Abstract
Theoretical biology is incomplete without a comprehensive theory of evolution, since evolution is at the core of biological thought. Evolution is visualized as a migration process in genotype or sequence space that is either an adaptive walk driven by some fitness gradient or a random walk in the absence of (sufficiently large) fitness differences. The Darwinian concept of natural selection consisting in the interplay of variation and selection is based on a dichotomy: All variations occur on genotypes whereas selection operates on phenotypes, and relations between genotypes and phenotypes, as encapsulated in a mapping from genotype space into phenotype space, are central to an understanding of evolution. Fitness is conceived as a function of the phenotype, represented by a second mapping from phenotype space into nonnegative real numbers. In the biology of organisms, genotype–phenotype maps are enormously complex and relevant information on them is exceedingly scarce. The situation is better in the case of viruses but so far only one example of a genotype–phenotype map, the mapping of RNA sequences into RNA secondary structures, has been investigated in sufficient detail. It provides direct information on RNA selection in vitro and test-tube evolution, and it is a basis for testing in silico evolution on a realistic fitness landscape. Most of the modeling efforts in theoretical and mathematical biology today are done by means of differential equations but stochastic effects are of undeniably great importance for evolution. Population sizes are much smaller than the numbers of genotypes constituting sequence space. Every mutant, after all, has to begin with a single copy. Evolution can be modeled by a chemical master equation, which (in principle) can be approximated by a stochastic differential equation. In addition, simulation tools are available that compute trajectories for master equations. The accessible population sizes in the range of \(10^7\le N\le 10^8\) molecules are commonly too small for problems in chemistry but sufficient for biology.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Mathematics and Biology
The beginning of modern science in the sixteenth century was initiated by the extremely fruitful marriage between physics and mathematics. Nobody has expressed the close relation between mathematics and physics more clearly than Galileo Galilei in his famous statement [1]: Philosophy (science) is written in this grand book, the universe, … . It is written in the language of mathematics, and its characters are triangles, circles and other geometric features … . Indeed, physics and mathematics have cross-fertilized each other from the beginnings of modern science until the present day. Theoretical physics and mathematical physics are highly respected disciplines and no physics journal will accept empirical observations without an attempt to bring it into a context that allows for quantification and interpretation by theory. General concepts and successful abstractions have a high reputation in physics and the reductionists’ program Footnote 1 is the accepted scientific approach towards complex systems . This view is common in almost all subdisciplines of contemporary physics and, in essence, is shared with chemistry and molecular biology.
Conventional biology, in this context, is very different: Great works of biology, such as Charles Darwin’s Origin of Species [2] or, in recent years, Ernst Mayr’s Growth of Biological Thought [3], do not contain a single mathematical expression; theoretical and mathematical biology had and still have a bad reputation among macroscopic biologists; special cases are preferred over generalizations, which are looked upon with scepticism; and holistic views are commonly more appreciated than reductionists’ explanations, whether or not they are in a position to provide insight into problems. A famous and unique exception among others is Charles Darwin’s theory of natural selection by reproduction and variation in finite populations. Although not cast in mathematical equations, the theory is based on a general concept whose plausibility is erected upon a wealth of collected and carefully interpreted empirical observations. Darwin’s strategy has something in common with the conventional mathematical approach based on observation, abstraction, conjecture, and proof: On different islands of the Galapagos archipelago Darwin observed similar-looking species in different habitats and concluded correctly that these different species are closely related and owe their existence to histories of adaptation to different environments on the individual islands. The occurrence of adaptations has been attributed to natural selection as a common mechanism through abstraction from specific cases. Darwin’s conjecture combines three facts known in his time:
-
1.
Multiplication : All organisms multiply by cell division, (parthenogenesis or sexual reproduction), multiplication is accompanied by inheritance – “progeny resembles parents”, and under the condition of unlimited resources multiplication results in exponential growth of population size.
-
2.
Variation : All natural populations show variance in phenotypic properties, either continuously varying features, such as body size, or discontinuously varying features, such as the number of limbs, the number of digits, color of flowers, skin patterns, or seed shapes, and it is straightforward to relate variation to inheritance.Footnote 2
-
3.
Selection : Exponential growth results in overpopulation of habitats,Footnote 3 only a small fraction of offspring can survive and have progeny of their own, and this stringent competition prevents less efficient variants from reproduction.
Taking together these three items and introducing the notion of fitness for the number of offspring that reach the age of fertility, the conjecture could be formulated in the following way:
Natural selection : In nonhomogeneous populations the frequencies of variants with fitness values below the population average will decrease, while those with fitness values above average will increase and consequently the population average itself will increase until it reaches the maximum value corresponding to a homogeneous population of the best adapted or fittest variant.
Darwin’s Origin of Species is an overwhelming collection of observations from nature, from animal breeders, and from nursery gardens that provide strong evidence for the correctness of Darwin’s conjecture. This enormous collection in a way is the empirical substitute for a mathematical proof.
Although Gregor Mendel analyzed his experiments on inheritance in peas by mathematical statistics and found thereby the explanatory regularities, mathematics did not become popular in biology. On the contrary, Mendel’s work was largely ignored by the biological community for more than 30 years. Then Mendel was rediscovered and genetics became an important discipline of biology. Population genetics was founded by the three scholars Ronald Fisher [4] , J.B.S. Haldane, [5] and Sewall Wright [6] . In the 1930s they succeeded in uniting Mendelian genetics and Darwin’s natural selection, and to cast evolution in a rigorous mathematical frame, but conventional geneticists and evolutionary biologists continued to fight until the completion of the synthetic theory almost 20 years later [3] .
Modeling in biology became an important tool for understanding complex dynamical phenomena. Representative for many other approaches we mention here only three: (i) Modeling of coevolution in a predator–prey system was introduced by Alfred Lotka [11] and Vito Volterra [12] by means of differential equations that were borrowed from chemical kinetics . In a way, they were the pioneers of theoretical ecology , which was developed by the brothers Howard and Eugene Odum [13] and became a respectable field of applied mathematics later [14]. (ii) A model for pattern formation based on the reaction–diffusion (partial differential) equation with a special chemical mechanism was suggested and analyzed by Alan Turing [15]. Twenty years later the Turing model was applied to biological morphogenesis [16, 17] and provided explanations for patterns formed during development [18, 19]. (iii) Based on experimental studies of nerve pulse propagation in the squid giant axon, Alan Hodgkin and Andrew Huxley formulated a mathematical model for nerve excitation and pulse propagation [20] that became the standard model for single nerve dynamics in neurobiology. They were both awarded the Nobel Prize in Medicine in 1963. A second breakthrough in understanding neutral systems came from modeling networks of neurons. John Hopfield conceived an exceedingly simple model of neurons in networks [21] that initiated a whole new area of scientific computing: computation with neutral networks , in particular modeling and optimization of complex systems. Despite these undeniable and apparent successes, the skepticism of biologists with respect to theory and mathematics nevertheless continued for almost the entire remainder of the twentieth century.
The advent of molecular biology in the 1950s brought biology closer to chemistry and physics, and changed the general understanding of nature in a dramatic way [22]. Inheritance received a profound basis in molecular genetics and reconstruction of phylogenies became possible through comparison of biopolymer sequences from present-day organisms. Structures of biomolecules at atomic resolution were determined by refined techniques from physical chemistry and they provided deep insights into biomolecular functions. Spectroscopic techniques, in particular nuclear magnetic resonance, require a solid background in mathematics and physics for conceiving and analyzing conclusive experiments. A novel era of biology was initiated in the 1970s when the highly efficient new methods for DNA sequencing developed by Walter Gilbert and Frederick Sanger became available [23, 24]. Sequencing whole genomes became technically within reach and financially affordable. The first two complete bacterial genomes were published in 1995 [25] and the following years saw a true explosion of sequencing data. High-throughput techniques using chip technology for genome-wide analysis of translation and transcription products known as proteomics and transcriptomics followed, and an amount of data was created that had never been seen before. In this context it is worth citing the Nobel laureate Sydney Brenner, [26] who made the following statement in 2002 to characterize the situation in molecular biology:
I was taught in the pre-genomic era to be a hunter. I learnt how to identify the wild beasts and how to go out, hunt them down and kill them. We are now, however, being urged to be gatherers. To collect everything lying about and put it into storehouses. Someday, it is assumed someone will come and sort through the storehouses, discard the junk and keep the rare finds. The only difficulty is how to recognize them.
Who else but a theorist should this “someone” be? The current development seems to indicate that “someday” is not too far away. The flood of data and the urgent need for a comprehensive theory have driven back the biologists’ aversion to computer science and mathematics. Modern genetics and genome analysis without bioinformatics are unthinkable, and understanding network dynamics without mathematics and computer modeling is impossible.
The new discipline of systems biology has the ambitious goal to find holistic descriptions for cells and organisms without giving up the roots in chemistry and physics. Although still in its infancy and falling into one trap after another, modeling in systems biology is progressing slowly towards larger and more detailed models for regulatory modules in cell biology. New techniques are being developed and applied. Examples are flux-balance analysis [27] and application of inverse methods [28], whereby the primary challenge is up-scaling to larger systems such as whole organisms. Recent advances in experimental evolution allow for an extension of detailed models to questions of evolution, which is of central importance in biology, as Theodosius Dobzhansky encapsulated in his famous sentence: “Nothing in biology makes sense except in the light of evolution” [29] . From a conceptional point of view, theoretical biology is in a better position than theoretical physics, where attempts at unification of two fundamental theories, quantum mechanics and relativity theory, have not been successful so far. Biology has one comprehensive theory, the theory of evolution, and present-day molecular biology is building the bridge to chemistry and physics. Lacking are a proper language and efficient techniques to handle the enormous complexity and to build proper models.
Fibonacci series, exponential functions, and limited resources. The Fibonacci series (black; upper plot) is embedded between two exponential functions in the range \(0 <i\le10\): \(n_{\mathrm{upper}}(t)=\exp\bigl(0.4453(t-1)\bigr)\) (red) and \(n_{\mathrm{lower}}(t)=\exp\bigl(0.5009(t-2)\bigr)\) (blue), where the time t is the continuous equivalent to the discrete (generation) index i. The lower plot compares the exponential function, \(y(t)=y_0\,\exp(r\,t)\) for unlimited growth (red; \(y_0=0.02\), \(r=0.1\)) with the normalized solution of the Verhulst equation (\(x(t)\), black; \(x_0=0.02\), \(r=0.1\), and \(C=1\) by definition)
2 Darwin’s Theory in Mathematical Language
If Charles Darwin had been a mathematician, how might he have formulated his theory of natural selection? Application of mathematics to problems in biology has a long history. The first example that is relevant to evolution dates back to medieval times. In the famous Liber Abaci written in the year 1202 by Leonardo Pisano, also known as Fibonacci (filius Bonacci), we find a counting example of the numbers of pairs of rabbits in subsequent time spans. Every adult pair is assumed to give birth to another pair, newborn rabbits have to wait one time interval before they become fertile adults. Starting from a single couple yields the following series:
Every number is the sum of its two precursors and the Fibonacci series is defined by the recursion
It is straightforward to show that the Fibonacci series can be approximated well by exponential functions as upper and lower limits (Fig. 2.1). The exponential function, however, was not known before the middle of the eighteenth century; it was introduced in the fundamental work of the Swiss mathematician Leonhard Euler [30]. Robert Malthus – although he lived 50 years later – still used a geometric progression, 2,4,8,16,… , for the unlimited growth of populations [10]. The consequences of unlimited growth for demography are disastrous and, as said, Malthus’s work was influential on Darwin’s thoughts.
A contemporary of Charles Darwin, the mathematician Pierre-François Verhulst [31], formulated a model based on differential equations combining exponential growth and limited resources (Fig. 2.1) :
with N(t) describing the number of individuals at time t, r being the Malthusian parameter, and C the carrying capacity of the ecosystem. Equation (2.2) consists of two terms: (i) the exponential growth term, rN, and (ii) the constraint to finite population size expressed by the term \(-rN^2/C\). In other words, the ecosystem can only support \(N=C\) individuals and \(\lim_{t\to\infty}N(t)=C\). The solution of the differential equation (2.2) is of the form
Here \(N_0=N(0)\) is the initial number of individuals. It is straightforward to normalize the variable to the carrying capacity, \(x(t)=N(t)/C\), yielding
with \(x_0=N_0/C\). It will turn out to be useful to write the term representing the constraint in the form \(N\,\phi(t)/C=x\,\phi(t)\). Then we obtain for the Verhulst equation
being the (mean) reproduction rate of the population.
Finally, we generalize to the evolution of n species or variantsFootnote 4 in the population \({\boldsymbol{\varXi}}=\{X_1,X_2,\hdots,X_n\}\). The numbers of individuals are now denoted by \([X_i]=N_i\) with \(\sum\nolimits_{i=1}^n N_i=N\) and the normalized variables \(x_i=N_i/N\) with \(\sum\nolimits_{i=1}^n x_i=1\). Each variant has its individual Malthus parameter or fitness value f i, and for the selection constraint leading to constant population size we find now \(\phi(t)=\sum\nolimits_{i=1}^n x_i\,f_i\), which is the mean reproduction rate of the entire population. The selection constraint \(\phi(t)\) can be used for modeling much more general situations than constant population size by means of the mean reproduction rate. As we shall see in Sect. 2.5, the proof for the occurrence of selection can be extended to very general selection constraints \(\phi(t)\) as long as the population size does not become zero, \(N>0\).
The kinetic differential equation in the multispecies case, denoted as the selection equation ,
can be solved exactly by the integrating factors transform ([32], pp. 322ff.)
Insertion into (2.4) yields
where we have used \(z_j(0)=x_j(0)\) and the condition \(\sum\nolimits_{i=1}^n x_i=1\). The solution finally is of the form
The interpretation is straightforward. The term with the largest fitness value, \(f_m=\max\{f_1,f_2,\hdots,f_n\}\), dominates the sum in the denominator after sufficiently long timeFootnote 5:
Optimization in the sense of Charles Darwin’s principle of selection of the fittest variant, X m , takes place.
The occurrence of selection in (2.4) can be verified also without knowing the solution (2.6). For this goal we consider the time dependence of the constraint ϕ, which is given by
Since a variance is always nonnegative, (2.7) implies that \(\phi(t)\) is a nondecreasing function of time. The value \(\mathrm{var}\{f\}=0\) implies a (local) maximum of ϕ and hence, ϕ is optimized during selection. Zero variance is tantamount to a homogeneous population containing only one variant. Since ϕ is at a maximum, this is the fittest variant X m .
3 Evolution in Genotype Space
Evolution can be visualized as a process in an abstract genotype or sequence space , \({\mathcal Q}\). At constant chain lengths ℓ of polynucleotides the sequence space is specified as \({\mathcal Q}_{\ell}^{\mathcal A}\), where \({\mathcal A}\) is the alphabet, for example \({\mathcal A}=\{\mathsf{\mathbf{0}},\mathsf{\mathbf{1}}\}\) or \({\mathcal A}=\{\mathsf{\mathbf{G}},\mathsf{\mathbf{C}}\}\) is the binary alphabet and \({\mathcal A}=\{\mathsf{\mathbf{A}},\mathsf{\mathbf{U}},\mathsf{\mathbf{G}},\mathsf{\mathbf{C}}\}\) the natural nucleotide alphabet. The gains of such a comprehensive view of genotypes are generality and the framework for reduction to the essential features; the shortcomings, obviously, are lack of detail. Building a model for evolution upon a space that fulfills all requirements required for the molecular view of biology and which may, eventually, bridge microscopic and macroscopic views, is precisely what we are aiming for here. The genotypes are DNA or RNA sequences and the proper genotype space is sequence space. The concept of a static sequence space [33, 34] was invented in the early 1970s in order to bring some ordering criteria into the enormous diversity of possible biopolymer sequences. Sequence space \({\mathcal Q}_{\ell}^{\mathcal A}\), as long we are only dealing with reproduction and mutation , is a metric space with the Hamming distanceFootnote 6 serving as the most useful metric for all practical purposes. Every possible sequence is a point in the discrete sequence space and in order to illustrate the space by a graph, sequences are represented by nodes and all pairs of sequences with Hamming distance one by edges (Fig. 2.2 shows a space of binary sequences as an example. Binary sequence spaces are hypercubes of dimension ℓ, ℓ being the length of the sequences).
Sequence space of binary sequences of chain length \(\ell=5\). The sequence space \({\mathcal Q}_5^{\{0,1\}}\) comprises 32 sequences. Each sequence is represented by a point. The numbers in the yellow balls are the decimal equivalents of the binary sequences and can be interpreted as sequences of two nucleotides, “ 0” ≡ “C” and “ 1” ≡ “G”. Examples are \(0\,\equiv\,\mathsf{\mathbf{00000}}\,\equiv\,\mathsf{\mathbf{CCCCC}}\), \(14\,\equiv\,\mathsf{\mathbf{01110}}\,\equiv\,\mathsf{\mathbf{CGGGC}}\) or \(29\,\equiv\,\mathsf{\mathbf{11101}}\,\equiv\,\mathsf{\mathbf{GGGCG}}\). All positions of a (binary) sequence space are equivalent in the sense that each sequence has ℓ nearest neighbors, \(\ell(\ell-1)/2\) next nearest neighbors, etc. Accordingly, sequences are properly grouped in mutant classes around the reference sequence, here 0
Two properties of sequence spaces are important: (i) All nodes in a sequence space are equivalent in the sense that every sequence has the same number of nearest neighbors with Hamming distance \(d_{\mathrm{H}}=1\), next nearest neighbors with Hamming distance \(d_{\mathrm{H}}=2\), and so on, which are grouped properly in mutant classes. (ii) All nodes of a sequence space are at the boundary of the space or, in other words, there is no interior. Both features are visualized easily by means of hypercubes Footnote 7: All points are positioned at equal distances from the origin of the (Cartesian) coordinate system. What makes sequence spaces difficult to handle are neither internal structures nor construction principles but the hyper-astronomically large numbers of points: \(|{\mathcal Q}_{\ell}^{\mathcal A}|=\kappa^\ell\) for sequences of length ℓ over an alphabet of size κ with \(\kappa=|{\mathcal A}|\).
The population \({\boldsymbol{\varXi}}=\{X_1,X_2,\hdots,X_n\}\) is represented by a vector with the numbers of species as elements \(\textbf{N}=(N_1,N_2,\hdots,N_n)\), the population size is the L1-norm:
where absolute values are dispensable since particle numbers are real and nonnegative by definition. Normalization of the variables yields \(\textbf{x}=\textbf{N}/\|\textbf{N}\|\) or \(x_i=N_i/N\) and \(\sum\nolimits_{i=1}^n x_i=1\), respectively. A population is thus represented by an L1-normalized vector x and the population size N. An important property of a population is its consensus sequence , \(\bar X\), consisting of a nucleotide distribution at each position of the sequence. This consensus sequence can be visualized as the center of the population in sequence space.
A sequence is conventionally understood as a string of ℓ symbols chosen from some predefined alphabet with κ letters, which can be written as
The natural nucleotide alphabet contains four letters: \({\mathcal A}=\{\mathsf{\mathbf{A}},\mathsf{\mathbf{U}},\mathsf{\mathbf{G}},\mathsf{\mathbf{C}}\}\), but RNA molecules with catalytic functions have been derived also from three- and two-letter alphabets [36, 37]. For the forthcoming considerations it is straightforward to adopt slightly different definitions: A sequence X j results from the multiplication of the alphabet vector \({\boldsymbol{\alpha}}=(\alpha_1,\hdots,\alpha_{\kappa})\) with a \(\kappa\times\ell\) matrix \({\mathcal X}_j\) having only 0 and 1 as entries:
In other words, the individual nucleotides in the sequence X j are replaced by products of two vectors, \(b_i^{(j)}={\boldsymbol{\alpha}}\cdot{\boldsymbol{\beta}}_i^{(j)}.\)
With the definition (2.8) it is straightforward to compute the consensus sequence of a population Ξ k :
and the distribution of nucleotides at position “i” is given by
It is important to note the difference between \(b_i^{(j)}\) and \(\textbf{\textit{b}}_i^{(k)}\): The former refers to the nucleotide at position “i” in a given sequence whereas the latter describes the nucleotide distribution at position “i” in the population. If one nucleotide is dominant at all positions, the distribution can be collapsed to a single sequence, the consensus sequence .
The internal structure of every sequence space \({\mathcal Q}_{\ell}^{\mathcal A}\) is induced by point mutation and this is essential for inheritance because it creates a hierarchy in the accessability of genotypes. Suppose we have a probability p of making one error in the reproduction of a sequence then, provided mutation at different positions is assumed to be independent, the probability of making two errors is p 2, of making three errors is p 3, etc. Inheritance requires sufficient accuracy of reproduction – otherwise children would not resemble their parents – and this implies p has to be sufficiently small. Then, p 2 is smaller and the power series \(p^{\,d_{\mathrm{H}}}\) decreases further with increasing distance from the reference sequence. This ordering of sequences according to a probability criterion that is intimately related to the Hamming metric (Sect. 2.5). As a matter of fact, mutation is indeed a fairly rare event in evolution and populations are commonly dominated by a well-defined single consensus sequence since single nucleotide exchanges that occur at many different positions do not contribute significantly to the average.
Evolutionary dynamics is understood as change of the population vectors in time: \({\textbf{N}}(t)\) or \({\textbf{x}}(t)\). This change can be modeled by means of differential equations (Sect. 2.5) or stochastic processes (Sect. 2.6). A practical problem concerns the representation of genotype space. Complete sequence space, \({\mathcal Q}_{\ell}^{\mathcal A}\) has the advantage of covering all possible genotypes but its extension is huge and, since the numbers of possible genotypes exceed even the largest populations by far, we are confronted with the problem that most degrees of freedom are empty and very likely will never be populated during the evolutionary process described. Alternatively the description could be restricted to those genotypes that are actually present in the population and that constitute the population support \({{\boldsymbol{\varPhi}}(t)}\), which is defined by
The obvious advantage is a drastic reduction in the degrees of freedom to a tractable size but one has to pay a price for this simplification: The population support is time dependent and changes whenever a new genotype is produced by mutation or an existing one goes extinct [38]. Depending on population size, population dynamics on the support can either be described by differential equations or modeled as a stochastic process. Support dynamics, on the other hand, are intrinsically stochastic since every mutant starts from a single copy.
Finally, it is important to mention that recombination without mutation can be modeled successfully as a process in an abstract recombination space [39, 40, 41] and plays a major role in the theory of genetic algorithms [42, 43]. A great challenge for theorists is the development of a genotype space for both mutation and recombination. Similarly, convenient sequence spaces for genotypes with variable chain lengths are not at hand.
4 Modeling Genotype–Phenotype Mappings
The unfolding of genotypes to yield phenotypes is studied in developmental biology and provides the key to understanding evolution and, in particular, the origin of species. For a long time it has been common knowledge that the same genotype can develop into different phenotypes, depending on differences in the environmental conditions and epigenetic effects.Footnote 8 Current molecular biology provides explanations for several epigenetic observations and reveals mechanisms for the inheritance of properties that are not encoded by the DNA of the individual. Genetics is still shaping the phenotypes – otherwise progeny would not resemble parents – but epigenetics and environmental influences provide additional effects that are indispensable for understanding and modeling the relations between genotypes and phenotypes. Here we shall adopt the conventional strategy of physicists and consider simple cases in which the genotypes unfolds unambiguously into a unique phenotype. This condition is fulfilled, for example, in evolution in vitro when biopolymer sequences form (the uniquely defined) minimum free energy structures as phenotypes. Bacteria in constant environments provide other cases of simple genotype–phenotype mappings (the long-term experiments of Richard Lenski [44–46] may serve as examples; see Sect. 2.6). Under this simplifying assumption genotype–phenotype relations can be modeled as mappings from an abstract genotype space into a space of phenotypes or shapes. A counter example in a simple system is provided by biopolymers with metastable suboptimal conformations, which can serve as models where a single genotype – a sequence – can give rise to several phenotypes – molecular structures [47].
Since only point mutations will be considered here, the choice of an appropriate genotype space is straightforward. It is the sequence space \({\mathcal Q}_{\ell}^{{\mathcal A}}\) with the Hamming distance d H as metric. The phenotype space or shape space \({\mathcal S}_{\ell}\) is the space of all phenotypes formed by all genotypes of chain length ℓ. Although the definition of a physically or biologically meaningful distance between phenotypes is not at all straightforward, some kind of metric can always be found. Accordingly the genotype–phenotype mapping ψ can be characterized by
where mfe indicates minimum free energy. The map ψ need not be invertible. In other words, several genotypes can be mapped onto the same phenotype when we are dealing with a case of neutrality.
An example of a genotype–phenotype mapping that can be handled straightforwardly by analytical tools is provided by in vitro evolution of RNA molecules [48–50]. RNA molecules are transferred to a solution containing activated monomers as well as a virus-specific RNA replicase. The material consumed by the replication reaction is replenished by serial transfer of a small sample into fresh solution. The replicating ensemble of RNA molecules optimizes the mean RNA replication rate of the population in the sense of Darwinian evolution [see (2.6)]. The interpretation of RNA evolution in vitro identifies the RNA sequence with the genotype. The RNA structure, the phenotype, is responsible for binding to the enzyme and for the progress of reproduction, since the structure of the template molecules has to open in order to allow replication [51–53]. In the case of RNA aptamer selectionFootnote 9 the binding affinity is a function of molecular structure, and sequence–structure mapping is an excellent model for the relation between genotype and phenotype.
Symbolic notation of RNA secondary structures. RNA molecules have two chemically different ends, the 5′- and the 3′-end. A general convention determines that all strings corresponding to RNA molecules (sequences, symbolic notation, etc.) start from the 5′-end and have the 3′-end at the right-hand side (rhs). The symbolic notation is equivalent to graphical representation of secondary structures. Base pairs are denoted by parentheses, where the opening parenthesis corresponds to the nucleotide closer to the 5′-end and the closing parenthesis to the nucleotide closer to the 3′-end of the sequence. In the figure we compare the symbolic notation with the conventional graphical representations for two structures formed by the same sequence
RNA sequences fold spontaneously into secondary structures consisting of double-helical stacks and single-stranded stretches. Within a stack, nucleotides form base pairs that are elements of a pairing logic \({\mathcal B}\), which consists of six allowed base pairs in the case of RNA structures: \({\mathcal B}=\{\mathsf{\mathbf{AU}},\mathsf{\mathbf{UA}},\mathsf{\mathbf{GC}},\mathsf{\mathbf{CG}},\mathsf{\mathbf{GU}},\mathsf{\mathbf{UG}}\}\). Further structure formation, very often initiated by the addition of two-valent cations, mostly Mg2+, folds secondary structure into three-dimensional structures by means of sequence specific tertiary interactions of nucleotide bases called motifs [54, 55]. Secondary structures have the advantage of computational and conceptional simplicity, allowing the application of combinatorics to global analysis of sequence–structure mappings [47, 56]. A conventional RNA secondary structure consists exclusively of base pairs and unpaired nucleotides and can be represented in a formal three-letter alphabet with the symbols ‘…’, ‘(‘,’)’ for unpaired nucleotides, downstream-bound, and upstream-bound nucleotides, respectively (Fig. 2.3). A straightforward way to annotate pairs in structures is given by the base pair count \(S_i=[\gamma_1^{(i)},\hdots,\gamma_{\ell}^{(i)}]\), which we illustrate here by means of the lower (blue) structure in the figure as an exampleFootnote 10:
Consecutive numbers are assigned to first nucleotides of base pairs corresponding to an opening parenthesis in the sequence, in which they appear in the structure, and the same number is assigned to the corresponding closing parenthesis lying downstream. Unpaired nucleotides are denoted by ‘0’. In total the structure contains n p base pairs and n s single nucleotides with \(2n_{\mathrm p}+n_{\mathrm s}=\ell\).
Molecular physics provides an excellent tool for modeling folding of molecules into structures, the concept of conformation space: A free energy is assigned to or calculated for each conformation of the molecule. Commonly, the variables of conformation space are continuous, bond lengths, valence angles or torsion angles may serve as examples. The free energy (hyper)surface or free energy landscape of a molecule presents the free energy as a function of the conformational variables. The mfe structure corresponds to the global minimum of the landscape, metastable states to local minima. In the case of RNA secondary structures conformation space and shape space are identical, and they are discrete spaces, since a nucleotide is either paired or unpaired. Whether a given conformation, a given base pairing pattern, is a local minimum or not depends also on the set of allowed moves in shape space \({\mathcal S}\). The move set defines the distance between structures, the metric \(d_S(S_i,S_j)\) in (2.11). An appropriate move set for RNA secondary structures comprises three moves: (i) base pair closure, (ii) base pair opening, and (iii) base pair shift [47, 62]. The first two moves need no further explanation; the shift move combines base pair opening and base pair formation with neighboring unpaired nucleotides. This set of three moves corresponds to a metric \(d_S(S_i,S_j)\), which is the Hamming distance between the symbolic notations of the two structures S i and S j .
Secondary structures of ribonucleic acid molecules (RNAs). Conventional RNA folding algorithms compute the mfe structure for a given sequence [58, 59]. Hairpin formation is shown as an example on the lhs of the figure. In addition, the sequence can fold also into a large number of suboptimal conformations (diagram in the middle of the figure), which are readily computed by efficient computer programs [60, 61]. If a suboptimal structure is separated from the mfe structure by a sufficiently high activation barrier, the structure is metastable. The metastable structure in the example shown here is a double hairpin (rhs of the figure). The activation energy of more than 20 kcal/mol does not allow interconversion of the two structures at room temperature. (For the calculation of kinetic structures see, for example, [62, 63])
Conventional structure prediction deals with single structures derived from single sequence inputs. Structure formation depends on external conditions such as temperature, pH value, ionic strength, and the nature of the counter-ions; in order to obtain a unique solution these conditions have to be specified. Commonly the search is for the most stable structure, the mfe structure, which corresponds to the global minimum of the conformational free energy landscape of the RNA molecule. In Fig. 2.4 the mfe structure \(S_0=\psi(X)\) is a single long hairpin shown (in red) at the lhs of the picture. A sequence that forms a stable mfe structure S 0 (free energy of foldingFootnote 11: \(\Delta G_{\mathrm{fold}}(S_0)<0\)) commonly forms almost always a set of suboptimal conformations \(\{S_1,S_2,\hdots,S_m\}\) with higher free energies of formation, \(\Delta G_{\mathrm{fold}}(S_i)>\Delta G_{\mathrm{fold}}(S_0)\ \mathrm{for}\ i\neq0\). In Fig. 2.4 (middle) the ten lowest suboptimal structures are listed; together with S 0 they represent the 11 lowest states of the spectrum of structures associated with the sequence X. Low-lying suboptimal conformations may influence the molecular properties, in particular when conformational changes are involved. The Boltzmann-weighted contributions of all suboptimal structures at temperature T are readily calculated by means of the partition function of RNA secondary structures [59, 64]. Instead of base pairs the analysis of the partition function yields base pairing probabilities that tell how likely it is to find two specific nucleotides forming a base pair in the ensemble of structures at thermal equilibrium.
Although folding RNA sequences into secondary structures is, presumably, the simplest conceivable case of a genotype–phenotype map, it is at the same time an example of the origin of complexity at the molecular level. The base pairing interaction is essentially nonlocal since a nucleotide can pair with another nucleotide from almost any position of the sequence.Footnote 12 The strongest stabilizing contributions to the free energy of structure formation come from neighboring base pairs and are therefore local. The combination of local and nonlocal effects is one of the most common sources of complex relations in mappings.
Mappings from sequence space onto shape space and back. The upper part of the figure sketchows schematically a mapping from sequence space onto structure or shape space. (Both sequence space and shape space are high-dimensional. The two-dimensional representation is used for the purpose of illustration only.) One structure is uniquely assigned to each sequence. The drawing shows the case of a mapping that is many-to-one and noninvertible: Many sequences fold into the same secondary structure and build a neutral network. The lower part of the figure illustrates the set of stable (sub)optimal structures that are formed by a single sequence. The mfe structure is indicated by a larger circle
The relation of an RNA sequence and its suboptimal structures is sketched in Fig. 2.5 (lower part). A single sequence X gives rise to a whole set of structures spread all over shape space. In principle, all structures that are compatible with the sequence appear in the spectrum of suboptimals but only a subset is stable in the sense that the structure S i (\(i=1,\hdots\)) corresponds to a local minimum of the conformational energy surface and the free energy of folding is negative (\(\Delta G_{\mathrm{fold}}(S_i) <0\)). Using the base pair count, the set of all structures that are compatible with the sequence X h can be defined straightforwardly:
In other words, a structure S i is compatible with a sequence X h if, and only if, two nucleotides that can form a base pair appear in the sequence at all pairs of positions that are joined by a base pair in the structure. For an arbitrary sequence the number of compatible structures is extremely large but the majority of them have either positive free energies of folding \((\Delta G_{\mathrm{fold}}(S_i)>0\)) and/or represent saddle points rather than local minima of the conformational energy surface. Figure 2.5 indicates the relation between an RNA sequence, its mfe structure, and its stable suboptimal conformations.
Studies of mfe structures or suboptimal structures refer to a certain set of conditions – for example, temperature T, pH, ionic strength – but time is missing because free energy differences (\(\Delta G\)) or partition functions are equilibrium properties. The structures that are determined and investigated experimentally, however, refer always to some time window – we are not dealing with equilibrium ensembles but with metastable states. The finite time structures of RNA are obtained by kinetic folding (see, e.g., [62, 63]). The RNA example shown in Fig. 2.4 represents the case of a bistable molecule: The most stable suboptimal structure S 1, a double hairpin conformation (blue), is the most stable representative of a whole family of double hairpin structures forming a broad basin of the free energy landscape of the molecule. This basin is separated from the basin of the single hairpin structure S 0 by a high energy barrier of about 20 kcal/mol and this implies that practically no interconversion of the two structures will take place at room temperature. We are dealing with an RNA molecule with one stable and one metastable conformation, a so-called RNA switch. RNA switches are frequent regulatory elements in prokaryotic regulation of translation [65].
5 Chemical Kinetics of Evolution
Provided population sizes N are sufficiently large, mutation rates are high enough, and stochastic effects are reduced by statistical compensation, evolution can be described properly by means of differential equations. In essence, we proceed as described in Sect. 2.2 and find for replication and mutation as an extension of the selection equation (2.4)
where x is an n-dimensional column vector and Q and F are \(n\times n\) matrices. The matrix Q contains the mutation probabilities Q ji , referring to the production of X j as an error copy of template X i , and F is a diagonal matrix whose elements are the replication rate parameters or fitness values f i.
Solutions of the mutation-selection equation (2.13) can be obtained in two steps: (i) integrating factor transformation allows the nonlinear term \(\phi(t)\) to be eliminated and (ii) the remaining linear equation is solved in terms of an eigenvalue problem [66–69]:
The new quantities in this equation, b jk and h kj , are the elements of two transformation matrices:
The columns of B and the rows of B−1 represent the right-hand and left-hand eigenvectors of the matrix \(\mathrm{W}=\mathrm{Q}\cdot\mathrm{F}\) with \(\mathrm{B}^{-1}\cdot\mathrm{W}\mathrm{B}=\boldsymbol{\Lambda}\) being a diagonal matrix containing the eigenvalues of W. The elements of the matrix W are nonnegative by definition since they are the product of a fitness value or replication rate parameter f i and a mutation probability Q ji , which are both nonnegative. If, in addition, W is a nonnegative primitive matrixFootnote 13 – implying that every sequence can be reached from every sequence by a finite chain of consecutive mutations – the conditions for the validity of the Perron–Frobenius theorem [70] are fulfilled. Two (out of six) properties of the eigenvalues and eigenvectors of W are important for replication-mutation dynamics:
-
(i).
The largest eigenvalue λ 1 is nondegenerate, \(\lambda_1>\lambda_2\ge\lambda_3\ge\hdots\ge\lambda_n\), and
-
(ii).
the unique eigenvector belonging to λ 1 denoted by ξ 1 has only positive elements, \(\xi_j^{(1)}>0\,\forall\,j=1,\hdots,n\).
After sufficiently long time the population converges to the largest eigenvector ξ 1, which is therefore the stationary state of (2.13). Since ξ 1 represents the genetic reservoir of an asexually replicating species it is called the quasispecies [68]. A quasispecies commonly consists of a fittest genotype, the master sequence, and a mutant distribution surrounding the master sequence in sequence space. Although the solution of the mutation-selection equation is straightforward, the experimental proof of the existence of a stationary mutant distribution and its analysis are quite involved [71]. The work has been conducted with relatively short RNA molecules (chain length: \(\ell=87\)). Genotypic heterogeneity in virus populations was first detected in the 1970s [72]. Later, the existence of quasispecies in nature was demonstrated for virus populations (For an overview and a collection of reviews see [73, 74]). Since it is very hard, if not impossible, to prove that a natural population is in a steady state, the notion virus quasispecies was coined for virus populations observed in vitro and in vivo.
In order to explore quasispecies as a function of the mutation rate p, a crude or zeroth-order approximation consisting of neglect of backward mutations has been adopted [33]. The differential equation for the master sequence is then of the form
with \(\bar f_{-m}=\bigl(\sum_{j=1,j\neq m}^n f_jx_j\bigr)/(1-x_m)\). We apply the uniform error approximation and assume that the mutation rate per nucleotide and replication event, p, is independent of the nature of the nucleotide (A, U, G or C) and the position along the sequence. We find for the elements of the mutation matrix Q
and obtain for the stationary concentration of the master sequence
where \(\sigma_m=f_m/\bar f_{-m}>1\) is the superiority of the master sequence and \(\bar f_{-m}\) is defined by
In this zeroth-order approximation the stationary concentration \(\bar x_m^{(0)}(p)\) vanishes at the critical value (Fig. 2.6)
The error threshold in RNA replication. The stationary frequency of the master sequence X m is shown as a function of the mutation rate p. In the zeroth-order approximation neglecting mutational backflow the function \(\bar x_m^{(0)}(p)\) is almost linear in the particular example shown here. In the inset the zeroth-order approximation (black) is shown together with the exact function (red) and an approximation applying the uniform distribution to the mutational cloud (\(\bar x_j=(1-\bar x_m)/(n-1)\) \(\forall\,j\neq m\); blue), which is exact at the mutation rate \(p=0.5\) for binary sequences. The error rate p has two natural limitations: (i) the physical accuracy limit of the replication process provides a lower bound for the mutation rate and (ii) the error threshold defines a minimum accuracy of replication that is required to sustain inheritance and sets an upper bound for the mutation rate. Parameters used in the calculations: binary sequences, \(\ell=6\), \(\sigma=1.4131\)
Needless to say, zero concentration of the master sequence is an artifact of the approximation, because the exact concentration of the master sequence cannot vanish by the Perron–Frobenius theorem as long as the population size is nonzero. In order to find out what really happens at the critical mutation rate p cr computer solutions of the complete equation (2.13) were calculated for the single peak fitness landscape.Footnote 14 These calculations [75] show a sharp transition from the ordered quasispecies to the uniform distribution, \(\bar x_j=\kappa^{-\ell}\,\forall\,j=1,\hdots,\kappa^{\ell}\). At the critical mutation rate p cr, replication errors accumulate and (independently of initial conditions) all sequences are present at the same frequency in the long-time limit, as is reflected by the uniform distribution. The uniform distribution is the exact solution of the eigenvalue problem at equal probabilities for all nucleotide incorporations (\(\mathsf{\mathbf{A}}\to\mathsf{\mathbf{A}}, \mathsf{\mathbf{A}}\to\mathsf{\mathbf{U}}, \mathsf{\mathbf{A}}\to\mathsf{\mathbf{G}}\), and \(\mathsf{\mathbf{A}}\to\mathsf{\mathbf{C}}\)) occurring at \(\tilde p=\kappa^{-1}\). The interesting aspect of the error threshold phenomenon consists in the fact that the quasispecies approaches the uniform distribution at a critical mutation rate p cr that is far below the random mutation value \(\tilde p\). As a matter of fact, the appearance of an error threshold and its shape depend on details of the fitness landscape [76, pp. 51–60]. Some landscapes show no error threshold at all but a smooth transition to the uniform distribution [77]. More realistic fitness landscapes with a distribution of fitness values reveal a much more complex situation: For constant superiority the value of p cr becomes smaller with increasing variance of fitness values. The error threshold phenomenon can be split into three different observations that coincide on the single peak landscape : (i) vanishing of the master sequence x m , (ii) phase-transition-like behavior, and (iii) transition to the uniform distribution. On suitable model landscapes the three observations do not coincide and thus can be separated [78, 79].
The error threshold on single peak fitness landscapes. The upper part of the figure shows the quasispecies as a function of the mutation rate p. The variables \(\bar y_k(p)\) (\(k=0,1,\hdots,\ell\)) represent the total concentrations of all sequences with Hamming distance \(d_{\mathrm{H}}=k\): \(\bar y_0=\bar x_m\) (black) is the concentration of the master sequence, \(\bar y_1=\sum\nolimits_{i=1,d_{\mathrm{H}}(X_i,X_m)=1}^n \bar x_i\) (red) is the concentration of the one-error class, \(\bar y_2=\sum\nolimits_{i=1,d_{\mathrm{H}}(X_i,X_m)=2}^n \bar x_i\) (yellow) that of the two-error class and, accordingly, we have \(\bar y_k=\sum\nolimits_{i=1,d_{\mathrm{H}}(X_i,X_m)=k}^n \bar x_i\) for the k-error class. The lower part shows an enlargement. The position of the error threshold computed from the zeroth-order approximation (2.16) is shown as by a dotted line (gray). Choice of parameters: \(\kappa=2\), \(\ell=100\), \(f_m=10\), \(f_0=1\) and hence \(\sigma_m=10\) and \(p_{\mathrm{cr}}=0.02276\)
How do populations behave at mutation rates above the error threshold? In reality a uniform distribution of variants as required for the stationary state cannot be realized. In RNA selection experiments population sizes hardly exceed 1015 molecules, the smallest aptamers have chain lengths of \(\ell=27\) nucleotides [80] and this implies \(4^{27}\approx18\times10^{15}\) different sequences. Even in this most favorable case we are dealing with more sequences than molecules in the population: a uniform distribution cannot exist. Although the origin of the lack of selective power is completely different – high mutation rates wiping out the differences in fitness values versus fitness differences being zero or too small for selection – the scenarios most likely to occur are migrating populations similar to evolution on a flat landscape [81]. Bernard Derrida and Luca Peliti find that the populations break up into clones, which migrate into different directions in sequence space. Migrating populations are unable to conserve a genotype over generations, and unless a large degree of neutrality allows a phenotype to be maintained despite changing genotypes, evolution becomes impossible because inheritance breaks down.
Because of high selection pressure resulting from the hosts’ defense systems, virus populations operate at mutation rates as high as possible in order to allow fast evolution, and this is just below the error threshold [82]. Increasing the mutation rate should drive the virus population beyond threshold, where sufficiently accurate replication is no longer possible. Therefore virus populations are doomed to die out at mutation rates above threshold, and this suggested a novel antiviral strategy that has led to the development of new drugs [83]. A more recent discussion of the error threshold phenomenon tries to separate the error accumulation phenomenon from mutation-caused fitness effects leading to virus extinction, known as lethal mutagenesis [84, 85]. In fact lethal mutagenesis describes the error threshold phenomenon for variable population size N as required for \(\lim N\to0\), but an analysis of population dynamics without and with stochastic effects at the onset of migration of populations is still lacking. In addition, more detailed kinetic studies on replication in vitro near the error threshold are required before the mechanism of virus extinction at high mutation rates can be understood.
Sequence–structure mappings of nucleic acid molecules (Sect. 2.4) and proteins provide ample evidence for neutrality in the sense that many genotypes give rise to the same phenotype and identical or almost identical fitness values that cannot be discriminated by natural selection. The possible occurrence of neutral variants was even discussed by Charles Darwin [2, chapter iv]. Based on the results of the first sequence data from molecular biology, Motoo Kimura formulated his neutral theory of evolution [86, 87]. In the absence of fitness differences between variants, random selection occurs because of stochastic enhancement through autocatalytic processes: more frequent variants are more likely to be replicated than less frequent ones. Ultimately a single genotype becomes fixated in the population. The average time of replacement for a dominant genotype is the reciprocal mutation rate, \(\nu^{-1}=(\ell p)^{-1}\), which, interestingly, is independent of the population size. Are Kimura’s results valid also for large population sizes and high mutation rates, as they occur, for example, with viruses? Mathematical analysis [88] together with recent computer studies [78] yields the answer: Random selection in the sense of Kimura occurs only for sufficiently distant (master) sequences. In full agreement with the exact result in the limit \(p\to0\) we find that two fittest sequences of Hamming distance \(d_{\mathrm{H}}=1\), two nearest neighbors in sequence space, are selected as a strongly coupled pair with equal frequency of the two members. Numerical results demonstrate that this strong coupling occurs not only for small mutation rates but extends over the whole range of p values from p = 0 to the error threshold \(p=p_{\mathrm{cr}}\). For clusters of more than two sequences with \(d_{\mathrm{H}}=1\), the frequencies of the individual members of the cluster are given by the components of the largest eigenvector of the adjacency matrix. Pairs of fittest sequences with Hamming distance \(d_{\mathrm{H}}=2\), i.e., two next-nearest neighbors with two sequences in between, are also selected together but the ratio of the two frequencies is different from one. Again coupling extends from zero mutation rates to the error threshold. Strong coupling of fittest sequences manifests itself in virology as systematic deviations from consensus sequences of populations, as indeed observed in nature. For two fittest sequences with \(d_{\mathrm{H}}\ge3\) random selection chooses arbitrarily one of the two and eliminates the other one, as predicted by the neutral theory.
The function \(\phi(t)\) was introduced as the mean fitness of a population in order to allow straightforward normalization of the population variables. A more general interpretation considers \(\phi(t)\) as a flux out of the system. Then the equation describing evolution of the column vector of particle numbers \(\textbf{\textit{N}}=(N_1,\hdots,N_n)\) is of the form [89]
where \(F_j(\textbf{\textit{N}})\) is the function of unconstrained reproduction. An example is provided by (2.13): \(F_j(\textbf{\textit{N}})=\sum\nolimits_{i=1}^n Q_{ji}f_i\,N_i\). Explicit insertion of the total concentration \(C(t)=\sum\nolimits_{i=1}^nN_i(t)\) yields
Either \(C(t)\) or \(\phi(t)\) can be chosen freely; the second function is then determined by the equation given above. For normalized variables we find
For a large number of examples and for most cases important in evolution, the functions \(F_j(\textbf{\textit{N}})\) are homogeneous functions in N. For homogeneity of degree λ we have \(F_j(\textbf{\textit{N}})=F_j(C\cdot\textbf{\textit{N}})=C^{\gamma}F_j(\textbf{x})\) and find
Two conclusions can be drawn from this equation: (i) For \(\gamma=1\), e.g., the selection equation (2.4) or the replication-mutation equation (2.13), the dependence on the total concentration C vanishes and the solution curves in normalized variables \(x_j(t)\) are the same in stationary (\(C=\mathrm{const}\)) and nonstationary systems as long as \(C(t)\) remains finite and does not vanish, and (ii) if \(\gamma\neq1\) the long-term behavior determined by \(\dot{\textbf{x}}=0\) is identical for stationary and nonstationary systems unless the population dies out \(C(t)\to0\) or explodes \(C(t)\to\infty\).
6 Evolution as a Stochastic Process
Stochastic phenomena are essential for evolution – each mutant after all starts out from a single copy – and a large number of studies have been conducted on stochastic effects in population genetics [90]. Not so much work, however, has been devoted so far to the development of a general stochastic theory of molecular evolution. We mention two examples representative for others [91, 92]. In the latter case the reaction network for replication and mutation was analyzed as a multi-type branching process and it was proven that the stochastic process converges to the deterministic equation (2.13) in the limit of large populations. What is still lacking is a comprehensive treatment, for example by means of chemical master equations [93]. Then the deterministic population variables \(x_j(t)\) are replaced by stochastic variables \({\mathcal X}_j(t)\) and the corresponding probabilities
The chemical master equation translates a mechanism into a set of differential equations for the probabilities. The pendant of (2.13), for example, is the master equation
The only quantity that has to be specified further in this equation is the flux term \(\phi(t)\). For the stochastic description it is not sufficient to have a term that just compensates the increase in population size due to replication , a detailed model of the process is required. Examples are (i) the Moran process [94–96] with strictly constant population size and (ii) the flow reactor (continuous stirred tank reactor, CSTR) with a population size fluctuating within the limits of a \(\sqrt N\) law [97, 98].Footnote 15 The Moran process assumes that for every newborn molecule one molecule is instantaneously eliminated. Strong coupling of otherwise completely independent processes has the advantage of mathematical simplicity but it lacks a physical background. The flow reactor, on the other hand, is harder to treat in the mathematical analysis but it is based on solid physical grounds and can be easily implemented experimentally. In computer simulation both models require comparable efforts and for molecular systems preference is given therefore to the flow reactor.
For evolution of RNA molecules through replication and mutation in the flow reactor , the following reaction mechanism has been implemented:
Stock solution flows into the reactor with a flow rate r and it feeds the reactor with the material required for polynucleotide synthesis – schematically denoted by A and consisting, for example, of activated nucleotides, \(\mathsf{\mathbf{ATP}}, \mathsf{\mathbf{UTP}}, \mathsf{\mathbf{GTP}}\), and CTP, as well as a replicating enzyme – into the system. The concentration of A in the stock solution is denoted by a 0. The molecules X j are produced by the second reaction either by correct copying or by mutation. The third and fourth reactions describe the outflux of material and compensate the increase in volume caused by the influx of stock solution. The reactor is assumed to be perfectly mixed at every instant (CSTR). For a targeted search the stochastic process in the reactor is constructed to have two absorbing states (Fig. 2.8): (i) extinction – all RNA molecules are diluted out of the reaction vessel – and (ii) survival – the predefined target structure has been produced in the reactor. The population size determines the outcome of the computer experiment: Below population sizes of \(N=13\) the reaction in the CSTR almost certainly goes extinct, but it reaches the target with a probability close to one for \(N>20\). The probability of extinction is very small for sufficiently large populations, and for population sizes \(N\ge1,000\), as reported here, extinction has been never observed.
Survival in the flow reactor. Replication and mutation in the flow reactor are implemented according to the mechanism (2.20). The stochastic process has two absorbing states: (i) extinction, \({\mathcal X}_j=0\,\forall\,j=1,\hdots,n\), and (ii) a predefined target state – here the structure of tRNAphe. A rather sharp transition in the long-time behavior of the population is shown in the lower plot: populations of natural sequences (AUGC) switch from almost certain extinction to almost certain survival in the range \(13\le N\le18\) and for binary sequences (GC) the transition is even sharper but requires slightly larger population sizes
In order to simulate the interplay between mutation acting on the RNA sequence and selection operating on RNA structures, the sequence–structure map has to be turned into an integral part of the model [97–99]. The simulation tool starts from a population of RNA molecules and simulates chemical reactions corresponding to replication and mutation in a CSTR according to (2.20) by using Gillespie’s algorithm [100–102]. Molecules replicate in the reactor and produce both correct copies and mutants, the materials to be consumed are supplied by the continuous influx of stock solution into the reactor, and excess volume is removed by means of the outflux of reactor solution. Two kinds of computer experiments were performed: Optimizations of properties on a landscape derived from the sequence–structure map and targeted searches in shape space where the target is some predefined structure.
Early simulations optimizing replication rates in populations of binary GC-sequences yielded two general results:
-
(i)
The progress in evolution is stepwise rather than continuous, as short adaptive phases are interrupted by long quasi-stationary epochs [97, 98].
-
(ii)
Different computer runs with identical initial conditionsFootnote 16
resulted in different structures with similar values of the optimized rate parameters. Despite identical initial conditions, the populations migrated in different – almost orthogonal – directions in sequence space and gave rise thereby to contingency in evolution [98].
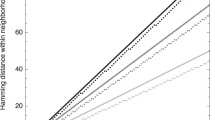
A trajectory of evolutionary optimization. The topmost plot presents the mean distance to the target structure of a population of 1,000 molecules, the plot in the middle shows the width of the population in Hamming distance between sequences, and the plot at the bottom is a measure of the velocity with which the center of the population migrates through sequence space. Diffusion on neutral networks causes spreading on the population in the sense of neutral evolution [105]). A remarkable synchronization is observed: At the end of each quasi-stationary plateau a new adaptive phase in the approach towards the target is initiated, which is accompanied by a drastic reduction in the population width and a jump in the population center. (The top of the peak at the end of the second long plateau is marked by an arrow.) A mutation rate of \(p=0.001\) was chosen, the replication rate parameter is defined in (2.21), and initial and target structures are shown in Table 2.1
In targeted search problems the replication rate of a sequence X k , representing its fitness f k , is chosen to be a function of the Hamming distanceFootnote 17 between the structure formed by the sequence, \(S_k=f(X_k)\), and the target structure, S T,
which increases when S k approaches the target (α is an empirically adjustable parameter that is commonly chosen to be 0.1). A trajectory is completed when the population reaches a sequence that folds into the target structure – appearance of the target structure in the population is defined as an absorbing state of the stochastic process. A typical trajectory is shown in Fig. 2.9. In this simulation a homogeneous population consisting of N molecules with the same random sequence and structure is chosen as the initial condition. The target structure is the well-known secondary structure of phenylalanyl-transfer RNA (tRNAphe). The mean distance to target of the population decreases in steps until the target is reached [99, 103, 104] and again the approach to the target is stepwise rather than continuous: Short adaptive phases are interrupted by long quasi-stationary epochs. In order to reconstruct optimization dynamics, a time-ordered series of structures is determined that leads from an initial structure S I to the target structure S T. This series, called the relay series, is a uniquely defined and uninterrupted sequence of shapes. It is retrieved through backtracking, that is, in the opposite direction, from the final structure to the initial shape. The procedure starts by highlighting the final structure and traces it back during its uninterrupted presence in the flow reactor until the time of its first appearance. At this point we search for the parent shape from which it descended by mutation. Now we record the time and structure, highlight the parent shape, and repeat the procedure. Recording further backwards yields a series of shapes and times of first appearance that ultimately ends in the initial population.Footnote 18 Use of the relay series and its theoretical background allows classification of transitions [99, 103, 106]. Inspection of the relay series together with the sequence record on the quasi-stationary plateaus provides strong hints for the distinction of two scenarios:
-
(i)
The structure is constant and we observe neutral evolution in the sense of Kimura’s theory of neutral evolution [87]. In particular, the numbers of neutral mutations accumulated are proportional to the number of replications in the population, and the evolution of the population can be understood as a diffusion process on the corresponding neutral network [105].
-
(ii)
The process during the quasi-stationary epoch involves several closely related structures with identical replication rates and the relay series reveals a kind of random walk in the space of these neutral structures.
The diffusion of the population on the neutral network is illustrated by the plot in the middle of Fig. 2.9, which shows the width of the population as a function of time [104]. The population width increases during the quasi-stationary epoch and sharpens almost instantaneously after a sequence has been created by mutation that allows the start of a new adaptive phase in the optimization process. The scenario at the end of the plateau corresponds to a bottleneck of evolution. The lower part of the figure shows a plot of the migration rate or drift of the population center and confirms this interpretation: Migration of the population center is almost always very slow unless the center “jumps” from one point in sequence space to a possibly distant point where the molecule initiating the new adaptive phase is located. A closer look at the three curves in Fig. 2.9 reveals coincidence of three events: (i) collapse-like narrowing of the population spread, (ii) jump-like migration of the population center, and (iii) beginning of a new adaptive phase.
It is worth mentioning that the optimization behavior observed in a long-term evolution experiment with Escherichia coli [46] can be readily interpreted in terms of random searches on a neutral network. Starting with twelve colonies in 1988, Lenski and his coworkers observed, after 31,500 generation or 20 years, a great adaptive innovation in one colony [45]: This colony developed a kind of membrane channel that allows uptake of citrate, which is used as a buffer in the medium. The colony thus conquered a new resource that led to a substantial increase in colony growth. The mutation providing citrate import into the cell is reproducible when earlier isolates of this particular colony are used for a restart of the evolutionary process. Apparently this particular colony has traveled through sequence space to a position from where the adaptive mutation allowing citrate uptake is within reach. None of the other eleven colonies gave rise to mutations with a similar function. The experiment is a nice demonstration of contingency in evolution: The conquest of the citrate resource does not happen through a single highly improbable mutation but by means of a mutation with standard probability from a particular region of sequence space where the population had traveled in one case out of twelve – history matters, or to repeat Theodosius Dobzhansky’s famous quote: “Nothing makes sense in biology except in the light of evolution” [29].
Table 2.1 collects some numerical data sampled from evolutionary trajectories of simulations repeated under identical conditions. Individual trajectories show enormous scatter in the time or the number of replications required to reach the target. The mean values and the standard deviations were obtained from statistics of trajectories under the assumption of log-normal distributions. Despite the scatter three features are unambiguously detectable:
-
(i)
The search in GC sequence space takes about five time as long as the corresponding process in AUGC sequence space, in agreement with the difference in neutral network structure.
-
(ii)
The time to target decreases with increasing population size.
-
(iii)
The number of replications required to reach target increases with population size.
Combination of the results (ii) and (iii) allows a clear conclusion concerning time and material requirements of the optimization process: Fast optimization requires large populations whereas economic use of material suggests working with small population sizes just sufficient to avoid extinction.
A study of parameter dependence of RNA evolution was reported in a recent simulation [107]. Increase in mutation rate leads to an error threshold phenomenon that is closely related to one observed with quasispecies on a single-peak landscape as described above [69, 75]. Evolutionary optimization becomes more efficientFootnote 19 with increasing error rate until the error threshold is reached. Further increase in error rates leads to a breakdown of the optimization process. As expected the distribution of replication rates or fitness values f k in sequence space is highly relevant too: Steep decrease of fitness with the distance to the master structure represented by the target, which has the highest fitness value, leads to sharp threshold behavior, as observed on single-peak landscapes, whereas flat landscapes show a broad maximum of optimization efficiency without an indication of threshold-like behavior.
7 Concluding Remarks
Biology developed differently from physics because it refrained from using mathematics as a tool to analyze and unfold theoretical concepts. Application of mathematics enforces clear definitions and reduction of observations to problems that can be managed. Over the years physics became the science of abstractions and generalizations, biology the science of encyclopedias of special cases with all their beauties and peculiarities. Among others there is one great exception to the rule: Charles Darwin presented a grand generalization derived from a wealth of personal and reported observations together with knowledge from economics concerning population dynamics. In the second half of the twentieth century the appearance of molecular biology on the stage changed the situation entirely. A bridge was built from physics and chemistry to biology, and mathematical models from biochemical kinetics or population genetics became presentable in biology. Nevertheless, the vast majority of biologists still smiled at the works of theorists. By the end of the twentieth century molecular genetics had created such a wealth of data that almost everybody feels nowadays that progress cannot be made without a comprehensive theoretical foundation and a rich box of suitable computational tools. Nothing like this is at hand but indications for attempts in the right direction are already visible. Biology is going to enter the grand union of science that started with physics and chemistry and is progressing fast. Molecular biology started out with biomolecules in isolation and deals now with cells, organs, and organisms. Hopefully, this spectacular success will end the so-far fruitless reductionism versus holism debate.
Insight into the mechanisms of evolution reduced to the simplest conceivable systems was provided here. These systems deal with evolvable molecules in cell-free assays and are accessible by rigorous mathematical analysis and physical experimentation. An extension to asexual species, in particular viruses and bacteria, is within reach. The molecular approach provides a simple explanation of why we have species for these organisms despite the fact that there is neither restricted recombination nor reproductive isolation. The sequence spaces are so large that populations, colonies, or clones can migrate for the age of the universe without coming close to another asexual species. We can give an answer to the question of the origin of complexity: Complexity in evolution results primarily from genotype–phenotype relations and from the influences of the environment. Evolutionary dynamics may be complicated in some cases but it is not complex at all. This has been reflected already by the sequence–structure map of our toy example. Conformation spaces depending on the internal folding kinetics as well as on environmental conditions and compatible sets are metaphors for more complex features in evolution proper.
Stochasticity is still an unsolved problem in molecular evolution. The mathematics of stochastic processes encounters difficulties in handling the equations of evolution in detail. A comprehensive stochastic theory is still not available and the simulations lack more systematic approaches since computer simulations of chemical kinetics of evolution are at an early stage too. Another fundamental problem concerns the spatial dimensions: Almost all treatments assume spatial homogeneity but we have evidence of the solid-particle-like structure of the chemical factories of the cell. In the future, any comprehensive theory of the cell will have to deal with these structurally rich supramolecular structures too.
Notes
- 1.
The reductionist program, also called methodological reductionism, aims at an exploration of complex objects through breaking them up into modular, preferentially molecular parts and studying the parts in isolation before reassembling the object. Emergent properties are assumed to be describable in terms of the phenomena from and the processes by which they emerge. The reductionist program is different from ontological reductionism, which denies the idea of ontological emergence by the claim that emergence is merely a result of the system’s description and does not exist on a fundamental level.
- 2.
Gregor Mendel was the first to investigate such relations experimentally [7–9] and discovered the transmittance of properties in discrete packages from the parents to offspring. His research objects were the pea (Pisum) from where he derived his rules of inheritance and the hawkweed (Hieracium), which was rather confusing for him, because it is apomictic, i.e., it reproduces asexually. Charles Darwin, on the other hand, had a mechanism of inheritance in mind that was entirely wrong. It was based on the idea of blending of the parents’ properties.
- 3.
According to his own records Charles Darwin was influenced strongly by Robert Malthus and his demographic theory [10].
- 4.
In this chapter we shall not consider sexual reproduction or other forms of recombination. In asexual reproduction a strict distinction between variants and species is neither required nor possible. We shall briefly come back to the problem of bacterial or viral species in Sect. 2.7.
- 5.
We assume here that the largest fitness value f m is non-degenerate, i.e., there is no second species having the same (largest) fitness value. In Sect. 2.5 we shall drop this restriction.
- 6.
The Hamming distance \(d_{\mathrm{H}}(X_i,X_j)\) [35] counts the number of positions at which two aligned sequences X i and X j differ. It fulfills the four criteria for a metric in sequence space: (i) \(d_{\mathrm{H}}(X_i,X_j)\ge0\) (nonnegativity), (ii) \(d_{\mathrm{H}}(X_i,X_ji)=0\) if and only if \(X_i=X_j\) (identity of indiscernibles), (iii) \(d_{\mathrm{H}}(X_i,X_j)=d_{\mathrm{H}}(X_i,X_j)\) (symmetry), and (iv) \(d_{\mathrm{H}}(X_i,X_j)\le d_{\mathrm{H}}(X_i,X_k)+d_{\mathrm{H}}(X_i,X_k)\) (triangle inequality). For sequences of equal chain length ℓ, end-to-end alignment is the most straightforward alignment, although it may miss close relatedness that is a consequence of deletions and insertions, which are mutations that alter sequence length.
- 7.
An ℓ-dimensional hypercube in the Cartesian space of dimension ℓ is the analogue of a (three-dimensional) cube. The ℓ-dimensional hypercube is constructed by drawing 2ℓ (hyper)planes of dimension \((\ell-1)\) perpendicular to the coordinate axes at the positions ±a. The corners of the hypercubes are the 2ℓ points where ℓ planes cross.
- 8.
Epigenetics was originally used as a term subsuming phenomena that could not be explained by conventional genetics.
- 9.
An aptamer is a molecule that binds to a predefined target molecule. Aptamers are commonly produced by an evolutionary selection process [57].
- 10.
The base pair count is another equivalent representation of RNA secondary structures. In the case of conventional secondary structures, the symbolic notation is converted into the base pair count by an exceedingly simple algorithm: Starting with zero at the 5′-end and proceeding from left to right a positive integer counting the number of open parenthesis is assigned to every position along the sequence. The base pair count is not only more convenient for base pair assignments but also more general. It is, for example, applicable to RNA structures with pseudoknots.
- 11.
The free energy of folding is the difference in free energy between the structure S i and the unfolded (open) chain \({\mathcal O}\): \(\Delta G_{\mathrm{fold}}(S_i)=G(S_i)-G({\mathcal O})\).
- 12.
Pairing with nearest neighbors is excluded for geometrical reasons. In other words, base pairs of two adjacent nucleotides have such a high positive free energy of formation that they are never observed.
- 13.
A square nonnegative matrix \(\textrm{W}=\{w_{ij};i,j=1,\hdots,n;\,w_{ij}\ge0\}\) is called primitive if there exists a positive integer m such that Wm is strictly positive: \(\textrm{W}^m>0\), which implies \(\textrm{W}^m=\{w_{ij}^{(m)};i,j=1,\hdots,n;w_{ij}^{(m)}>0\}\).
- 14.
The single peak fitness landscape is a kind of mean field approximation: A fitness value f m is assigned to the master sequence, whereas all other variants have the same fitness f 0. For this particular landscape the position \(\bar x_m^{(0)}=0\) calculated within the zeroth-order approximation almost coincides with the position of the critical change in the population structure (Fig. 2.7).
- 15.
All thermodynamically admissible processes obey a so-called \(\sqrt N\) law: For a mean population size of N the actual population size fluctuates with a standard deviation proportional to \(\sqrt N\).
- 16.
Identical means here that everything in the computer runs was the same except the seeds for the random number generators and this implies different series of random events.
- 17.
The distance between two structures is defined here as the Hamming distance between the two symbolic notations of the structures.
- 18.
It is important to stress two facts about relay series: (i) The same shape may appear two or more times in a given relay series series. Then, it was extinct between two consecutive appearances. (ii) A relay series is not a genealogy, which is the full recording of parent–offspring relations in a time-ordered series of genotypes.
- 19.
Efficiency of evolutionary optimization is measured by average and best fitness values obtained in populations after a predefined number of generations.
References
G. Galilei, Il Saggiatore, vol. 6 (Edition Nationale, Florence, Italy, 1896), p. 232. English translation from Italian original
C. Darwin, On the Origin of Species by Means of Natural Selection or the Preservation of Favoured Races in the Struggle for Life (John Murray, London, 1859)
E. Mayr, The Growth of Biological Thought. Diversity, Evolution, and Inhertiance (The Belknap Press of Harvard University Press, Cambridge, MA, 1982)
R.A. Fisher, The Genetical Theory of Natural Selection (Oxford University Press, Oxford, 1930)
J.B.S. Haldane, The Causes of Evolution (Longmans Green, London, 1932). Reprinted 1990 in the Princeton Science Library by Princeton University Press, Princeton, NJ
S. Wright, Evolution and the Genetics of Populations, vol. 1–4 (University of Chicago Press, Chicago, IL, 1968, 1969, 1977, 1978)
G. Mendel, Verhandlungen des naturforschenden Vereins in Brünn 4, 3 (1866)
G. Mendel, Verhandlungen des naturforschenden Vereins in Brünn 8, 26 (1870)
G.A. Nogler, Genetics 172, 1 (2006)
T.R. Malthus, An Essay on the Principle of Population as it Affects the Future Improvement of Society (J. Johnson, London, 1798)
A.J. Lotka, Elements of Physical Biology (Williams & Wilkins, Baltimore, MD, 1925)
V. Volterra, Mem. R. Accad. Naz. Lincei Ser.VI/2, 31 (1926)
E.P. Odum, Fundamentals of Ecology (W. B. Saunders, Philadelphia, PA, 1953)
R.M. May (ed.), Theoretical Ecology. Principles and Applications (Blackwell Scientific, Oxford, 1976)
A.M. Turing, Philos. Trans. R. Soc. Lond. Ser. B 237, 37 (1952)
A. Gierer, H. Meinhardt, Kybernetik 12, 30 (1972)
H. Meinhardt, Models of Biological Pattern Formation (Academic, London, 1982)
J.D. Murray, Sci. Am. 258(3), 62 (1988)
J.D. Murray, Mathematical Biology II: Spatial Models and Biomedical Applications, 3rd edn. (Springer, New York, NY, 2003)
A.L. Hodgkin, A.F. Huxley, J. Physiology 117, 500 (1952)
J.J. Hopfield, Proc. Natl. Acad. Sci. USA 79, 2554 (1982)
H. Judson, The Eighth Day of Creation. The Makers of the Revolution in Biology (Jonathan Cape, London, 1979)
A. Maxam, W. Gilbert, Proc. Natl. Acad. Sci. USA 74, 560 (1977)
F. Sanger, S. Nicklen, A. Coulson, Proc. Natl. Acad. Sci. USA 74, 5463 (1977)
J. Dale, M. von Schantz, From Genes to Genomes: Concepts and Applications of DNA Technology, 2nd edn. (Wiley, Chichester, 2007)
S. Brenner, Scientist 16(4), 14 (2002)
J.S. Edwards, M. Covert, B.O. Palsson, Environ. Microbiol. 4, 133 (2002)
H.W. Engl, C. Flamm, P. K¨nugler, J. Lu, S. Müller, P. Schuster, Inverse Probl. 25, 123014 (2009)
T. Dobzhansky, F.J. Ayala, G.L. Stebbins, J.W. Valentine, Evolution (W.H. Freeman, San Francisco, CA, 1977)
L. Euler, Introductio in Analysin Infinitorum, 1748. English Translation: John Blanton, Introduction to Analysis of the Infinite, vol. I, II (Springer, Berlin, Heidelberg, 1988)
P. Verhulst, Corresp. Math. Phys. 10, 113 (1838)
D. Zwillinger, Handbook of Differential Equations, 3rd edn. (Academic, San Diego, CA, 1998)
M. Eigen, Naturwissenschaften 58, 465 (1971)
J. Maynard-Smith, Nature 225, 563 (1970)
R.W. Hamming, Coding and Information Theory, 2nd edn. (Prentice-Hall, Englewood Cliffs, NJ, 1986)
J. Rogers, G. Joyce, Nature 402, 323 (1999)
J.S. Reader, G.F. Joyce, Nature 420, 841 (2002)
P. Schuster, Physica D 107, 351 (1997)
P. Gitchoff, G.P. Wagner, Complexity 2(1), 37 (1998)
P.F. Stadler, R. Seitz, G.P. Wagner, Bull. Math. Biol. 62, 399 (2000)
B.R.M. Stadler, P.F. Stadler, M. Shpak, G.P. Wagner, Z. Phys. Chem. 216, 217 (2002)
D.E. Goldberg, Genetic Algorithms in Search, Optimization, and Machine Learning (Addison-Wesley, Reading, MA, 1989)
L.M. Schmitt, Theor. Comput. Sci. 259, 1 (2001)
J.E. Barrick, D.S. Yu, H. Jeong, T.K. Oh, D. Schneider, R.E. Lenski, J.F. Kim, Nature 441, 1243 (2009)
Z.D. Blount, Z. Christina, R.E. Lenski, Proc. Natl. Acad. Sci. USA 105, 7898 (2008)
R.E. Lenski, M.R. Rose, S.C. Simpson, S.C. Tadler, Am. Nat. 38, 1315 (1991)
P. Schuster, Rep. Prog. Phys. 69, 1419 (2006)
D.R. Mills, R.L. Peterson, S. Spiegelman, Proc. Natl. Acad. Sci. USA 58, 217 (1967)
S. Spiegelman, Q. Rev. Biophys. 4, 213 (1971)
G.F. Joyce, Angew. Chem. Int. Ed. 46, 6420 (2007)
C.K. Biebricher, M. Eigen, W.C. Gardiner, Jr., Biochemistry 22, 2544 (1983)
C.K. Biebricher, M. Eigen, W.C. Gardiner, Jr., Biochemistry 23, 3186 (1984)
C.K. Biebricher, M. Eigen, W.C. Gardiner, Jr., Biochemistry 24, 6550 (1985)
A. Lescoute, N.B. Leontis, C. Massire, E. Westhof, Nucl. Acids Res. 33, 2395 (2005)
N.B. Leontis, A. Lescoute, E. Westhof, Curr. Opin. Struct. Biol. 16, 279 (2006)
I.L. Hofacker, P. Schuster, P.F. Stadler, Discr. Appl. Math. 89, 177 (1998)
S. Klussmann (ed.), The Aptamer Handbook. Functional Oligonucleotides and Their Applications (Wiley-VCH, Weinheim, Germany, 2006)
M. Zuker, P. Stiegler, Nucl. Acids Res. 9, 133 (1981)
I.L. Hofacker, W. Fontana, P.F. Stadler, L.S. Bonhoeffer, M. Tacker, P. Schuster, Monatsh. Chem. 125, 167 (1994)
M. Zuker, Science 244, 48 (1989)
S. Wuchty, W. Fontana, I.L. Hofacker, P. Schuster, Biopolymers 49, 145 (1999)
C. Flamm, W. Fontana, I.L. Hofacker, P. Schuster, RNA 6, 325 (1999)
M.T. Wolfinger, W.A. Svrcek-Seiler, C. Flamm, I.L. Hofacker, P.F. Stadler, J. Phys. A Math. Gen. 37, 4731 (2004)
J.S. McCaskill, Biopolymers 29, 1105 (1990)
M. Mandal, B. Boese, J.E. Barrick, W.C. Winkler, R.R. Breaker, Cell 113, 577 (2003)
C.J. Thompson, J.L. McBride, Math. Biosci. 21, 127 (1974)
B.L. Jones, R.H. Enns, S.S. Rangnekar, Bull. Math. Biol. 38, 15 (1976)
M. Eigen, P. Schuster, Naturwissenschaften 64, 541 (1977)
M. Eigen, J. McCaskill, P. Schuster, Adv. Chem. Phys. 75, 149 (1989)
E. Seneta, Non-negative Matrices and Markov Chains, 2nd edn. (Springer, New York, NY, 1981)
N. Rohde, H. Daum, C.K. Biebricher, J. Mol. Biol. 249, 754 (1995)
E. Domingo, D. Szabo, T. Taniguchi, C. Weissmann, Cell 13, 735 (1978)
E. Domingo, J. Holland, in RNA Genetics. Vol. III: Variability of Virus Genomes, ed. by E. Domingo, J. Holland, P. Ahlquist (CRC Press, Boca Raton, FL, 1988), pp. 3–36
E. Domingo (ed.), Quasispecies: Concepts and Implications for Virology (Springer, Berlin, Heidelberg, 2006)
J. Swetina, P. Schuster, Biophys. Chem. 16, 329 (1982)
P.E. Phillipson, P. Schuster, Modeling by Nonlinear Differential Equations. Dissipative and Conservative Processes, World Scientific Series on Nonlinear Science A, vol. 69 (World Scientific, Singapore, 2009)
T. Wiehe, Genet. Res. Camb. 69, 127 (1997)
P. Schuster, Theory Biosci. 130, 17 (2011)
P. Schuster. Quasispecies and error thresholds on realistic fitness landscapes (2010). Preprint
L. Jiang, A.K. Suri, R. Fiala, D.J. Patel, Chem. Biol. 4, 35 (1997)
B. Derrida, L. Peliti, Bull. Math. Biol. 53, 355 (1991)
J.W. Drake, Proc. Natl. Acad. Sci. USA 90, 4171 (1993)
E. Domingo (ed.), Virus Res. 107(2), 115 (2005)
J.J. Bull, L. Ancel Myers, M. Lachmann, PLoS Comput. Biol. 1, 450 (2005)
J.J. Bull, R. Sanjuán, C.O. Wilke, J. Virol. 81, 2930 (2007)
M. Kimura, Nature 217, 624 (1968)
M. Kimura, The Neutral Theory of Molecular Evolution (Cambridge University Press, Cambridge, 1983)
P. Schuster, J. Swetina, Bull. Math. Biol. 50, 635 (1988)
M. Eigen, P. Schuster, Naturwissenschaften 65, 7 (1978)
R.A. Blythe, A. McKane, J. Stat. Mech. Theor. Exp. P07018 (2007). doi 10.1088/1742-5468/2007/07/P07018
B.L. Jones, H.K. Leung, Bull. Math. Biol. 43, 665 (1981)
L. Demetrius, P. Schuster, K. Sigmund, Bull. Math. Biol. 47, 239 (1985)
C.W. Gardiner, Stochastic Methods. A Handbook for the Natural and Social Sciences, 4th edn. Springer Series in Synergetics (Springer, Berlin, Heidelberg, 2009)
P. Moran, Proc. Camb. Philos. Soc. 54, 60 (1958)
P. Moran, The Statistical Processes of Evolutionary Theory (Clarendon Press, Oxford, 1962)
M.A. Nowak, Evolutionary Dynamics: Exploring the Equations of Life (The Belknap Press of Harvard University Press, Cambridge, MA, 2006)
W. Fontana, P. Schuster, Biophys. Chem. 26, 123 (1987)
W. Fontana, W. Schnabl, P. Schuster, Phys. Rev. A 40, 3301 (1989)
W. Fontana, P. Schuster, Science 280, 1451 (1998)
D.T. Gillespie, J. Comput. Phys. 22, 403 (1976)
D.T. Gillespie, J. Phys. Chem. 81, 2340 (1977)
D.T. Gillespie, Annu. Rev. Phys. Chem. 58, 35 (2007)
W. Fontana, P. Schuster, J. Theor. Biol. 194, 491 (1998)
P. Schuster, in Evolutionary Dynamics – Exploring the Interplay of Accident, Selection, Neutrality, and Function, ed. by J.P. Crutchfield, P. Schuster (Oxford University Press, New York, NY, 2003), pp. 163–215
M.A. Huynen, P.F. Stadler, W. Fontana, Proc. Natl. Acad. Sci. USA 93, 397 (1996)
B.R.M. Stadler, P.F. Stadler, G.P. Wagner, W. Fontana, J. Theor. Biol. 213, 241 (2001)
A. Kupczok, P. Dittrich, J. Theor. Biol. 238, 726 (2006)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Schuster, P. (2011). Physical Principles of Evolution. In: Meyer-Ortmanns, H., Thurner, S. (eds) Principles of Evolution. The Frontiers Collection. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-18137-5_2
Download citation
DOI: https://doi.org/10.1007/978-3-642-18137-5_2
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-18136-8
Online ISBN: 978-3-642-18137-5
eBook Packages: Physics and AstronomyPhysics and Astronomy (R0)