). In addition to sugar metabolism, nitrogen and amino acid accumulation and metabolism might also play important roles in DT. In S. stapfianus, total amino acid content increased during the latter stages of water loss (>80% RWC) likely derived from insoluble protein breakdown (Whittaker et al. 2007). Specifically, the accumulation of large amounts of arginine and asparagine as nitrogen reserves might serve as essential nitrogen and carbon resources useful for successful rehydration (Martinelli et al. 2007).
11.3 Reactive Oxygen Scavenging
In addition to sugar metabolism, a critical stress adaptive mechanism for DT involves the detoxification of reactive oxygen intermediates (ROIs), whose production and accumulation increase as a result of environmental stresses including dehydration stress (Mittler 2002). Detoxification can be brought about by free-radical scavenging enzyme systems, which are active only under partially hydrated conditions, and by molecular antioxidants (e.g., ascorbate, glutathione, polyols, carbohydrates, proteins such as peroxiredoxin, and amphiphilic compounds, such as tocopherols, quinones, flavonoids, and phenolics), which can operate under dry conditions to alleviate oxidative stress damage (Vertucci and Farrant 1995). ROI scavenging enzymes such as ascorbate peroxidase (AP), glutathione reductase (GR), and superoxide dismutase (SOD) increased during early or late stages of drying in both C. wilmsii and X. viscosa (Sherwin and Farrant 1998). GR and SOD activities also increased during the early stages of rehydration, indicating that these enzymes likely afford critical free-radical protection until full rehydration and metabolic recovery has been achieved (Sherwin and Farrant 1998). However, antioxidant enzyme responses can vary depending on the species. GR activity increased upon drying in S. stapfianus (Sgherri et al. 1994a, b) and B. hygroscopica (Sgherri et al. 1994a, b), whereas AP activity declined or remained constant during dehydration. In T. ruralis, AP and catalase activity decreased during drying, whereas SOD activity remained constant (Seel et al. 1992). In C. plantagineum, only AP increased, whereas AP, GR, and SOD activity increased to varying extents following dehydration of M. flabellifolia and X. humilis (Farrant 2000). Comparison of ROI scavenging enzyme activities (e.g., AP, GR, and SOD) among three Eragrostis grass species showed that while these activities were elevated in all species during the early stages of dehydration, they remained elevated only in the DT species (E. nindensis) and declined in the closely related desiccation-sensitive species at RWC <70% (Illing et al. 2005). While these ROI scavenging enzymes are not considered unique to DT, their retention in this DT species is likely a consequence of the general enzyme protection mechanisms of resurrection species. However, some enzymes, such as 1-Cys peroxiredoxin, whose expression is induced upon drying in the resurrection plant X. viscosa (Mowla et al. 2002) and expressed during rehydration in the DT moss T. ruralis (Oliver 1996), might be considered an evolutionary prerequisite for DT (Illing et al. 2005). Polyphenol oxidase (PPO), which catalyzes the oxidation of mono- and o-diphenols to o-diquinones, showed increased protein abundance and enzyme activity in dehydrating leaves of B. hygroscopica (Jiang et al. 2007). PPO activity has also been shown to increase several-fold in leaves of R. serbica during dehydration stress (Veljovic-Jovanovic et al. 2008). Polyphenols are powerful detoxifiers of toxic reactive oxygen species (Rice-Evans et al. 1997), and might function as antioxidants during the first few hours of rehydration (Veljovic-Jovanovic et al. 2008).
In addition to enzymes, various metabolites afford protection to resurrection plants in the dried state. Anthocyanidins protect against light stress during desiccation by providing sunscreen to reduce damage to photosynthetic pigments chlorophyll and carotenoids and/or free-radical quenching. Anthocyanin content increased six-fold in the homoiochlorophyllous species C. wilmsii, but then declined rapidly following rehydration (Sherwin and Farrant 1998). Anthocyanin content also increases in the related species C. pumilum (Hoekstra et al. 2001). The resurrection fern Polypodium polypodioides (Muslin and Homann 1992) and the moss T. ruralis (Seel et al. 1992) are able to prevent and/or repair photooxidative damage of the photosynthetic apparatus despite the retention of chlorophyll in the dried state. In contrast, the poikilochlorophyllous species X. viscosa also displayed a sixfold increase in anthocyanin content upon drying, and this level was retained during rehydration (Sherwin and Farrant 1998). Because poikilochlorophyllous species lose their chlorophyll and dismantle their thylakoid membranes during dehydration, this retention of anthocyanin during rehydration might protect against photooxidation while the photosynthetic apparatus is being reassembled (Sherwin and Farrant 1998). Carotenoid content was retained during drying in C. wilmsii, but declined in X. viscosa consistent with the dismantling of its photosynthetic apparatus. An increase in reduced glutathione content, consistent with increased abundance of the GR enzyme itself and presumably the result of increased GR activity, has been reported in B. hygrometrica undergoing dehydration (Jiang et al. 2007). Increases in reduced glutathione content have also been reported in other resurrection species undergoing dehydration including B. hygrometrica (Navari-Izzo et al. 1997) and R. serbica (Augusti et al. 2001). A novel member of the vicinal oxygenase chelate (VOC) superfamily of metalloenzymes was cloned and characterized from X. humilis (Mulako et al. 2008). This gene is induced during dehydration in both vegetative and seed tissues of X. humilis and in mature, dry seeds of A. thaliana and is thought to play a role in the detoxification of methylglyoxal during desiccation (Mulako et al. 2008). Methylglyoxal is a cytotoxic by-product of glycolysis that accumulates following a variety of abiotic stresses.
Certain resurrection plants, such as the woody, medicinal plant, M. flabellifolia, can serve as sources for unique polyphenols including procyanidins (Anke et al. 2008). The predominant polyphenol in the leaves of this species is 3,4,5 tri-O-galloylquinic acid, a compound shown to stabilize artificial membranes (liposomes) against desiccation damage presumably by preserving the liquid crystalline phase of the membrane (Moore et al. 2005). This compound can also protect linoleic acid against free-radical-induced oxidation in vitro by itself becoming oxidized (Moore et al. 2005).
11.4 Membranes and Lipids
A common response to water deficit is a sharp reduction in lipid content, and similar reductions in lipids have been observed to occur in many resurrection plant species including R. serbica (Quartacci et al. 2002), S. stapfianus (Quartacci et al. 1997), and B. hygroscopica (Navari-Izzo et al. 1995, 2000). Such a reduction might aid membrane integrity. Another consequence of desiccation is a sharp increase in the free-sterol content, particularly cholesterol and cerebrosides, of the membranes in R. serbica (Quartacci et al. 2002). Sterol enrichment is thought to increase membrane rigidity, thereby reducing water permeation rates, which might play an important role in stress tolerance. Reductions in acyl chain unsaturation were also observed in R. serbica (Quartacci et al. 2002). Decreased fatty acid unsaturation results in decreased membrane fluidity, resulting in a more rigid, tighter lipid bilayer that might reduce solute leakage. However, such reductions in acyl chain unsaturation have not been observed after desiccation in other species including B. hygroscopica (Navari-Izzo et al. 1995), S. stapfianus (Quartacci et al. 1997), and S. tamariscina (Liu et al. 2008), indicating that species-specific adaptive mechanisms are likely to exist.
12 Signaling Pathways
The signaling mechanisms that coordinate the constitutive or inducible DT are not well understood (see also Chap. 13). Several studies have investigated the presence and alterations in endogenous concentrations of ABA in response to dehydration as evidence for its involvement in the maintenance or acquisition of DT (see Chaps. 9, 12, and 16). In the moss T. ruralis, ABA is not detectable upon drying and exogenous application of ABA does not appear to trigger the synthesis of mRNA or proteins (Bewley et al. 1993). Instead, bryophytes appear to rely on alterations in translational controls to mount a response to desiccation in contrast to the well-established signaling pathways associated with abiotic stress responses in angiosperms. The recent characterization of significantly accumulating transcripts in the polysomal mRNA pools of dehydrated T. ruralis invokes the likely participation of gamma-aminobutyric acid (GABA) and jasmonic acid signaling in DT (Oliver et al. 2009).
In vascular resurrection species, however, ABA appears to play a key role in the acquisition of DT. ABA concentrations increase threefold in dehydrated fronds of S. tamariscina relative to fully hydrated fronds, indicating that ABA probably plays a role in the acquisition of DT (Liu et al. 2008). A subset (11) of the increased abundance clones were also verified to be ABA-responsive genes verifying that ABA mediates dehydration stress responses in S. tamariscina (Liu et al. 2008). Further evidence for the production of ABA in a Sellaginella species was provided by the discovery of cDNAs encoding 9-cis-epoxycarotenoid dioxyenase, a key dehydration stress-inducible enzyme of the ABA biosynthetic pathway from S. lepidophylla (Iturriaga et al. 2006). In C. plantagineum, treatment of desiccation-sensitive callus with ABA renders it DT and induces a set of mRNAs comparable with that activated by whole plant dehydration (Bartels et al. 1990). ABA content increased in S. stapfianus leaves in response to dehydration and peaks between 40 and 15% RWC (Gaff and Loveys 1992). Exogenous application of ABA to leaves of S. stapfianus also results in the elevated mRNA accumulation of four dehydration-induced cDNA clones (Blomstedt et al. 1998b). However, exogenous ABA application cannot rescue the survival of detached leaves undergoing desiccation, so non-ABA-dependent processes are also likely to be involved.
T-DNA-mediated activation tagging screens of transgenic C. plantagineum callus leading to the creation of dominant mutants has led to the discovery of calli, which are capable of surviving desiccation without prior ABA treatment (Furini et al. 1997; Smith-Espinoza et al. 2005). The genes targeted by the T-DNA include two Craterostigma desiccation-tolerant (CDT) gene family members that encode naturally occurring siRNA molecules with features of a short interspersed element retrotransposon (SINE) that likely plays important roles in ABA signal transduction, because both activation mutant lines exhibit increased expression of ABA-induced LEA proteins (Phillips et al. 2007; Hilbricht et al. 2008). However, their involvement in ABA-independent DT pathways cannot be excluded (Furini et al. 1997; Smith-Espinoza et al. 2005).
In addition to ABA signaling, phospholipid-based signaling is also likely to play an important role in the acquisition of DT. In C. plantagineum, activity of phospholipase D (PLD), which catalyzes the hydrolysis of phosphatidylcholine and other phospholipids to phosphatidic acid (PA), which in turn regulates protein kinases or small GTP-binding proteins, is induced within minutes of dehydration and is not induced by exogenous ABA treatment (Frank et al. 2000). One member of the PLD family (CpPLD-1) in C. plantagineum is constitutively expressed, whereas the other (CpPLD-2) is responsive to dehydration and ABA (Frank et al. 2000). Furthermore, upon water-deficit stress in C. plantagineum, PLD and diacylglycerol kinase (DAG kinase) activities can lead to the increased accumulation of PA and diacylglycerol pyrophosphate (DGPP), which itself can serve as a second messenger (Munnik et al. 2000). The recent identification of multiple, rehydration-reversible phosphorylation events of a functionally diverse set of C. plantagineum proteins triggered by dehydration reinforces the importance of protein kinase and protein phosphatase activity in metabolic regulation during the dehydration/rehydration cycle (Röhrig et al. 2008). Furthermore, the identification of EBP1, a key regulator of cell growth and differentiation in plants as a phosphoprotein (Horváth et al. 2006), or 14-3-3 GF14 omega protein, which undergoes phosphorylation only during rehydration and might be involved in cell cycle checkpoint regulation (Sorrell et al. 2003), implicates their involvement in critical signaling and regulatory events required for DT.
13 Developmental Pathways of Seeds and DT Vegetative Tissues
Given the similarities of DT in orthodox seeds and DT in the vegetative tissues of resurrection plants (Illing et al. 2005), one might postulate that DT is simply a reiteration of seed desiccation. The molecular networks that control seed maturation in Arabidopsis during ripening, dormancy, and germination have been well characterized (Holdsworth et al. 2007, 2008). Therefore, one might expect that a comparison of the molecular networks in both seed dormancy and DT in a phylogenetic context would reveal evolutionarily conserved control pathways/networks that are common to both networks. Ongoing integrated transcriptomic, proteomic, and metabolomic studies are expected to result in characterizing these molecular networks in T. ruralis, S. lepidophylla, C. plantagineum, and S. stapfianus in the near future (see also Chap. 16). However, given that very different developmental and tissue-specific controls likely exist between the two systems, one might also expect fundamental differences to exist. One striking example of these differences is illustrated by a recent study in which a seed-specific transcription factor from sunflower, HaHSFA9, was constitutively expressed in transgenic tobacco (Prieto-Dapena et al. 2008). Overexpression of this transcription factor conferred improved (~40%) survival to 3-week-old seedling to severe (−40 MPa) dehydration. The overexpression of HaHSFA9 was correlated with the ectopic expression of small, seed-specific heat shock proteins, but neither LEA protein expression, nor elevated levels of glucose, sucrose, trehalose, RFOs, or proline. The HSPs are thought to confer the observed severe dehydration tolerance by the expected chaperone functions of preventing stress-induced denaturation of protein structure or aggregation of proteins as well as protecting membrane integrity (Prieto-Dapena et al. 2008). While the levels of dehydration tolerance are not like those exhibited by resurrection plants (e.g., −50 to > −100 MPa), they are better than all previous reports of engineered dehydration tolerance in sensitive species in the literature. Therefore, additional regulators of genes encoding different desiccation protectants, such as LEA proteins, are likely to exist that further contribute to the severe DT of orthodox seeds and resurrection plants. An example of other possible components that contribute to DT in seeds, and possibly in resurrection plants, is group 1 LEA proteins. Loss of these proteins in an Arabidopsis T-DNA knockout line caused seeds to dry out more rapidly than wild-type seeds resulting in more rapid acquisition of DT (Manfre et al. 2008). In addition to transcriptional regulators, components of signaling pathways can also be expected to play critical roles in seed development as well as DT. For example, a triple T-DNA knockout of three SNF1 (sucrose nonfermenting 1)-related protein kinases (SnRKs), SnRK2.2, SnRK2.3, and SnRK2.6, resulted in severe defects during seed development and seed dormancy by disruption of ABA signaling pathways via control of gene expression programs through the phosphorylation of ABI5 and other transcription factors (Nakashima et al. 2009).
14 Conclusion
The last decade has resulted in a dramatic increase in the volume of gene discovery efforts and large-scale transcriptomic and proteomic studies performed in a variety of model resurrection species. To date, the vast majority of transcript data have been obtained using traditional Sanger sequencing of ESTs from only a very limited number of varieties and species. However, transcriptome analysis in resurrection species is rapidly entering a new phase of discovery that will embrace the use of a variety of next-generation sequencing platforms. In the near future, large amounts of gene sequence information and transcriptome-scale mRNA expression data will be available using one or more of these platforms, resulting in the rapid characterization of key signaling and regulatory components required for the control of the dehydration/rehydration cycle of DT. Integration of this information with other “omics” datasets including proteomics and metabolomics from closely related species that are desiccation-sensitive versus desiccation-tolerant will help to elucidate the discrete differences in gene content, signaling, and regulatory mechanisms that are responsible for the development of DT. Such information will also inform the development of novel strategies to engineer improvements in dehydration tolerance in crops.
References
Abdalla K, Thomson J, Rafudeen M (2009) Protocols for nuclei isolation and nuclear protein extraction from the resurrection plant Xerophyta viscosa for proteomic studies. Anal Biochem 384:365–367
Adams R, Kendall E, Kartha K (1990) Comparison of free sugars in growing and desiccated plants of Selaginella lepidophylla. Biochem Syst Ecol 18:107–110
Alamillo J, Bartels D (2001) Effects of desiccation on photosynthesis pigments and the ELIP-like dsp 22 protein complexes in the resurrection plant Craterostigma plantagineum. Plant Sci 160:1161–1170
Alamillo J, Almoguera C, Bartels D, Jordano J (1995) Constitutive expression of small heat shock proteins in vegetative tissues of the resurrection plant Craterostigma plantagineum. Plant Mol Biol 29:1093–1099
Alba R, Fei Z, Payton P, Liu Y, Moore S, Debbie P, Cohn J, D’Ascenzo M, Gordon J, Rose J, Martin G, Tanksley S, Bouzayen M, Jahn M, Giovannoni J (2004) ESTs, cDNA microarrays, and gene expression profiling: tools for dissecting plant physiology and development. Plant J 39:697–714
Alpert P (2006) Constraints of tolerance: why are desiccation-tolerant organisms so small or rare? J Exp Biol 209:1575–1584
Amuti K, Pollard C (1977) Soluble carbohydrates of dry and developing seeds. Phytochemistry 156:529–532
Anisimov S (2008) Serial Analysis of Gene Expression (SAGE): 13 years of application in research. Curr Pharm Biotechnol 9:338–350
Anke J, Petereit F, Engelhardt C, Hensel A (2008) Procyanidins from Myrothamnus flabellifolia. Nat Prod Res 22:1243–1254
Arezi B, Hogrefe H (2007) Escherichia coli DNA polymerase III epsilon subunit increases Moloney murine leukemia virus reverse transcriptase fidelity and accuracy of RT-PCR procedures. Anal Biochem 360:84–91
Audic S, Claverie J (1997) The significance of digital gene expression profiles. Genome Res 7:986–995
Augusti A, Scartazza A, Navari-Izzo F, Sgherri C, Stevenovic B, Brugnoli E (2001) Photosystem II photochemical efficiency, zeaxanthin, and antioxidant contents in the poikilohydric Ramonda serbica during dehydration and rehydration. Photosyn Res 67:79–88
Bachem C, van der Hoeven R, de Bruijn S, Vreugdenhil D, Zabeau M, Visser R (1996) Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J 9:745–753
Bartels D, Schneider K, Terstappen G, Piatkowski D, Salamini F (1990) Molecular cloning of abscisic acid-modulated genes which are induced during desiccation of the resurrection plant Craterostigma plantagineum. Planta 181:27–34
Bartels D, Hanke C, Schneider K, Michel D, Salamini F (1992) A desiccation-related Elip-like gene from the resurrection plant Craterostigma plantagineum is regulated by light and ABA. EMBO J 11:2771–2778
Bentley D (2006) Whole-genome re-sequencing. Curr Opin Genet Dev 16:545–552
Berjak P, Farrant J, Pammenter N (2007) Plant desiccation tolerance. In: Jenks M, Woods A (eds) Seed desiccation-tolerance mechanisms. Blackwell, Oxford, UK, pp 151–192
Bernacchia G, Schwall G, Lottspeich F, Salamini F, Bartels D (1995) The transketolase gene family of the resurrection plant Craterostigma plantagineum: differential expression during the rehydration phase. EMBO J 14:610–618
Bernacchia G, Salamini F, Bartels D (1996) Molecular characterization of the rehydration process in the resurrection plant Craterostigma plantagineum. Plant Physiol 111:1043–1050
Bewley J, Reynolds T, Oliver M (1993) Evolving strategies in the adaptation to desiccation. In: Close T, Bray E (eds) Plant responses to cellular dehydration during environmental stress, current topics in plant physiology, vol 10, Am Soc Plant Physiol Series. American Society of Plant Physiologists, Rockville, MD, pp 193–201
Bianchi G, Gamba A, Murelli C, Salamini F, Bartels D (1991) Novel carbohydrate metabolism in the resurrection plant Craterostigma plantagineum. Plant J 1:355–359
Blomstedt C, Gianello R, Gaff D, Hamill J, Neale A (1998a) Differential gene expression in desiccation-tolerant and desiccation-sensitive tissue of the resurrection grass, Sporobolus stapfianus. Aust J Plant Physiol 25:937–946
Blomstedt C, Gianello R, Hamill J, Neale A, Gaff D (1998b) Drought-stimulated genes correlated with desiccation tolerance of the resurrection grass Sporobolus stapfianus. Plant Growth Regul 24:153–161
Bochicchio A, Vazzana C, Puliga S, Alberti A, Cinganelli S, Vernieri P (1998) Moisture content of the dried leaf is critical to desiccation tolerance in detached leaves of the resurrection plant Boea hygroscopica. Plant Growth Regul 24:163–170
Bockel C, Salamini F, Bartels D (1998) Isolation and characterization of genes expressed during early events of the dehydration process in the resurrection plant Craterostigma plantagineum. J Plant Physiol 152:158–166
Bonaldo M, Lennon G, Soares M (1996) Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res 6:791–806
Breyne P, Dreesen R, Cannoot B, Rombaut D, Vandepoele K, Rombauts S, Vanderhaeghen R, Inzé D, Zabeau M (2003) Quantitative cDNA-AFLP analysis for genome-wide expression studies. Mol Genet Genomics 269:173–179
Buitink J, Mireilleet M, Claessens M, Marcus A, Hemminga A, Hoekstra F (1998) Influence of water content and temperature on molecular mobility and intracellular glasses in seeds and pollen. Plant Physiol 118:531–541
Cascante M, Marin S (2008) Metabolomics and fluxomics approaches. Essays Biochem 45:67–81
Chandler J, Bartels D (1997) Structure and function of the vp1 gene homologue from the resurrection plant Craterostigma plantagineum Hochst. Mol Gen Genet 256:539–546
Chen X, Kanokporn T, Zeng Q, Wilkins T, Wood A (2002) Characterization of the V-type H((+))−ATPase in the resurrection plant Tortula ruralis: accumulation and polysomal recruitment of the proteolipid c subunit in response to salt-stress. J Exp Bot 53:225–232
Clugston S, Daub E, Honek J (1998) Identification of glyoxalase I sequences in Brassica oleracea and Sporobolus stapfianus: evidence for gene duplication events. J Mol Evol 47:230–234
Collett H, Butowt R, Smith J, Farrant J, Illing N (2003) Photosynthetic genes are differentially transcribed during the dehydration-rehydration cycle in the resurrection plant, Xerophyta humilis. J Exp Bot 54:2593–2595
Collett H, Shen A, Gardner M, Farrant J, Denby K, Illing N (2004) Towards transcript profiling of desiccation tolerance in Xerophyta humilis: construction of a normalized 11 k X. humilis cDNA set and microarray expression analysis of 424 cDNAs in response to dehydration. Physiol Plant 122:39–53
Cooper K, Farrant J (2002) Recovery of the resurrection plant Craterostigma wilmsii from desiccation: protection versus repair. J Exp Bot 53:1805–1813
Deeba F, Pandey V, Pathre U, Kanojiya S (2009) Proteome analysis of detached fronds from a resurrection plant Selaginella bryopteris – response to dehydration and rehydration. J Proteomics Bioinform 2:108–116
Degl’Innocenti E, Guidi L, Stevanovic B, Navari F (2008) CO2 fixation and chlorophyll a fluorescence in leaves of Ramonda serbica during a dehydration-rehydration cycle. J Plant Physiol 165:723–733
Deng X, Hu Z, Wang H (1999) mRNA differential display visualized by silver staining tested on gene expression in resurrection plant Boea hygrometrica. Plant Mol Biol Rep 17:279
Deng X, Phillips J, Meijer A, Salamini F, Bartels D (2002) Characterization of five novel dehydration-responsive homeodomain leucine zipper genes from the resurrection plant Craterostigma plantagineum. Plant Mol Biol 49:601–610
Deng X, Phillips J, Bräutigam A, Engström P, Johannesson H, Ouwerkerk P, Ruberti I, Salinas J, Vera P, Iannacone R, Meijer A, Bartels D (2006) A homeodomain leucine zipper gene from Craterostigma plantagineum regulates abscisic acid responsive gene expression and physiological responses. Plant Mol Biol 61:469–489
Diatchenko L, Lau Y, Campbell A, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E, Siebert P (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA 93:6025–6030
Ditzer A, Bartels D (2006) Identification of a dehydration and ABA-responsive promoter regulon and isolation of corresponding DNA binding proteins for the group 4 LEA gene CpC2 from C. plantagineum. Plant Mol Biol 61:643–663
Droege M, Hill B (2008) The Genome Sequencer FLX System–longer reads, more applications, straight forward bioinformatics and more complete data sets. J Biotechnol 136:3–10
Eickmeier W (1982) Protein synthesis and photosynthetic recovery in the resurrection plant, Selaginella lepidophylla. Plant Physiol 69:135–138
Farrant J (2000) A comparison of mechanisms of desiccation tolerance among three angiosperm resurrection plant species. Plant Ecol 151:29–39
Frank W, Phillips J, Salamini F, Bartels D (1998) Two dehydration-inducible transcripts from the resurrection plant Craterostigma plantagineum encode interacting homeodomain-leucine zipper proteins. Plant J 15:413–421
Frank W, Munnik T, Kerkmann K, Salamini F, Bartels D (2000) Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. Plant Cell 12:111–124
Furini A, Koncz C, Salamini F, Bartels D (1997) High level transcription of a member of a repeated gene family confers dehydration tolerance to callus tissue of Craterostigma plantagineum. EMBO J 16:3599–3608
Gaff D, Loveys B (1992) Abscisic acid levels in drying plants of a resurrection grass. Trans Malays Soc Plant Physiol 3:286–287
Gaff D, Bartels D, Gaff J (1997) Changes in gene expression during drying in a desiccation-tolerant grass Sporobolus stapfianus and a desiccation-sensitive grass Sporobolus pyramidalis. Aust J Plant Physiol 24:617–622
Garwe D, Thomson J, Mundree S (2003) Molecular characterization of XVSAP1, a stress-responsive gene from the resurrection plant Xerophyta viscosa Baker. J Exp Bot 54:191–201
Garwe D, Thomson J, Mundree S (2006) XVSAP1 from Xerophyta viscosa improves osmotic-, salinity- and high-temperature-stress tolerance in Arabidopsis. Biotechnol J 1:1137–1146
Georgieva K, Szigeti Z, Sarvari E, Gaspar L, Maslenkova L, Peeva V, Peli E, Tuba Z (2007) Photosynthetic activity of homoiochlorophyllous desiccation tolerant plant Haberlea rhodopensis during dehydration and rehydration. Planta 225:955–964
Georgieva K, Röding A, Büchel C (2009) Changes in some thylakoid membrane proteins and pigments upon desiccation of the resurrection plant Haberlea rhodopensis. J Plant Physiol 166(14):1520–1528
Ghasempour H, Kianian J (2007) The study of desiccation-tolerance in drying leaves of the desiccation-tolerant grass Sporobolus elongatus and the desiccation-sensitive grass Sporobolus pyramidalis. Pak J Biol Sci 10:797–801
Ghasempour H, Gaff D, Williams R, BGianellow R (1998) Contents of sugars in leaves of drying desiccation tolerant flowering plants, particularly grasses. Plant Growth Regul 24:185–191
Gygi S, Rochon Y, Fransz B, Aebersold R (1999) Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19:1720–1730
Harten J, Eickmeier W (1986) Enzyme dynamics of the resurrection plant Selaginella lepidophylla (Hook. & Grev.) spring during rehydration. Plant Physiol 82:61–64
Hilbricht T, Salamini F, Bartels D (2002) CpR18, a novel SAP-domain plant transcription factor, binds to a promoter region necessary for ABA mediated expression of the CDeT27-45 gene from the resurrection plant Craterostigma plantagineum Hochst. Plant J 31:293–303
Hilbricht T, Varotto S, Sgaramella V, Bartels D, Salamini F, Furini A (2008) Retrotransposons and siRNA have a role in the evolution of desiccation tolerance leading to resurrection of the plant Craterostigma plantagineum. New Phytol 179:877–887
Hillier L, Lennon G, Becker M, Bonaldo M, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, Hawkins M, Hultman M, Kucaba T, Lacy M, Le M, Le N, Mardis E, Moore B, Morris M, Parsons J, Prange C, Rifkin L, Rohlfing T, Schellenberg K, Bento Soares M, Tan F, Thierry-Meg J, Trevaskis E, Underwood K, Wohldman P, Waterston R, Wilson R, Marra M (1996) Generation and analysis of 280,000 human expressed sequence tags. Genome Res 6:807–828
Hoekstra F, Golovina E (1999) Membrane behavior during dehydration: implications for desiccation tolerance. Russ J Plant Physiol 46:295–306
Hoekstra F, Golvina E, Buitink J (2001) Mechanisms of plant desiccation tolerance. Trends Plant Sci 6:431–438
Holdsworth M, Rinch-Savage W, Grappin P, Job D (2007) Post-genomics dissection of seed dormancy and germination. Trends Plant Sci 13:7–13
Holdsworth M, Bentsink L, Soppe W (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179:33–54
Horváth B, Magyar Z, Zhang Y, Hamburger A, Bakó L, Visser R, Bachem C, Bögre L (2006) EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J 25:4909–4920
Ichikawa T, Nakazawa M, Kawashima M, Iizumi H, Kuroda H, Kondou Y, Tsuhara Y, Suzuki K, Ishikawa A, Seki M, Fujita M, Motohashi R, Nagata N, Takagi T, Shinozaki K, Matsui M (2006) The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J 48:974–985
Ideker T, Thorsson V, Ranish J, Christmas R, Buhler J, Eng J, Bumgarner R, Goodlett D, Aebersold R, Hood L (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292:929–934
Illing N, Denby K, Collett H, Shen A, Farrant J (2005) The signature of seeds in resurrection plants: a molecular and physiological comparison of desiccation tolerance in seeds and vegetative tissues. Integr Comp Biol 45:771–787
Ingle R, Schmidt U, Farrant J, Thomson J, Mundree S (2007) Proteomic analysis of leaf proteins during dehydration of the resurrection plant Xerophyta viscosa. Plant Cell Environ 30:435–446
Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47:377–403
Ingram J, Chandler J, Gallagher L, Salamini F, Bartels D (1997) Analysis of cDNA clones encoding sucrose-phosphate synthase in relation to sugar interconversions associated with dehydration in the resurrection plant Craterostigma plantagineum Hochst. Plant Physiol 115:113–121
Iturriaga G, Schneider K, Salamini F, Bartels D (1992) Expression of desiccation-related proteins from the resurrection plant Craterostigma plantagineum in transgenic tobacco. Plant Mol Biol 20:555–558
Iturriaga G, Leyns L, Villegas A, Gharaibeh R, Salamini F, Bartels D (1996) A family of novel myb-related genes from the resurrection plant Craterostigma plantagineum are specifically expressed in callus and roots in response to ABA or desiccation. Plant Mol Biol 32:707–716
Iturriaga G, Gaff D, Zentella R (2000) New desiccation-tolerant plants, including a grass, in the central highlands of Mexico, accumulate trehalose. Aust J Bot 48:153–158
Iturriaga G, Cushman M, Cushman J (2006) An EST catalogue from the resurrection plant Selaginella lepidophylla reveals abiotic stress-adaptive genes. Plant Biol 170:1173–1184
Jiang G, Wang Z, Shang H, Yang W, Hu Z, Phillips J, Deng X (2007) Proteome analysis of leaves from the resurrection plant Boea hygrometrica in response to dehydration and rehydration. Planta 225:1405–1420
Kirch H, Nair A, Bartels D (2001) Novel ABA- and dehydration-inducible aldehyde dehydrogenase genes isolated from the resurrection plant Craterostigma plantagineum and Arabidopsis thaliana. Plant J 28:555–567
Kleines M, Elster R, Rodrigo M, Blervacq A, Salamini F, Bartels D (1999) Isolation and expression analysis of two stress-responsive sucrose-synthase genes from the resurrection plant Craterostigma plantagineum (Hochst.). Planta 209:13–24
Kondou Y, Higuchi M, Takahashi S, Sakurai T, Ichikawa T, Kuroda H, Yoshizumi T, Tsumoto Y, Horii Y, Kawashima M, Hasegawa Y, Kuriyama T, Matsui K, Kusano M, Albinsky D, Takahashi H, Nakamura Y, Suzuki M, Sakakibara H, Kojima M, Akiyama K, Kurotani A, Seki M, Fujita M, Enju A, Yokotani N, Saitou T, Ashidate K, Fujimoto N, Ishikawa Y, Mori Y, Nanba R, Takata K, Uno K, Sugano S, Natsuki J, Dubouzet J, Maeda S, Ohtake M, Mori M, Oda K, Takatsuji H, Hirochika H, Matsui M (2009) Systematic approaches to using the FOX hunting system to identify useful rice genes. Plant J 57(5):883–894
Kuang J, Gaff D, Gianello R, Blomstedt C, Neale A, Hamill J (1995) Desiccation-tolerant grass Sporobolus stapfianus and a desiccation-sensitive grass Sporobolus pyramidalis. Aust J Plant Physiol 22:1027–1034
Le T, Blomstedt C, Kuang J, Tenlen J, Gaff G, Hamill J, Neale A (2007) Desiccation-tolerance specific gene expression in leaf tissue of the resurrection plant Sporobolus stapfianus. Funct Plant Biol 34:589–600
Lee B, Shin G (2009) CleanEST: a database of cleansed EST libraries. Nucleic Acids Res 37:D686–D689
Lehner A, Chopera D, Peters S, Keller F, Mundree S, Thomson J, Farrant J (2008) Protection mechanisms in the resurrection plant Xerophyta viscosa: cloning, expression, characterisation and role of XvINO1, a gene coding for a myo-inositol 1-phosphate synthase. Funct Plant Biol 35:26–39
LePrince O, Buitink J (2007) The glassy state in dry seeds and pollen. In: Jenks M, Woods A (eds) Plant desiccation tolerance. Blackwell, Oxford, UK, pp 193–214
Li M, Nordborg M, Li L (2004) Adjust quality scores from alignment and improve sequencing accuracy. Nucleic Acids Res 32:5183–5191
Liu F, Jenssen T, Trimarchi J, Punzo C, Cepko C, Ohno-Machado L, Hovig E, Kuo W (2007) Comparison of hybridization-based and sequencing-based gene expression technologies on biological replicates. BMC Genomics 8:153
Liu M-S, Chien C-T, Lin T-P (2008) Constitutive components and induced gene expression are involved in the desiccation tolerance of Selaginella tamariscina. Plant Cell Physiol 49:653–663
Manfre A, LaHatte G, Climer C, Marcotte WJ (2008) Seed dehydration and the establishment of desiccation tolerance during seed maturation is altered in the Arabidopsis thaliana mutant atem6-1. Plant Cell Physiol 50:243–253
Mardis E (2008a) Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet 9:387–402
Mardis E (2008b) The impact of next-generation sequencing technology on genetics. Trends Genet 24:133–141
Margulies M, Egholm M, Altman W, Attiya S, Bader J, Bemben L, Berka J, Braverman M, Chen Y, Chen Z, Dewell S, Du L, Fierro J, Gomes X, Godwin B, He W, Helgesen S, Ho C, Irzyk G, Jando S, Alenquer M, Jarvie T, Jirage K, Kim J, Knight J, Lanza J, Leamon J, Lefkowitz S, Lei M, Li J, Lohman K, Lu H, Makhijani V, McDade K, McKenna M, Myers E, Nickerson E, Nobile J, Plant R, Puc B, Ronan M, Roth G, Sarkis G, Simons J, Simpson J, Srinivasan M, Tartaro K, Tomasz A, Vogt K, Volkmer G, Wang S, Wang Y, Weiner M, Yu P, Begley R, Rothberg J (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380
Mariaux J, Bockel C, Salamini F, Bartels D (1998) Desiccation- and abscisic acid-responsive genes encoding major intrinsic proteins (MIPs) from the resurrection plant Craterostigma plantagineum. Plant Mol Biol 38:1089–1099
Martinelli T (2008) In situ localization of glucose and sucrose in dehydrating leaves of Sporobolus stapfianus. J Plant Physiol 165:580–587
Martinelli T, Whittaker A, Bochicchio A, Vazzana C, Suzuki A, Masclaux-Daubresse C (2007) Amino acid pattern and glutamate metabolism during dehydration stress in the ‘resurrection’ plant Sporobolus stapfianus: a comparison between desiccation-sensitive and desiccation-tolerant leaves. J Exp Bot 58:3037–3046
Matsumura H, Reich S, Ito A, Saitoh H, Kamoun S, Winter P, Kahl G, Reuter M, Kruger D, Terauchi R (2003) Gene expression analysis of plant host-pathogen interactions by SuperSAGE. Proc Natl Acad Sci USA 100:15718–15723
Matsumura H, Krüger D, Kahl G, Terauchi R (2008) SuperSAGE: a modern platform for genome-wide quantitative transcript profiling. Curr Pharm Biotechnol 9:368–374
Michel D, Furini A, Salamini F, Bartels D (1994) Structure and regulation of an ABA- and desiccation-responsive gene from the resurrection plant Craterostigma plantagineum. Plant Mol Biol 24:549–560
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Moore J, Westall K, Ravenscroft N, Farrant J, Lindsey G, Brandt W (2005) The predominant polyphenol in the leaves of the resurrection plant Myrothamnus flabellifolius, 3, 4, 5 tri-O-galloylquinic acid, protects membranes against desiccation and free radical-induced oxidation. Biochem J 385:301–308
Moore J, Nguema-Ona E, Chevalier L, Lindsey G, Brandt W, Lerouge P, Farrant J, Driouich A (2006) Response of the leaf cell wall to desiccation in the resurrection plant Myrothamnus flabellifolius. Plant Physiol 141:651–662
Moore J, Lindsey G, Farrant J, Brandt W (2007) An overview of the biology of the desiccation-tolerant resurrection plant Myrothamnus flabellifolia. Ann Bot Lond 99:211–217
Moore J, Le N, Brandt W, Driouich A, Farrant J (2009) Towards a systems-based understanding of plant desiccation tolerance. Trends Plant Sci 14:110–117
Mowla S, Thomson J, Farrant J, Mundree S (2002) A novel stress-inducible antioxidant enzyme identified from the resurrection plant Xerophyta viscosa Baker. Planta 215:716–726
Mulako I, Farrant J, Collett H, Illing N (2008) Expression of Xhdsi-1VOC, a novel member of the vicinal oxygen chelate (VOC) metalloenzyme superfamily, is up-regulated in leaves and roots during desiccation in the resurrection plant Xerophyta humilis (Bak) Dur and Schinz. J Exp Bot 59:3885–3901
Mundree S, Whittaker A, Thomson J, Farrant J (2000) An aldose reductase homolog from the resurrection plant Xerophyta viscosa Baker. Planta 211:693–700
Munnik T, Meijer H, Ter Riet B, Hirt H, Frank W, Bartels D, Musgrave A (2000) Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J 22:147–154
Muslin E, Homann P (1992) Light as a hazard for the desiccation-resistant “resurrection” fern Polypodium polypodioides L. Plant Cell Enivon 15:81–89
Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50:1345–1363
Navari-Izzo F, Ricci F, Vazzana C, Quartacci M (1995) Unusual composition of thylakoid membranes of the resurrection plant Boea hygroscopica: changes in lipids upon dehydration and rehydration. Physiol Plant 94:135–142
Navari-Izzo F, Meneguzzo S, Loggini B, Vazzana C, Sgherri C (1997) The role of the glutathione system during dehyration of Boea hygroscopica. Physiol Plant 99:23–30
Navari-Izzo F, Quartacci M, Pinzino C, Rascio N, Vazzana C, Sgherri C (2000) Protein dynamics in thylakoids of the desiccation-tolerant plant Boea hygroscopica during dehydration and rehydration. Plant Physiol 124:1427–1436
Neale A, Blomstedt C, Bronson P, Le T-N, Guthridge K, Evans J, Gaff D, Hamill J (2000) The isolation of genes from the resurrection grass Sporobolus stapfianus which are induced during severe drought stress. Plant Cell Enivon 23:265–277
Norwood M, Truesdale M, Richter A, Scott P (2000) Photosynthetic carbohydrate metabolism in the resurrection plant Craterostigma plantagineum. J Exp Biol 51:159–165
Nygaard V, Liu F, Holden M, Kuo W, Trimarchi J, Ohno-Machado L, CL C, Frigessi A, Glad I, Wiel M, Hovig E, Lyng H (2008) Validation of oligoarrays for quantitative exploration of the transcriptome. BMC Genomics 9:258
O’Mahony P, Oliver M (1999a) The involvement of ubiquitin in vegetative desiccation tolerance. Plant Mol Biol 41:657–667
O'Mahony P, Oliver M (1999b) Characterization of a desiccation-responsive small GTP-binding protein (Rab2) from the desiccation-tolerant grass Sporobolus stapfianus. Plant Mol Biol 39:809–821
Oliver M (1991) Influence of protoplasmic water loss on the control of protein synthesis in the desiccation-tolerant moss Tortula ruralis: ramifications for a repair-based mechanism of desiccation tolerance. Plant Physiol 97:1501–1511
Oliver M (1996) Desiccation tolerance in vegetative plant cells. Physiol Plant 97:779–787
Oliver M, Tuba Z, Mishler B (2000) The evolution of vegetative desiccation tolerance in land plants. Plant Ecol 151:85–100
Oliver M, Dowd S, Zaragoza J, Mauget S, Payton P (2004) The rehydration transcriptome of the desiccation-tolerant bryophyte Tortula ruralis: transcript classification and analysis. BMC Genomics 5:89
Oliver M, Velten J, Mishler B (2005) Desiccation tolerance in bryophytes: a reflection of the primitive strategy for plant survival in dehydrating habitats? Integr Comp Biol 45:788–799
Oliver M, Hudgeons J, Dowd S, Payton P (2009) A combined subtractive suppression hybridization and expression profiling strategy to identify novel desiccation response transcripts from Tortula ruralis gametophytes. Physiol Plant 136:437–460
Peters S, Mundree S, Thomson J, Farrant J, Keller F (2007) Protection mechanisms in the resurrection plant Xerophyta viscosa (Baker): both sucrose and raffinose family oligosaccharides (RFOs) accumulate in leaves in response to water deficit. J Exp Bot 58:1947–1956
Phillips J, Hilbricht T, Salamini F, Bartels D (2002a) A novel abscisic acid- and dehydration-responsive gene family from the resurrection plant Craterostigma plantagineum encodes a plastid-targeted protein with DNA-binding activity. Planta 215:258–266
Phillips J, Oliver M, Bartels D (2002b) Molecular genetics of desiccation and tolerant systems. In: Black HWPM (ed) Desiccation and plant survival. CABI, Wallingford, UK
Phillips J, Dalmay T, Bartels D (2007) The role of small RNAs in abiotic stress. FEBS Lett 581:3592–3597
Phillips J, Fischer E, Baron M, van den Dries N, Facchinelli F, Kutzer M, Rahmanzadeh R, Remus D, Bartels D (2008) Lindernia brevidens: a novel desiccation-tolerant vascular plant, endemic to ancient tropical rainforests. Plant J 54:938–948
Phizicky E, Bastiaens P, Zhu H, Snyder M, Fields S (2003) Protein analysis on a proteomic scale. Nature 422:208–215
Porembski S, Barthlott W (2000) Granitic and gneissic outcrops (inselbergs) as center of diversity for desiccation-tolerant vascular plants. Plant Ecol 151:19–28
Prieto-Dapena P, Castaño R, Almoguera C, Jordano J (2008) The ectopic overexpression of a seed-specific transcription factor, HaHSFA9, confers tolerance to severe dehydration in vegetative organs. Plant J 54:1004–1014
Proctor M, Pence V (2002) Vegetative tissues: bryophytes, vascular resurrection plants and vegetative propagules. In: Black M, Pritchard H (eds) Desiccation and survival in plants: drying without dying. CABI, Oxford
Proctor M, Ligrone R, Duckett J (2007a) Desiccation tolerance in the moss Polytrichum formosum: physiological and fine-structural changes during desiccation and recovery. Ann Bot Lond 99:75–93
Proctor M, Oliver M, Wood A, Alpert P, Stark L, Cleavitt N, Mishler B (2007b) Desiccation-tolerance in bryophytes: a review. Bryologist 110:595–621
Prosdocimi F, Lopes D, Peixoto F, Mourão M, Pacífico L, Ribeiro R, Ortega J (2007) Effects of sample re-sequencing and trimming on the quality and size of assembled consensus sequences. Genet Mol Res 6:756–765
Quartacci M, Forli M, Rascio N, Dalla Vecchia F, Bochicchio A, Navari-Izzo F (1997) Desiccation-tolerant Sporobolus stapfianus: lipid composition and cellular ultrastructure during dehydration and rehydration. J Exp Bot 48:1269–1279
Quartacci M, Glisić O, Stevanović B, Navari-Izzo F (2002) Plasma membrane lipids in the resurrection plant Ramonda serbica following dehydration and rehydration. J Exp Bot 53:2159–2166
Reynolds T, Bewley J (1993) Characterization of protein synthetic changes in a desiccation-tolerant fern, Polypodium virginianum. Comparison of the effects of drying, rehydration and abscisic acid. J Exp Bot 44:921–928
Rice-Evans C, Miller N, Paganga G (1997) Antioxidant properties of phenolic compounds. Trends Plant Sci 2:152–159
Rodrigo M, Bockel C, Blervacq A, Bartels D (2004) The novel gene CpEdi-9 from the resurrection plant C. plantagineum encodes a hydrophilic protein and is expressed in mature seeds as well as in response to dehydration in leaf phloem tissues. Planta 219:579–589
Rodriguez M, Edsgard D, Hussain SS, Alquezar A, Rasmussen M, Gilbert T, Nielsen B, Bartels D, Mundy J (2010) Transcriptomes of the desiccation tolerant resurrection plant Craterostigma plantagineum. Plant J 63:212–228
Röhrig H, Schmidt J, Colby T, Bräutigam A, Hufnagel P, Bartels D (2006) Desiccation of the resurrection plant Craterostigma plantagineum induces dynamic changes in protein phosphorylation. Plant Cell Environ 29:1606–1617
Röhrig H, Colby T, Schmidt J, Harzen A, Facchinelli F, Bartels D (2008) Analysis of desiccation-induced candidate phosphoproteins from Craterostigma plantagineum isolated with a modified metal oxide affinity chromatography procedure. Proteomics 8:3548–3560
Ronaghi M, Uhlén M, Nyrén P (1998) A sequencing method based on real-time pyrophosphate. Science 281:363–365
Rudd S (2003) Expressed sequence tags: alternative or complement to whole genome sequences? Trends Plant Sci 8:321–329
Saha S, Sparks A, Rago C, Akmaev V, Wang C, Vogelstein B, Kinzler K, Velculescu V (2002) Using the transcriptome to annotate the genome. Nat Biotechnol 20:508–512
Scott H, Oliver M (1994) Accumulation and polysomal recruitment of transcripts in response to desiccation and rehydration of the moss Tortula ruralis. J Exp Bot 45:577–583
Seel W, Hendry G, Lee J (1992) Effects of desiccation on some activated oxygen processing enzymes and antioxidants in mosses. J Exp Bot 43:1031–1037
Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, Muramatsu M, Hayashizaki Y, Kawai J, Carninci P, Itoh M, Ishii Y, Arakawa T, Shibata K, Shinagawa A, Shinozaki K (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296:141–145
Seki M, Satou M, Sakurai T, Akiyama K, Iida K, Ishida J, Nakajima M, Enju A, Narusaka M, Fujita M, Oono Y, Kamei A, Yamaguchi-Shinozaki K, Shinozaki K (2004) RIKEN Arabidopsis full-length (RAFL) cDNA and its applications for expression profiling under abiotic stress conditions. J Exp Bot 55:213–223
Sen Gupta A (1977) Non-autotrophic CO2 fixation by mosses. University of Calgary, Calgary
Sgherri C, Loggini B, Bochicchio A, Navari-Izzo F (1994a) Antioxidant system in Boea hygroscopica: changes in response to desiccation and rehydration. Phytochemistry 35:377–381
Sgherri C, Loggini B, Puliga S, Navari-Izzo F (1994b) Antioxidant system in Sporobolus stapfianus: changes in response to desiccation and rehydration. Phytochemistry 35:561–565
Sherwin H, Farrant J (1998) Protection mechanisms against excess light in the resurrection plants Craterostigma wilmsii and Xerophyta viscosa. Plant Growth Regul 24:203–210
Simon S, Zhai J, Nandety R, McCormick K, Zeng J, Mejia D, Meyers B (2009) Short-read sequencing technologies for transcriptional analyses. Annu Rev Plant Biol 60:305–333
Smirnoff N (1992) The carbohydrates of bryophytes in relation to desiccation-tolerance. J Bryol 17:185–191
Smith A, Xuan Z, Zhang M (2008) Using quality scores and longer reads improves accuracy of Solexa read mapping. BMC Bioinform 9:128
Smith-Espinoza C, Phillips J, Salamini F, Bartels D (2005) Identification of further Craterostigma plantagineum cdt mutants affected in abscisic acid mediated desiccation tolerance. Mol Genet Genomics 274:364–372
Smith-Espinoza C, Bartels D, Phillips J (2007) Analysis of a LEA gene promoter via Agrobacterium-mediated transformation of the desiccation tolerant plant Lindernia brevidens. Plant Cell Rep 26:1681–1688
Soltis P, Soltis D, Savolainen V, Crane P, Barraclough T (2002) Rate heterogeneity among lineages of tracheophytes: integration of molecular and fossil data and evidence for molecular living fossils. Proc Natl Acad Sci USA 99:4430–4435
Sorrell D, Marchbank A, Chrimes D, Dickinson J, Rogers H, Francis D, Grierson C, Halford N (2003) The Arabidopsis 14-3-3 protein, GF14omega, binds to the Schizosaccharomyces pombe Cdc25 phosphatase and rescues checkpoint defects in the rad24- mutant. Planta 218:50–57
Stewart G, Lee J (1972) Desiccation injury in mosses II: the effects of moisture stress on enzyme levels. New Phytol 71:461–466
Taji T, Sakurai T, Mochida K, Ishiwata A, Kurotani A, Totoki Y, Toyoda A, Sakaki Y, Seki M, Ono H, Sakata Y, Tanaka S, Shinozaki K (2008) Large-scale collection and annotation of full-length enriched cDNAs from a model halophyte, Thellungiella halophila. BMC Plant Biol 8:115
Tuba Z, Proctor M, Csintalan Z (1998) Ecological responses of homoiochlorophyllous and poikilochlorophyllous desiccation tolerance plants: a comparison and an ecological perspective. Plant Growth Regul 24:211–217
Umezawa T, Sakurai T, Totoki Y, Toyoda A, Seki M, Ishiwata A, Akiyama K, Kurotani A, Yoshida T, Mochida K, Kasuga M, Todaka D, Maruyama K, Nakashima K, Enju A, Mizukado S, Ahmed S, Yoshiwara K, Harada K, Tsubokura Y, Hayashi M, Sato S, Ana T, Ishimoto M, Funatsuki H, Teraishi M, Osaki M, Shinano T, Akashi R, Sakaki Y, Yamaguch-Shinozaki K, Shinozaki K (2008) Sequencing and analysis of approximately 40,000 soybean cDNA clones from a full-length-enriched cDNA library. DNA Res 15:333–346
Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, Sakurai N, Suzuki H, Saito K, Shibata D, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2009) Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J 57:1065–1078
Vander Willigen C, Pammenter N, Jaffer M, Mundree S, Farrant J (2003) An ultrastructural study using anhydrous fixation of Eragrostis nindensis, a resurrection grass with both desiccation-tolerant and -sensitive tissues. Funct Plant Biol 30:281–290
Vega-Sanchez M, Gowda M, Wang G (2007) Tag-based approaches for deep transcriptome analysis in plants. Plant Sci 173:371–380
Velasco R, Salamini F, Bartels D (1994) Dehydration and ABA increase mRNA levels and enzyme activity of cytosolic GAPDH in the resurrection plant Craterostigma plantagineum. Plant Mol Biol 26:541–546
Velasco R, Salamini F, Bartels D (1998) Gene structure and expression analysis of the drought- and abscisic acid-responsive CDeT11-24 gene family from the resurrection plant Craterostigma plantagineum Hochst. Planta 204:459–471
Velculescu V, Zhang L, Vogelstein B, Kinzler K (1995) Serial analysis of gene expression. Science 270:484–487
Veljovic-Jovanovic S, Kukavica B, Navari-Izzo F (2008) Characterization of polyphenol oxidase changes induced by desiccation of Ramonda serbica leaves. Physiol Plant 132:407–416
Vertucci C, Farrant J (1995) Acquisition and loss of desiccation tolerance. In: Kigel J, Galili G (eds) Seed development and germination. Marcel Dekker, New York, pp 237–271
Vicré M, Lerouxel O, Farrant J, Lerouge P, Driouich A (2004) Composition and desiccation-induced alterations of the cell wall in the resurrection plant Craterostigma wilmsii. Physiol Plant 120:229–239
Vuylsteke M, Peleman J, van Eijk M (2007) AFLP-based transcript profiling (cDNA-AFLP) for genome-wide expression analysis. Nat Protoc 2:1399–1413
Wasinger V, Humphery-Smith I (1998) Small genes/gene-products in Escherichia coli K-12. FEMS Microbiol Lett 169:375–382
Weng J, Tanurdzic M, Chapple C (2005) Functional analysis and comparative genomics of expressed sequence tags from the lycophyte Selaginella moellendorffii. BMC Genomics 6:85–99
White E, Towers G (1967) Comparative biochemisty of the lycopods. Phytochemistry 6:663–667
Whittaker A, Bochicchio A, Vazzana C, Lindsey G, Farrant J (2001) Changes in leaf hexokinase activity and metabolite levels in response to drying in the desiccation-tolerant species Sporobolus stapfianus and Xerophyta viscosa. J Exp Bot 52:961–969
Whittaker A, Martinelli T, Farrant J, Bochicchio A, Vazzana C (2007) Sucrose phosphate synthase activity and the co-ordination of carbon partitioning during sucrose and amino acid accumulation in desiccation-tolerant leaf material of the C4 resurrection plant Sporobolus stapfianus during dehydration. J Exp Bot 58:3775–3787
Wiechert W, Schweissgut O, Takanaga H, Frommer W (2007) Fluxomics: mass spectrometry versus quantitative imaging. Curr Opin Plant Biol 10:323–330
Wood A (2007) New Frontiers in Bryology and Lichenology. The Nature and Distribution of Vegetative Desiccation-tolerance in Hornworts, Liverworts and Mosses. Bryologist 110:163–177
Wood A, Duff R, Oliver M (1999) Expressed sequence Tags (ESTs) from desiccated Tortula ruralis identify a large number of novel plant genes. Plant Cell Physiol 40:361–368
Wood A, Joel Duff R, Oliver M (2000) The translational apparatus of Tortula ruralis: polysomal retention of transcripts encoding the ribosomal proteins RPS14, RPS16 and RPL23 in desiccated and rehydrated gametophytes. J Exp Bot 51:1655–1662
Zeng Q, Wood A (2000) A cDNA encoding ribosomal protein RPL15 from the desiccation-tolerant bryophyte Tortula ruralis: mRNA transcripts are stably maintained in desiccated and rehydrated gametophytes. Biosci Biotechnol Biochem 64:2221–2224
Zeng Q, Chen X, Wood A (2002) Two early light-inducible protein (ELIP) cDNAs from the resurrection plant Tortula ruralis are differentially expressed in response to desiccation, rehydration, salinity, and high light. J Exp Bot 53:1197–1205
Zentella R, Mascorro-Gallardo J, Van Dijck P, Folch-Mallol J, Bonini B, Van Vaeck C, Gaxiola R, Covarrubias A, Nieto-Sotelo J, Thevelein J, Iturriaga G (1999) A Selaginella lepidophylla trehalose-6-phosphate synthase complements growth and stress-tolerance defects in a yeast tps1 mutant. Plant Physiol 119:1473–1482
Živković T, Quartacci M, Stevanović B, Marinone F, Navari-Izzo F (2005) Low-molecular weight substances in the poikilohydric plant Ramonda serbica during dehydration and rehydration. Plant Sci 168:105–111
Acknowledgments
This work was supported by grants from the United States Department of Agriculture (USDA) National Research Initiative (NRI) (CREES-NRI-2007-02007) to MJ and JCC, and the University of Nevada Agricultural Experiment Station. Support for the Nevada Proteomics Center was made possible by NIH Grant Number P20 RR-016464 from the INBRE-BRIN Program of the National Center for Research Resources and the NIH IDeA Network of Biomedical Research Excellence (INBRE, RR-03-008).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Cushman, J.C., Oliver, M.J. (2011). Understanding Vegetative Desiccation Tolerance Using Integrated Functional Genomics Approaches Within a Comparative Evolutionary Framework. In: Lüttge, U., Beck, E., Bartels, D. (eds) Plant Desiccation Tolerance. Ecological Studies, vol 215. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-19106-0_15
Download citation
DOI: https://doi.org/10.1007/978-3-642-19106-0_15
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-19105-3
Online ISBN: 978-3-642-19106-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)