Abstract
Although extant data favour centromere being an epigenetic structure, it is also clear that centromere formation is based on DNA, in particular, tandemly repeated satellite DNA and its transcripts. Presence of conserved structural motifs within satellite DNAs such as periodically distributed AT tracts, protein binding sites, or promoter elements indicate that despite sequence flexibility, there are structural determinants that are prerequisite for centromere function. In addition, existence of functional centromeric DNA transcripts indicates possible importance of structural elements at the level of RNA secondary or tertiary structure. Rapid centromere evolution is explained by homologous recombination followed by extrachromosomal rolling circle replication. This could lead to amplification of different satellite sequences within a genome. However, only those satellites that have inherent centromere-competence in the form of structural requirements necessary for centromere function are after amplification fixed in a population as a new centromere.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
3.1 Introduction
The centromere is a region of the chromosome that enables the accurate partition of newly replicated sister chromatids between daughter cells during mitosis and meiosis. It holds sister chromatids together and through its centromere DNA–protein complex known as the kinetochore binds spindle microtubules to bring about accurate chromosome movements (Dobie et al. 1999). In addition, centromere regulates progression of cell cycle and is critical in sensing completion of metaphase and triggers the onset of anaphase (Nasmyth 2002). It is visible as the primary constriction on metaphase chromosome.
Centromeric DNA sequences and proteins have been characterized in different organisms, ranging from yeast to human. While a number of proteins shares homology among evolutionarily distant organisms, centromeric DNA sequences differ significantly even among closely related species and evolve rapidly during speciation (Malik and Henikoff 2002). The lack of conservation of centromere DNA could be the characteristic of a single organism as illustrated by neocentromere formation from different genomic sequences in humans (Marshall et al. 2008). Formation of a neocentromere occurs as a result of chromosomal rearrangement that leads to the loss of normal centromere. Most neocentromeres, however, share no sequence homology to normal centromere. Such a plasticity of centromeric DNA could be explained by epigenetic control of centromere function, which does not depend absolutely on primary DNA sequence (Dawe and Henikoff 2006). According to such concept, centromere activation or inactivation might be caused by modifications of chromatin. Such acquired chromatin epigenetic modifications are then inherited from one cell division to the next. Concerning centromere-specific chromatin modification, it is now evident that all centromeres contain a centromere specific histone H3 variant, CenH3, which replaces histone H3 in centromeric nucleosomes and provides a structural basis that differentiates the centromere from the surrounding chromatin. This modified histone H3 is known under different names such as CENP-A (humans), Cid (Drosophila melanogaster), or Cse4 (Saccharomyces cerevisiae) (reviewed in Black and Bassett 2008; see Chap. 1 in this book). CenH3 is characteristic not only for normal centromeres but also for neocentromeres and is essential for the establishment and maintenance of centromere function. Centromeric nucleosomes are specific not only by the presence of CenH3, but also by their internal organization. They seem to be organized as a tetramer composed of one molecule each of CenH3, H2A, H2B, and H4, different from the octamer found in bulk nucleosomes (Dalal et al. 2007). CenH3 chromatin is localized in the inner kinetochore plate and it seems that it exhibits greater conformational rigidity necessary to maintain the architecture during metaphase when tension pulls the kinetochore towards the poles (reviewed in Vagnarelli et al. 2008).
3.2 Types of Centromere
Although extant data favour centromere being an epigenetic structure, it is also clear that centromere formation is based on DNA, and as new results suggest, also very probably on RNA. A most simple centromere characteristic for budding yeast S. cerevisiae is referred as a point centromere, as it encompasses a short distinct DNA sequence of approximately 125 bp, which contains no repetitive DNA. This sequence specifies a kinetochore formation and such simple centromere binds a single microtubule (Kalitsis 2008). More complex, regional centromeres are common for higher eukaryotes, including fission yeast Schizosaccharomyces pombe. They encompass longer, usually Mb size arrays composed of repetitive sequences and form a larger kinetochore that interacts with a number of microtubules. The common feature of regional centromeres across the wide species range, which includes Arabidopsis thaliana, rice, maize, D. melanogaster, and humans, is the presence of satellite DNA as their predominant component (Schueler et al. 2001; Kumekawa et al. 2001; Sun et al. 2003; Jin et al. 2004; Zhang et al. 2004). In the case of human chromosomes, the main centromeric component is alpha satellite DNA. Human alpha satellite DNA makes up 3–5% of each chromosome and the fundamental repeat unit is based on diverged 171 bp monomers. Monomers are tandemly arranged into long homogenous arrays of 250 kb to more than 4 Mb per chromosome (Ugarković 2008a). Alpha satellite DNA is not absolutely necessary for centromere formation, because in its absence euchromatic DNA is capable of being activated to form a neocentromere (Amor and Choo 2002). However, studies of de novo chromosome formation have revealed the preferential formation of centromere on stretches composed of tandemly repeated satellite DNA (Grimes et al. 2002). For example, de novo assembly of human centromere occurs on alpha satellite DNA array, which contains a 17 bp binding motif for centromeric protein B (CENP-B) known as CENP-B box (Grimes et al. 2002; Masumoto et al. 2004). The studies show that alpha satellite is a preferred substrate for centromere formation and that CENP-B box plays an essential role in centromere establishment. However, once established, centromere seems to be further propagated and maintained without CENP-B protein (Okada et al. 2007).
These examples reveal that point centromeres are restricted completely to particular DNA sequence, while in regional centromere this restriction is a partial one. On the other hand, there are examples when centromeres are not localized to any particular chromosomal region. Such diffuse centromeres of holocentric chromosomes of nematodes are distributed along the lengths of the chromosomes attaching to microtubules at many sites (Maddox et al. 2004). The character of DNA sequences that are responsible for the establishment of diffuse centromeres is not defined. However, sequencing of genome of nematode Caenorhabditis elegans revealed the presence of many families of short interspersed repeats. Some of them, after cloning into suitable vectors and introduction into yeast S. cerevisiae are shown to contribute to increased mitotic stability of plasmids, indicative of centromeric role (Kalitsis 2008).
In addition to DNA and proteins, RNA seems also to be a structural component of centromere. Transcripts of alpha satellite DNAs have been shown to be a functional component of the kinetochore, participating in recruitment of kinetochore proteins (Wong et al. 2007). In addition, ribonucleoprotein complexes are required for mitotic spindle assembly (Blower et al. 2005). All these data point to an important role for DNA and RNA, in particular, tandemly repeated satellite DNA and its transcripts in centromere/kinetochore establishment and function. New findings related to evolutionary constraints on centromeric satellite DNAs also shed more light on the possible role of these sequences. Despite sequence heterogeneity among species, the common pattern of DNA structural motifs required for centromere specification is beginning to be discerned.
3.3 Evolutionary Mechanisms Affecting Centromeric DNA
3.3.1 Role of Stochastic Processes
In general, centromeric regions are considered the most rapidly evolving compartments in the eukaryotic genome. In the case of point centromere, high mutation rate seems to be responsible for such a rapid sequence change (Bensasson et al. 2008). Regional centromeres, however, which are characterized by repetitive structure, mostly in the form of tandem satellite DNA repeats exhibit change not only in sequence but also in repeat copy number. Therefore, evolution of regional centromere proceeds not only by mutations but also by recombination. Recombinational mechanisms such as gene conversion and unequal crossingover affect repetitive DNAs and are responsible for the rapid horizontal spread of newly occurring mutations among monomers within a repetitive family. This results ultimately in homogenization of changes among repeats within the genome and their subsequent fixation in members of reproductive populations in a process known as molecular drive (Dover 1986). This mode of horizontal evolution, characteristic for repetitive families, is known as concerted evolution. The process of homogenization occurs at species-specific rates but is faster and independent of the mutation rate. As a result of concerted evolution, repeats of a satellite DNA within regional centromere exhibit high homology within a species. However, because of the same process, different mutations are randomly fixed in reproductively isolated populations, causing rapid divergence of centromere sequence among species.
Besides being responsible for the spreading of mutations horizontally through members of the repetitive family, unequal crossingover is also responsible for changes in repetitive DNA copy number, affecting in this way the length of centromere arrays (Smith 1976). Theoretical studies on satellite DNA dynamics explain its loss from the genome by unequal crossingover, demonstrating an inverse correlation between the rate of unequal crossingover and the preservation time of the satellite DNA (Stephan 1986). Satellite DNAs can also increase in copy number either by replication slippage, rolling circle replication, and conversion-like mechanisms in a relatively short evolutionary time (reviewed in Ugarković and Plohl 2002). The outcome of all these mechanisms affecting satellite DNA arrays is a high turnover of centromeric and pericentromeric regions of the eukaryotic genome. On the model of mouse cells, it has been shown that centromere mitotic recombination occurs at a much higher frequency than chromosome arm recombination, and is controlled by the epigenetic state of centromeric heterochromatin, in particular by centromeric DNA methylation (Jaco et al. 2008). Methylation of centromeric DNA represses illicit recombination at repeated satellite DNA and is suggested to be important for the maintenance of centromere integrity. On the other hand, the reduced frequency of recombination in the neighborhood of centromeres during meiosis, relative to the rest of chromosome, has been documented in D. melanogaster and many other organisms (Charlesworth et al. 1986; Stephan 2007). It has been proposed that the reduced meiotic recombination could be the consequence of natural selection, which lowers the unequal exchange between repeats and in this way prevents significant change in repetitive array lengths. Repeat length change could lead to the variation in the number of microtubule binding sites per chromosome, which can further result in nondisjunction events and aneuploidy.
3.3.2 Role of Natural Selection
In addition to stochastic, random processes that affect centromeric DNA and induce its rapid sequence evolution, there are indications for the natural selection shaping evolution of centromeric DNA sequence (Ugarković 2005). This indication is based on the extreme sequence preservation and wide evolutionary distribution of some satellite DNAs as well as on the conservation of particular structural motifs. Selection was first thought to influence satellite DNA sequences following the observation of nonrandom distribution of variability along the satellite monomers, resulting in constant and variable regions in Arabidopsis thaliana and human alpha satellite DNA (Romanova et al. 1996; Heslop-Harrison et al. 1999). Nonrandom pattern of variability was subsequently detected in many centromeric satellites (Hall et al. 2003; Mravinac et al. 2004; 2005), as well as preservation of variability at particular positions within a satellite in different populations (Feliciello et al. 2005). Restricted variability could be probably related to interaction of satellite DNAs with specific proteins necessary for heterochromatin and centromere formation as well as to the role of satellite DNAs in controlling gene expression.
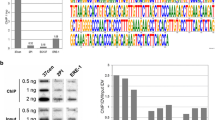
The best characterized satellite DNA-binding protein is human centromere protein B (CENP-B), which binds to a 17 bp motif in human alpha satellite DNA known as the CENP-B box (Masumoto et al. 1989). Proteins homologous to CENP-B have been found in many eukaryotes, including the fission yeast S. pombe, and motifs that are 60–70% similar to the CENP-B box have been detected in diverse centromeric repeats of mammals and insects (Kipling and Warburton 1997; Mravinac et al. 2004; Fig. 3.1). Although only 23% of repeats in human α satellite DNA have a functional CENP-B box, it seems to be essential for the assembly of centromere-specific chromatin and centromere establishment, but not for the centromere maintenance (Ohzeki et al. 2002; Basu et al. 2005; Okada et al. 2007).
Evolutionary constraints on centromeric satellite DNAs. Structural requirements posed on satellite DNAs which enable them to be retained in the genome as members of satellite library and to be potentially expanded into a “new” centromere might include periodic clusters of A + Ts, binding sites for centromeric proteins such as CENP-B box, or promoter elements necessary for active transcription. Periodic distribution of AT tracts leads to curvature of the DNA helix axis and formation of superhelical tertiary structure thought to be important for heterochromatin establishment. Transcription of satellite DNAs proceeds in the form of either double-stranded RNA (dsRNA) or single-stranded RNA (ssRNA). Long ssRNAs are required for the association of kinetochore proteins, while dsRNA is processed into small interfering RNAs (siRNAs) that participate in heterochromatin formation. Constraints on satellite RNA secondary and/or tertiary structure could exist in order to preserve its ability to bind kinetochore proteins
Satellite DNAs are usually AT rich but A’s or T’s are not randomly distributed within the sequence. Clustering of A or T and regular phasing of A or T ≥3 tracts has been reported for many different satellite DNAs, including human alpha satellite DNA (Martinez-Balbas et al. 1990; Ugarković et al. 1996a; Fig. 3.1). Periodic distribution of AT tracts usually induces curvature of the DNA helix axis and formation of tertiary structure in the form of a superhelix (Fitzgerald et al. 1994). Such a structure is thought to be important for the tight packing of DNA and proteins in heterochromatin (Ugarković et al. 1992).
Palindromic sequences that could potentially lead to the formation of dyad structures are common elements of centromeric and pericentromeric satellite DNAs in budding yeast, insects, and human (Tal et al. 1994; Ugarković et al. 1996b; Zhu et al. 1996). It is not clear if they perform some function, but it can be hypothesized that some palindromic sequences could be recognized by DNA binding proteins, such as transcription factors. Some homeodomain proteins like Pax3, which is known to play an important role during neurogenesis, bind short palindromes present within major mouse satellite DNA (personal communication). The recent investigation has revealed that the topoisomerase II recognizes and cleaves a specific hairpin structure formed by alpha satellite DNA (Jonstrup et al. 2008). It has been suggested that a subpopulation of the cellular topoisomerase II located at centromeres plays a role for sister chromatid cohesion in the centromeric region. The hairpin cleavage therefore could be connected to a cohesion role of topoisomerase II at centromeres.
Other functional motifs and regulatory elements for RNA polymerase (pol) II and RNA pol III are predicted in some satellite sequences (Renault et al. 1999; Fig. 3.1). Human satellite III, which is specifically expressed under stress, has a binding motif for the heat shock transcription factor 1 that drives RNA pol II transcription (Metz et al. 2004). In schistosome satellite DNA, which encodes an active ribozyme, a functional RNA pol III promoter is present (Ferbeyre et al. 1998). The sequence of satellite 2 found in the newts Notophthalmus viridescens and Triturus vulgaris meridionalis contains a functional analogue of the vertebrate small nuclear RNA (snRNA) promoter that is responsible for RNA pol II transcription (Coats et al. 1994). Promoters for RNA Pol II are also the characteristic of centromeric satellite DNAs from beetle species Palorus ratzeburgii and Palorus subdepressus (Pezer and Ugarković 2008a; 2009). In general, the presence of functional elements within centromeric satellite DNA sequences points to the role of natural selection in preserving such motifs.
Some centromeric satellites, however, exhibit sequence conservation of the whole monomer sequence for long evolutionary periods. Extreme sequence conservation of two satellite DNAs that represent major pericentromeric repeats in the coleopteran insect species Palorus ratzeburgii and Palorus subdepressus has been reported (Mravinac et al. 2002; 2005). These satellites are present in many coleopteran species at a low copy number and their sequences have remained unchanged for 60 million years. This remarkable antiquity and sequence conservation are also characteristic of human alpha satellite DNA, which has been detected as a rare, highly conserved repeat in evolutionary distant species such as chicken and zebrafish (Li and Kirby 2003). This complete sequence conservation and the wide evolutionary distribution of some satellite sequences has led to the assumption that, in addition to participating in centromere formation, they could perform some other role possibly acting as cis-regulatory elements of gene expression.
In addition to relatively conserved regions found in diverse centromeric satellites, other more variable regions also exist. Variable regions might also be functionally important owing to their interaction with rapidly evolving proteins. Such an example is the centromere-specific histone, CenH3, which replaces histone H3 in centromeric nucleosomes and is required for proper chromosome distribution during cell division (Henikoff and Dalal 2005). Unlike the highly conserved histone H3, CenH3 is divergent and subject to the influence of positive selection, which particularly affects the sites that potentially interact with satellite DNA (Cooper and Henikoff 2004; see Chap. 2 in this book, Sect. 3.2.2). It has been proposed that variable regions within satellite DNA sequence drive the adaptive evolution of specific centromeric histones. In addition to CenH3, other kinetochore proteins exhibit rapid sequence evolution in fly D. melanogaster as well as in worm C. elegans, while in mammals, plants, and fungi the rate of evolution is much lower (Meraldi et al. 2006).
3.4 Point Centromere DNA and Its Evolution
While in most animals and plants species, centromeres are complex and regional, encompassing long Mb size arrays of highly repetitive, satellite DNA, centromeres in Saccharomyces yeast and several other budding yeasts such as Candida glabrate and Kluyvermyces lactis occupy a very small region of approximately 120 bp and are referred to as point centromere. The centromeric sequence contains no repetitive DNA and consists of three functionally distinct regions: CDEI and CDEIII, which are 8 bp and approximately 25 bp long, respectively, and represent protein binding sites, as well as of CDEII, approximately 90 bp long, which binds centromere-specific histone Cse4 (Hegemann and Fleig 1993). CDEI and CDEIII elements exhibit sequence conservation among different budding yeast species. Mutations in CDEI impair but do not abolish function in mitosis and meiosis, while single base change or short deletions within CDEIII completely inactivate the centromere. CDEII from different chromosomes within same species are highly divergent, up to 60%, but functionally interchangeable (Clarke and Carbon 1983), suggesting that binding of Cse4 is not sequence specific. However, changes in AT content, which is averaging 90%, pattern of homopolymer runs of A’s and T’s, and length can disrupt centromere function (Baker and Rogers 2005). This indicates that DNA curvature or flexibility which depends on the pattern of distribution of A and T tracts could be related to centromere function. It has been shown that bent and unbent CDEII DNAs, differing at only six nucleotides, displayed a 60-fold difference in mitotic chromosome loss rates. Since AT rich sequences that exhibit homopolymer bias such as CDEII are found predominantly at centromeres of various species, this seems to represent a type of «code» that partially can explain centromere identity.
Periodic distribution of A and T tracts represents a commonality between point Saccharomyces centromere and complex regional centromeres of higher organisms. Survey of more than hundred different satellite DNAs revealed that approximately 50% of them exhibit DNA curvature induced by periodic distribution of A or T tracts (Fitzgerald et al. 1994). Such highly nonrandom patterns of A’s and T’s characterized by homopolymer runs of 5–7 nucleotides might imply influence of selection to preserve mitotic centromere function in Saccharomyces as well as in many higher eukaryotes (Baker and Rogers 2005).
Comparison of near-complete sequences of chromosome III from three closely related lineages of the wild yeast Saccharomyces paradoxus, which is a relative of S. cerevisiae, has shown that the centromere region CDEII is the most rapidly evolving part of the chromosome (Bensasson et al. 2008). This centromere region is evolving faster than sequences that are not under selective constraint. Such rapid evolution could result from elevated mutation rate or influence of positive selection. It has been proposed that positive selection drives rapid fixation of mutations in centromeric regions by imposing a bias in favour of retaining mutations. The positive selection might be due to the advantage conferred to mutated centromere during female meiosis known as «centromere drive hypothesis» (Malik and Henikoff 2002; see Chap. 2 in this book). However, in the case of point Saccharomyces centromere, it seems that elevated mutation rate within CDEII is responsible for the rapid evolution and not positive selection. What on the other hand could induce such a high substitution rate in the yeast centromere region is not clear.
While elevated mutation rate is considered as a major contributor to rapid evolution of point centromere, recombinational mechanisms such as unequal crossing over and gene conversion that preferentially affect segments of repetitive DNA are major genetic mechanisms governing evolution of complex regional centromeres (Ugarković and Plohl 2002). Comprehensive phylogenetic and structural analysis of centromere/kinetochore proteins from different species revealed that organisms with regional and point centromeres have a common ancestor, a fungus containing a regional centromere, implying that simple, point centromere arose from complex, regional centromere (Meraldi et al. 2006).
Different from the regional centromeres that generally have no transcribed genes in their vicinity, transcribed genes are found very close to point S. cerevisiae centromeres (Westermann et al. 2007). It is, however, not known if transcripts are structural component of point Saccharomyces centromere, as found for complex regional centromeres (Wong et al. 2007).
3.5 Regional Centromere DNA and Its Evolution
Regional centromere encompasses from 1 kb in budding yeast Candida albicans (Sanyal et al. 2004) to few megabases in human (Schueler et al. 2001), and is typically composed of repetitive DNA elements, mostly in the form of tandemly repeated satellite DNAs. A single satellite DNA can predominate at the centromeric regions such as the case of alpha satellite DNA at human centromeres (Schueler et al. 2001). In D. melanogaster and beetle species Tribolium madens, two or more different satellites are interspersed within centromeric regions (Durajlija-Žinić et al. 2000; Sun et al. 2003).
Different centromeric satellite DNAs may persist in the genome usually at centromeric or pericentromeric locations for long evolutionary time forming a collection or library of satellite sequences shared among related lineages (Fry and Salser 1977). The amount of satellite DNAs in a single centromere can be increased or reduced dramatically in a short time frame. Such rapid turnover characteristic for regional centromere evolution can be explained by differential amplification or expansion of satellite DNAs from the library in any species (Ugarković and Plohl 2002). The first experimental demonstration of a satellite DNA library is found in the insect genus Palorus (Coleoptera), where all examined species posses a common collection of centromeric satellite DNAs (Meštrović et al. 1998). A different single satellite is significantly amplified or expanded in each of the different species, resulting in species-specific satellite DNA profiles. The existence of satellite libraries is supported for different groups of species, including plants, nematodes, insects, and mammals, as well as their preferential localization within pericentromeric and centromeric regions (King et al. 1995; Vershinin et al. 1996; Cesari et al. 2003; Lin and Li 2006; Meštrović et al. 2006; Bruvo-Mad¯arić et al. 2007; Kawabe and Charlesworth 2007). In the marsupial genus Macropus, three satellite DNAs are involved in the creation of centromeric arrays in nine examined species (Bulazel et al. 2007; see Chap. 4 in this book). Each species, however, has experienced different expansion and contraction of individual satellites. In Bovini, six related centromeric satellite DNAs are shared among species fluctuating considerably in relative amounts (Nijman and Lenstra 2001).
3.5.1 Human Centromeric DNA
Different satellite DNAs that coexist in the same species can vary significantly in their sequence homogeneity and are considered as independent evolutionary units. In addition, each satellite DNA can exist in the form of different, usually chromosome-specific satellite subfamilies (reviewed in Ugarković and Plohl 2002). All primate species share alpha satellite DNA, which in the form of different subfamilies represents the major component of all centromeres (Lee et al. 1997). Alpha satellite is composed of two basic types of repeat units: a 171 bp monomer and higher order repeats (HOR). Higher order repeats have complex repeat units composed of up to 30 diverged 171 bp monomers (Alexandrov et al. 2001) and are characteristic of centromeres of higher primates, while in the genomes of lower primates, monomeric alpha satellite repeats prevail and comprise long centromeric arrays.
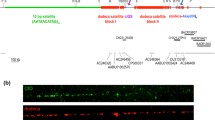
The centromeric region has been characterized in detail for the human X chromosome (Fig. 3.2; Schueler et al. 2001). Two evolutionarily distinct classes of alpha satellite are present within the centromeric region of the X chromosome. One class encompasses an approximately 3 Mb array of alpha satellite DNA known as DXZ1, which is present at the primary constriction and is X chromosome specific. This region is defined by a 2.0 kb higher-order repeat, which consists of twelve 171 bp monomers. The canonical higher order repeats are highly homogenous, showing an average of 1–2% divergence on the same or different X chromosome. Mapping of deletion chromosomes has delimited the functional centromere of the X chromosome to the higher order alpha satellite array in the DXZ1 region. The other class is composed of ∼450 kb region located between DXZ1 and expressed sequences on the short arm of chromosome X, also highly enriched in alpha satellite. The 450 kb junction region is characterized by tandemly repeated monomeric repeat structure and the monomers exhibit higher mutual divergence relative to higher order repeats within DXZ1 region.
Organization of alpha satellite DNA within centromere of human X-chromosome based on data from Schueler et al. (2005). DXZ1 region of 3 Mb in which primary constriction is located is composed of tandemly repeated higher order repeats (HORs). HORs are mutually highly homologous exhibiting 1–2% divergence. DXZ1 array is flanked on both sites by region of approximate size of 450 kb, which is composed mostly of alpha satellite monomers. Alpha satellite monomers within 450 kb array exhibit divergence between 20% and 30% and are interspersed with transposable elements such as LINE and SINE. Higher order repeats participate in kinetochore formation while diverged monomers contribute to heterochromatin establishment. Phylogenetic analysis resolves alpha satellite monomers within 450 kb region into four subfamilies, while monomers within DXZ1 array form distinct, fifths alpha satellite subfamily. Adjacent to 450 kb region is euchromatic DNA
Based on the presence of interspersed LINE elements within arrays of alpha satellite DNA as well as on the phylogenetic analysis of primate species, particular alpha satellite subdomains can be defined and their age can be estimated. According to such analyses, human X chromosome monomeric alpha satellite arrays are divided into four age groups: 35–65 million years (Myr), 25–35 Myr, 15–25 Myr, 7–15 Myr, while the DXZ1 region which is based on higher order repeats is the most recent one with an approximate age between 2 and 7 Myr (Schueler and Sullivan 2006). Monomeric alpha satellite DNA predates higher order arrays of alpha satellite and may represent direct descendants of the ancestral primate centromere sequence. Comparison with centromeric alpha satellite DNA sequences in other primate species revealed that alpha satellite DNA has evolved through proximal expansion events occurring within the central active region of the centromere (Fig. 3.3; Schueler et al. 2005). Each addition of new material splits the previous centromeric DNA and moves it distally onto each arm, while the newly added sequence confers centromere function. The alpha satellite region immediately proximal to the euchromatin chromosome arm is a remnant of the ancestral primate X centromere. A higher order satellite array located within the DXZ1 domain evolved as a replacement for the monomeric alpha satellite repeat. Highly homogenous arrays of higher-order alpha satellite represent a relatively recent addition to the primate genome, emerging near the orangutan/gorilla split. Based on the molecular analysis of the human X-chromosome centromere, it becomes evident that alpha satellite regions have evolved through a series of events, resulting in the addition and amplification of “new” subfamilies that have partially replaced the “old” ones (Fig. 3.3).
Model of evolution of primate centromeric region from the ancestral primate to humans. The series of amplification events are responsible for the spreading of “new” alpha satellite subfamilies and replacement of “old” ones, which however remain preserved in genome in lower number of copies (differently dashed rectangles). In each round of amplification, the “old” centromere is split and moved distally onto each arm while the newly added sequence confers centromere function. The “old” subfamilies are based on tandemly repeated monomers, but the most recently amplified subfamily is based on tandemly repeated HOR. This subfamily comprises centromeric regions in humans and other great ape. The model is based on data on human X chromosome centromere structure (Schueler et al. 2005)
The kinetochore domain composed of higher order repeats comprises one half to two thirds of the alpha satellite DNA located at human centromeres. The remainder of alpha satellite arrays composed predominantly of diverged tandemly repeated monomers contributes to pericentromeric heterochromatin establishment, which is necessary for chromatid cohesion.
3.5.2 Model of Centromere Evolution Based on Satellite DNA Library
Rapid sequence evolution is characteristic of complex regional centromeres. Comparison of alpha satellite arrays from orthologous chromosomes of chimps and human revealed higher divergence of centromeric regions relative to the pericentromeric ones (Rudd et al. 2006). To explain rapid evolution of centromeric DNA, a «centromere drive hypothesis» has been introduced (Malik and Henikoff 2002; see Chap. 2 in this book). According to it, rapid evolution of centromeric DNA is caused by positive selection that imposes a bias in favour of retaining mutations in centromere region. The positive selection is proposed to be due to the advantage conferred to mutated centromere during female meiosis. Such centromere has a higher affinity for centromeric chromatin proteins and is the most successful at being incorporated into the functional germ cells (i.e., the oocyte). Other centromeres are then forced to adopt the same sequence and protein variants to segregate efficiently. According to the “centromere drive hypothesis,” evolution of the centromere proceeds through «de novo» adoption of «new», previously noncentromeric sequences that are repeatedly introduced into the genome (Dawe and Henikoff 2006).
On the other hand, based on the library hypothesis, it can be proposed that centromere is formed from already adapted sequences with certain structural characteristics that enable them to confer a centromeric role or to perform some other function such as regulation of gene expression (Ugarković 2005; 2008b; Fig. 3.1). Such sequences after exaptation, that is, after becoming functional, can reside within the genome for long evolutionary periods and create a satellite DNA library. The content of the library is constantly evolving, and new sequences can be generated and added into the library such as the case of alpha satellite complex HORs, which appear later in the evolution of primate lineage (Alexandrov et al. 2001). On the other hand, some «old» centromeric satellite repeats can be lost in particular lineages as shown for centromeric satellites in species of grass (Lee et al. 2005). Removal of centromeric satellites from the library is probably a stochastic process mediated by mechanisms of unequal crossing over and illegitimate recombination (Stephan 1986; Ma and Jackson 2006).
Centromeric and pericentromeric satellite sequences from the library can undergo recurrent repeat copy number expansion and contraction in divergent lineages (Fig. 3.4). Such changes in copy number seem to be random and do not correlate with phylogeny of the species as shown for the insect genus Pimelia, the marsupial genus Macropus, and the grass species (Pons et al. 2004; Lee et al. 2005; Bulazel et al. 2007). The same satellite sequences can undergo convergent expansion on all chromosomes in different lineages. Although the evolution of centromeric satellite DNA composition does not follow species phylogeny, it parallels chromosome evolution in some karyotypically divergent lineages (Slamovits et al. 2001; Bulazel et al. 2007; see Chap. 4 in this book). The rate of turnover of centromere differs among species ranging from abrupt-saltatory amplification and replacement of “old” centromere in relatively short periods of time, through gradual changes, while in some instances no apparent change occurs for long evolutionary time (Pons et al. 2004). Amplification of a satellite sequences could occur due to unequal crossingover or duplicative transposition (Smith 1976; Ma and Jackson 2006), while the spreading and fixation in population can be influenced by stochastic process of molecular drive (Dover 1986) and by natural selection. The discovery of human extrachromosomal elements originating from satellite DNA arrays in cultured human cells indicates the possible existence of other amplification mechanisms based on extrachromosomal rolling-circle replication (Assum et al. 1993). Satellite DNA-derived extrachromosomal circular DNA is common in plant genomes and is considered as an intermediate in process driving satellite expansion and evolution (Navratilova et al. 2008). It has been proposed that satellite sequences excised from their chromosomal loci via intrastrand homologous recombination could be amplified in this way, followed by reintegration of tandem arrays into the genome (Feliciello et al. 2006). Mechanistic processes inherent to chromosome fusion and translocation have also been supposed to be responsible for contraction and expansion of centromeric satellite DNA arrays (Bulazel et al. 2007).
Model of satellite DNA evolution and centromere formation based on satellite DNA library. Satellite DNAs possessing certain structural features which enable them to become functional are retained in the genome in the form of satellite DNA library. Satellite DNA could have dual function in the genome: either it can be extended into long array and together with its transcripts participates in centromere/kinetochore establishment, or satellite transcripts could act as regulators of gene expression, probably through RNAi mechanism. A stochastic process of differential amplification of satellite DNAs from the library in two related species induced by unequal crossingover, duplicative transposition or extrachromosomal rolling circle replication can lead to the formation of long, uninterrupted arrays. An expanded arrays can replace the previous centromere if it has some selective advantage relative to the «old» centromere, e.g., transmission advantage at meiosis due to some structural characteristic or just due to the higher homogeneity of newly amplified array relative to the «old» one. Such “new” centromere can then be spread through the population by processes of natural selection and molecular drive
A newly expanded satellite array can replace the previous centromere and prevail in the population if it has some selective advantage relative to the «old» centromere, for example, transmission advantage at meiosis due to some sequence or structural characteristic of newly amplified satellite DNA or just due to the higher homogeneity of newly amplified array relative to the «old» one (Fig. 3.4). Based on the structure of the human X chromosome centromere, it can be proposed that high homogeneity and integrity of newly expanded satellite arrays might represent an additional requirement imposed on the centromere. In addition, it seems that a newly expanded array has to be of certain length to become a preferred substrate for centromere formation. This could be related to the number of microtubule binding sites per chromosome necessary to ensure the proper chromosome segregation.
The repetitiveness of satellite DNA has been proposed to be important for orderly packing of nucleosomes (Vogt 1990), and nucleosome crystallization on reverse repeats of alpha satellite DNA support this assumption (Harp et al. 1996; Luger et al. 1997). There is strong indication that a specific set of periodic DNA motifs encoded in tandemly repeated satellite DNA provides signals for specific chromatin organization in the form of distinctive nucleosome arrays characteristic for centromere (Takasuka et al. 2008). It is known that centromeric nucleosomes are organized as a heterotypic tetramer composed of one molecule each of CenH3, H2A, H2B, and H4, different from the octamer found in bulk nucleosomes (Dalal et al. 2007). It is suggested that such nucleosome tetramers distributed orderly on homogenous and uninterrupted satellite arrays represent an accessible surface for kinetochore assembly. Therefore, extension of satellite repeat from the library by stochastic recombinational processes and/or extrachromosomal rolling circle replication might create uninterrupted homogenous array, which could be a favoured substrate for centromere chromatin establishment and microtubule binding relative to the “old” nonhomogenous array interspersed with different transposable elements. Such centromere array exhibiting a slight advantage relative to the “old” one could then be fixed in a population (Fig. 3.4).
3.6 RNA in Centromere Establishment
3.6.1 RNAs as Epigenetic Regulator of Heterochromatin Establishment
Transcripts of centromeric satellite DNAs have been reported in several organisms, including vertebrates, invertebrates, and plants. Transcripts are usually heterogeneous in size and are in some cases strand-specific, while in others transcription proceeds from both DNA strands. Most transcripts are present as polyadenylated RNA in the cytoplasm but some are found exclusively in the nucleus (reviewed in Ugarković 2005). Recently, it has been shown that transcripts derived from tandemly repeated centromeric DNA of the fission yeast S. pombe exist in the form of small 20–25 bp long RNAs that are involved in chromatin modifications and establishment of heterochromatin (Volpe et al. 2002). The chromatin silencing mechanism is initiated by long double-stranded RNA (dsRNA) that arises from bidirectional transcription of repeated centromeric DNA and is further processed by the RNAse III-like ribonuclease Dicer into small interfering RNAs (siRNAs). siRNAs are then loaded into the RNA-induced transcriptional silencing complex (RITS) through their association with the Argonaute protein. RITS also interacts with the RNA-directed RNA polymerase complex (RDRC), which is required for the production of secondary dsRNA and amplification of the silencing signal (Verdel et al. 2004). Both RITS and RDRC associate with the nascent noncoding centromeric RNA transcript, and binding to RITS is probably achieved through the base-pairing of siRNA molecules with nascent RNA and by direct contact with the RNA pol II elongation complex. In addition to siRNAs, the association of RITS with chromatin also requires a histone methyltransferase. Histone H3 methylation at lysine 9 is essential for the recruitment of heterochromatin protein 1 (HP1). This represents an initial step in the formation of heterochromatin. HP1 has several functions at centromere such as silencing gene expression and recombination, promotion of kinetochore assembly, and prevention of erroneous microtubule attachment to the kinetochores (Yamagishi et al. 2008).
Mutations in components of the RNAi pathway lead to the loss of pericentromeric heterochromatin in fission yeast, resulting in mis-segregation of chromosomes (Allshire et al. 1995; Volpe et al. 2002; Fig. 3.5). S. pombe cells deficient in pericentromeric heterochromatin are unable to recruit the chromosome cohesin to centromeres and fail to maintain centromere cohesion (Bernard et al. 2001). It was recently revealed that heterochromatic proteins and RNAi machinery promote CENP-A deposition and kinetochore assembly over the central domain of the fission yeast centromere (Folco et al. 2008). However, absence of these factors does not affect CENP-A deposition on endogenous centromeres or on minichromosome centromeres, which have incorporated CENP-A in previous generation. In general, pericentromeric heterochromatin appears to be an absolute requirement for the establishment of centromere in fission yeast together with central DNA region, which binds CENP-A (cnt region) as well as otr region which contains dg-dh repeats (Folco et al. 2008). In addition to fission yeast, pericentromeric heterochromatin seems to be required for the accurate segregation of chromosomes during mitosis in many eukaryotes, including Drosophila and mammals (Kellum and Alberts 1995; Peters et al. 2001).

Fig. 3.5 Link between centromeric RNA and aneuploidy. Aberrant expression of centromeric satellite DNA affects centromere/kinetochore function and causes abnormality in chromosome segregation. Defects in RNA metabolism could affect heterochromatin maintenance and fidelity in mitosis
RNA interference (RNAi) machinery has been shown to be evolutionary conserved and is proposed to be responsible for pericentromeric heterochromatin formation in different animal species. In addition to S. pombe, siRNAs cognate to satellite DNAs are involved in the epigenetic process of chromatin modification in Arabidopsis and C. elegans (Bernstein and Allis 2005; Grewal and Elgin 2007). In D. melanogaster RNAi seems to be involved in the establishment of heterochromatin in early embryo. Once set, heterochromatin can be maintained in the absence of RNAi in somatic tissues (Huisinga and Elgin 2008). In mammals, however, siRNAs seem not to elicit chromatin modification, although an unidentified RNA component appears to be required for maintaining pericentric heterochromatin (Maison et al. 2002; Wang et al. 2006). In mouse pericentromeric heterochromatin, γ satellite DNA as its major constituent is transcribed as small, approximately 200-nt-long RNA during mitosis, while during G1 and S phase, transcription occurs in the form of long, heterogeneous RNAs (Lu and Gilbert 2007). The transcription is cell-cycle regulated with the highest rate in early S phase and in mitosis, similar to regulation in fission yeast where the peak of transcription occurs at S phase (Chen et al. 2008). Besides being cell-cycle regulated, transcription of mouse pericentromeric heterochromatin is also linked to cellular proliferation.
3.6.2 RNAs as Structural Component of Centromere
Recently it has been shown that long, single-stranded alpha satellite DNA transcripts encompassing a few satellite monomers are functional components of the human kinetochore (Wong et al. 2007; Fig. 3.1). Centromere alpha satellite RNA is required for the assembly of CENPC1, INCENP (inner centromere protein), and survivin (an INCENP-interacting protein) at the metaphase centromere. It also directly facilitates the accumulation and assembly of centromere-specific nucleoprotein components at the interphase nucleolus. The nucleolus sequesters centromeric components such as alpha satellite RNA and centromere proteins for timely delivery to the chromosomes for kinetochore assembly at mitosis. CENP-C has been shown to be an RNA-associating protein that binds alpha satellite RNA, as revealed by in vitro binding assay. The same protein also binds alpha satellite DNA in vivo and obviously has dual RNA- and DNA-binding function (Politi et al. 2002). In mammals, CENP-C evolving rapidly and different from CENP-A (vertebrate CenH3) shows evidence of positive selection (Talbert et al. 2004; see Chap. 2 in this book). It is possible that a pool of CENP-C has a centromere DNA-binding role that persists throughout the cell cycle. The other pool of CENP-C is involved in relocation of alpha satellite RNA and centromere proteins from the nucleolus onto the mitotic centromere.
CENP-B and CENP-C recognize the same subfamilies of alpha satellite DNA, but it is not clear whether CENP-C preferentially recognizes a specific sequence within satellite DNA or RNA. In vitro experiments indicate that CENP-C does not bind a specific DNA sequence, similar to CENP-A which also seems to be a sequence nonspecific binding protein (Politi et al. 2002). However, the existence of binding sites for different proteins in alpha satellite DNA could explain the nonrandom distribution of mutations within a sequence and can give strong support for the influence of selection on the evolution of this satellite DNA sequence.
Numerous examples illustrate the involvement and possible importance of longer RNAs for the formation of centromeric chromatin and for centromere function. RNA encoded by centromeric satellite DNA and retrotransposons, ranging in size between 40 and 200 nt, has been shown to be an integral component of the kinetochore in maize, tightly bound to centromeric histone H3 (Topp et al. 2004). Murine minor satellite DNA associated with the centromeric region is transcribed from both strands, and transcripts are processed into 120 nt RNA, which localizes to the centromere (Bouzinba-Segard et al. 2006). The overexpression of satellite transcripts is impaired by mislocalization of centromere-associated proteins essential for the formation of centromeric heterochromatin. In addition, forced accumulation of transcripts leads to defects in chromosome segregation and impaired centromere function, resulting in aneuploidy (Fig. 3.5). The absence of siRNAs homologous to murine minor satellite indicates that the longer noncoding RNA plays a role in heterochromatin formation and centromere establishment in the murine system. Long, stable transcripts of centromeric satellite DNAs are also the characteristics of some beetle species (Pezer and Ugarković 2008a; 2009). Functional studies reveal that in this animal system an increase in the amount of centromeric satellite DNA transcripts coincides with the irregular chromosome segregation and often leads to aneuploidy. Since functional promoters for RNA polymerase II are detected within satellite DNAs from coleopteran genera Tribolium and Palorus, it is proposed that constitutive expression of centromeric satellites is necessary for proper centromere establishment (Pezer and Ugarković 2008b).
Mitotic and chromosome segregation defects have been reported for fission yeast mutants defective in RNA metabolism (Win et al. 2006). RNase activity of Dis3, a core component of the exosome that is required for the processing of different RNAs, is shown to be required for heterochromatin silencing within the centromere as well as for proper kinetochore formation and establishment of kinetochore–microtubule interactions (Murakami et al. 2007; Buhler et al. 2007). Thus, RNAi-independent degradation of centromeric transcripts also contributes to heterochromatin formation and proper centromere function.
All these examples demonstrate the importance of cellular RNA metabolism for proper chromosome segregation during mitosis (Fig. 3.5). In addition to the relatively well understood RNAi mechanism that moderates heterochromatin establishment in different eukaryotic systems, other mechanisms involving longer RNAs also operate in centromeric chromatin assembly and kinetochore formation. Although these mechanisms are poorly understood, it can be proposed that centromere-encoded longer RNAs could serve as a scaffold for chromatin-remodeling complexes at centromere as well as structural component of kinetochore (Fig. 3.1). It can be proposed that specific secondary and tertiary structures of centromeric RNAs are important for assembly of such complexes.
Based on studies in mammalian and insect systems, it appears that aberrant transcription of noncoding centromeric satellite DNA affects heterochromatin maintenance and fidelity of mitosis (Pezer and Ugarković 2008b; Frescas et al. 2008). This indicates that centromeric RNA is an important functional component of the centromere/kinetochore complex, probably tightly bound to proteins, and subtle changes in centromeric RNA/kinetochore protein ratio affect chromosome stability and segregation (Fig. 3.5). Stoichiometric expression of all kinetochore components including proteins and noncoding centromeric RNA seems to be important for normal kinetochore assembly and function.
Overexpression of noncoding satellite DNAs is characteristic of some tumours. Analysis of transcription of human satellite 2 and α-satellite, which are located in pericentromeric and centromeric heterochromatin, respectively, revealed an elevated level of their expression in ovarian epithelial carcinomas and Wilms tumours, relative to the control (Alexiadis et al. 2007). It can be hypothesized that increased accumulation of noncoding RNA deriving from the two satellite DNAs interferes with heterochromatin formation and kinetochore establishment, affecting in this way mitotic segregation.
3.7 Conclusion
It can be proposed that the occurrence of new centromere results from a stochastic process affecting repetitive DNA, which is induced by homologous recombination followed probably by extrachromosomal rolling circle replication. As a result of such process, amplification of different satellite sequences already present within a genome occurs. However, only those satellites that have inherent centromere-competence in the form of some structural requirements necessary for centromere function are after amplification fixed in a population as a new centromere.
Presence of some conserved structural motifs within satellite DNAs such as periodically distributed AT tracts or protein binding sites indicates that despite centromere sequence flexibility, there are structural determinants that are prerequisite for centromere function. In addition, detection of transcripts from centromeric DNA that represent structural component of centromere indicates possible importance of structural elements at the level of RNA secondary or tertiary structures.
References
Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y (2001) Alpha-satellite DNA of primates: old and new families. Chromosoma 110:253–266
Alexiadis V, Ballestas ME, Sanchez C, Winokur S, Vedanarayanan V, Warren M, Ehrilch M (2007) RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA. Biochim Biophys Acta 1796:29–40
Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9:218–233
Amor DJ, Choo KH (2002) Neocentromeres: role in human disease, evolution and centromere studies. Am J Hum Genet 71:695–714
Assum G, Fink T, Steinbeisser T, Fisel KJ (1993) Analysis of human extrachromosomal DNA elements originating from different beta-satellite subfamilies. Hum Genet 91:489–495
Baker RE, Rogers K (2005) Genetic and genomic analysis of the AT-rich centromere DNA element II of Saccharomyces cerevisiae. Genetics 171:1463–1475
Basu J, Stromberg G, Compitello G, Willard HF, Van Bokkelen G (2005) Rapid creation of BAC-based human artificial chromosome vectors by transposition with synthetic alpha-satellite arrays. Nucleic Acids Res 33:587–596
Bensasson D, Zarowiecki M, Burt A, Koufopanou V (2008) Rapid evolution of yeast centromeres in the abscence of drive. Genetics 178:2161–2167
Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC (2001) Requirement of heterochromatin for cohesion at centromeres. Science 21:2539–2542
Bernstein E, Allis CD (2005) RNA meets heterochromatin. Genes Dev 19:1635–1655
Black BE, Bassett EA (2008) The histone variant CENP-A and centromere specification. Curr Opin Cell Biol 20:91–100
Blower MD, Nachury M, Heald R, Weis K (2005) A Rac-1 containing ribonucleoprotein is required for mitotic spindle assemby. Cell 121:223–234
Bouzinba-Segard H, Guais A, Francastel C (2006) Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci USA 103:8709–8714
Bruvo-Mad¯arić B, Plohl M, Ugarković Đ (2007) Wide distribution of related satellite DNA families within the genus Pimelia (Tenebrionidae). Genetica 130:35–42
Bulazel KV, Ferreri GC, Eldridge MD, O’ Neill RJ (2007) Species-specific shifts in centromere sequence composition are coincident with breakpoint reuse in karyotypically divergent lineages. Genome Biol 8:R170
Bühler M, Haas W, Gygi SP, Moazed D (2007) RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129:707–721
Cesari M, Luchetti A, Passamonti M, Scali V, Mantovani B (2003) Polymerase chain reaction amplification of the Bag320 satellite family reveals the ancestral library and past gene conversion events in Bacillus rossius (Insecta Phasmatodea). Gene 312:289–295
Charlesworth B, Langley CH, Stephan W (1986) The evolution of restricted recombination and the accumulation of repeated DNA sequences. Genetics 112:947–962
Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI (2008) Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451:734–737
Clarke L, Carbon J (1983) Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature 305:23–28
Coats SR, Zhang Y, Epstein LM (1994) Transcription of satellite 2 DNA from the newt is driven by a snRNA type of promoter. Nucleic Acids Res 22:4697–4704
Cooper JL, Henikoff S (2004) Adaptive evolution of the histone fold domain in centromeric histones. Mol Biol Evol 21:1712–1718
Dalal Y, Furuyama T, Vermaak D, Henikoff S (2007) Structure, dynamics, and evolution of centromeric nucleosomes. Proc Natl Acad Sci USA. 104:15974–15981
Dawe RK, Henikoff S (2006) Centromeres put epigenetics in the driver’s seat. Trends Biochem Sci 31:662–669
Dobie KW, Hari KL, Maggert KA, Karpen GH (1999) Centromere proteins and chromosome inheritance: a complex affair. Curr Opin Genes Dev 9:206–217
Dover GA (1986) Molecular drive in multigene families: how biological novelties arise, spread and are assimilated. Trends Genet 2:159–165
Durajlija žinić S, Ugarković Đ, Cornudella L, Plohl M (2000) A novel interspersed type of organization of satellite DNAs in Tribolium madens heterochromatin. Chromosome Res 8:201–212
Feliciello I, Picariello O, Chinali G (2005) The first characterisation of the overall variability of repetitive units in a species reveals unexpected features of satelite DNA. Gene 349:153–164
Feliciello I, Picariello O, Chinali G (2006) Intra-specific variability and unusual organization of the repetitive units in a satellite DNA from Rana dalmatina: molecular evidence of a new mechanism of DNA repair acting on satellite DNA. Gene 383:81–92
Ferbeyre G, Smith JM, Cedergren R (1998) Schistosome satellite DNA encodes active hammerhead-ribozymes. Mol Cell Biol 18:3880–3888
Fitzgerald DJ, Dryden GL, Bronson EC, Williams JS, Anderson JN (1994) Conserved pattern of bending in satellite and nucleosome positioning DNA. J Biol Chem 269:21303–21314
Folco HD, Pidoux AL, Urano T, Allshire RC (2008) Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319:94–97
Frescas D, Guardavaccaro D, Kuchay SM, Kato H, Poleshko A, Basrur V, Elenitoba-Johnson KS, Katz RA, Pagano M (2008) KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state. Cell Cycle 7(29):3539–3547
Fry K, Salser W (1977) Nucleotide sequences of HS-α satellite DNA from kangaroo rat Dipodomys ordii and characterisation of similar sequences in other rodents. Cell 12:1069–1084
Grewal SI, Elgin SC (2007) Transcription and RNA interference in the formation of heterochromatin. Nature 447:399–406
Grimes BR, Rhoades AA, Willard HF (2002) Alpha-satellite DNA and vector composition influence rates of human artificial chromosome formation. Mol Ther 5:798–805
Hall SE, Kettler G, Preuss D (2003) Centromere satellites from Arabidopsis populations: maintenance of conserved and variable domains. Genome Res 13:195–205
Harp JM, Uberbacher EC, Roberson AE, Palmer EL, Gewiess A, Bunick GJ (1996) X-ray diffraction analysis of crystals containing twofold symmetric nucleosome core particles. Acta Crystallogr D 52:283–288
Hegemann JH, Fleig UN (1993) The centromere of budding yeast. Bioessays 15:451–460
Henikoff S, Dalal Y (2005) Centromeric heterochromatin: what makes it unique. Curr Opin Genet Dev 15:177–184
Heslop-Harrison JS, Murata M, Ogura Y, Schwarzacher T, Motoyoshi F (1999) Polymorphisms and genomic organization of repetitive DNA from centromeric regions of Arabidopsis chromosomes. Plant Cell 11:31–42
Huisinga KL, Elgin SCR (2009) Small RNA-directed heterochromatin formation in the context of development: What flies might learn from fission yeast. Biochim Biophys Acta 1789:3–16
Jaco I, Canela A, Vera E, Blasco MA (2008) Centromere mitotic recombination in mammalian cells. J Cell Biol 181:885–92
Jin W, Melo JR, Nagaki K, Talbert PB, Henikoff S, Dawe RK, Jiang J (2004) Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell 16:571–581
Jonstrup AT, Thomsen T, Wang Y, Knudsen BR, Koch J, Andersen AH (2008) Hairpin structures formed by alpha satellite DNA of human centromeres are cleaved by human topoisomerase II α. Nucleic Acids Res 36:6165–6175
Kalitsis P (2008) Centromeres. In: Encyclopedia of life sciences (ELS). Wiley, Chichester
Kawabe A, Charlesworth D (2007) Patterns of DNA variation among three centromere satellite families in Arabidopsis halleri and A. lyrata. J Mol Evol 64:237–247
Kellum R, Alberts BM (1995) Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J Cell Sci 108:1419–1431
King K, Jobst J, Hemleben V (1995) Differential homogenisation and amplification of two satellite DNAs in the genus Cucurbita (Cucurbitaceae). J Mol Evol 4:996–1005
Kipling D, Warburton PE (1997) Centromeres, CENP-B and Tigger too. Trends Genet 13:141–145
Kumekawa N, Hosouchi T, Tsuruoka H, Kotani H (2001) The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 4. DNA Res 8:285–290
Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC (1997) Human centromeric DNAs. Hum Genet 100:291–304
Lee HR, Zhang W, Langdon T, Jin W, Yan H, Cheng Z, Jiang J (2005) Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci USA 102:11793–117998
Li YX, Kirby ML (2003) Coordinated and conserved expression of alphoid repeat and alphoid repeat-tagged coding sequences. Dev Dynamics 228:72–81
Lin CC, Li YC (2006) Chromosomal distribution and organization of three cervid satellite DNAs in Chinese water deer (Hydropotes inermis). Cytogenet Genome Res 114:147–154
Lu J, Gilbert DM (2007) Proliferation-dependent and cell cycle-regulated transcription of mouse pericentromeric heterochromatin. J Cell Biol 179:411–421
Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8-angstrom resolution. Nature 389:251–260
Ma J, Jackson SA (2006) Retrotransposon accumulation and satellite amplification mediated by segmental duplication facilitate centromere expansion in rice. Genome Res 16:251–259
Maddox PS, Oegema K, Desai A, Cheesman IM (2004) “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res 12:641–653
Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G (2002) Higher-order structure in pericentromeric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet 30:329–334
Malik HS, Henikoff S (2002) Conflict begets complexity: the evolution of centromeres. Curr Opin Genet Dev 12:711–718
Marshall OJ, Chuch AC, Wong LH, Choo KH (2008) Neocentromeres. new insights into centromere structure, disease, development, and karyotype evolution. Am J Hum Genet 82:261–282
Martinez-Balbas A, Rodriguez-Campos A, Gracia-Ramirez M, Sainz J, Carrera P, Aymami J, Azorin F (1990) Satellite DNAs contain sequences that induce curvature. Biochemistry 29:2342–2348
Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromere satellite. J Cell Biol 109:1963–1973
Masumoto H, Nakano M, Ohzeki J (2004) The role of CENP-B and alpha-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres. Chromosome Res 12:543–556
Meraldi P, McAinsh AD, Rheinbay E, Sorger PK (2006) Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol 7:R23
Meštrović N, Plohl M, Mravinac B, Ugarković, Đ (1998). Evolution of satellite DNAs from the genus Palorus- experimental evidence for the “library” hypothesis. Mol Biol Evol 15:1062–1068
Meštrović N, Castagnone-Sereno P, Plohl M (2006) Interplay of selective pressure and stochastic events directs evolution of the MEL172 satellite DNA library in root-knot nematodes. Mol Biol Evol 23:2316–2325
Metz A, Soret J, Vourc’h C, Tazi J, Jolly C (2004) A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. J Cell Sci 117:4551–4558
Mravinac B, Plohl M, Meštrović N, Ugarković Đ (2002) Sequence of PRAT satellite DNA “frozen” in some coleopteran species. J Mol Evol 54:774–783
Mravinac B, Plohl M, Ugarković Đ (2004) Conserved patterns in the evolution of Tribolium satellite DNAs. Gene 332:169–177
Mravinac B, Plohl M, Ugarković Đ (2005) Preservation and high sequence conservation of satellite DNAs suggest functional constraints. J Mol Evol 61:542–550
Murakami H, Goto DB, Toda T, Chen ES, Grewal SI, Martienssen RA, Yanagida M (2007) Ribonuclease Activity of Dis3 is required for mitotic progression and provides a possible link between heterochromatin and kinetochore function. PLoS ONE 3:e317
Nasmyth K (2002) Segregating sister genomes: the molecular biology of chromosome separation. Science 288:559–565
Navratilova A, Koblizkova A, Macas J (2008) Survey of extrachromosomal circular DNA derived from plant satellite repeats. BMC Plant Biol 8:90
Nijman IJ, Lenstra JA (2001) Mutation and recombination in cattle satellite DNA: a feedback model for the evolution of satellite DNA repeats. J Mol Evol 52:361–371
Ohzeki J, Nakano M, Okada T, Masumoto H (2002) CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159:765–775
Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H (2007) CENP-B controls centromere formation depending on the chromatin context. Cell 131:187–1300
Pezer Ž, Ugarković Đ (2008a) RNA Pol II promotes transcription of centromeric satellite DNA in Beetles. PLoS ONE 3:e1594
Pezer Ž, Ugarković Đ (2008b) Role of non-coding RNA and heterochromatin in aneuploidy and cancer. Semin Cancer Biol 18:123–130
Pezer Ž, Ugarković Đ (2009) Transcription of pericentromeric heterochromatin in beetles – satellite DNAs as active regulatory elements. Cytogenet Genome Res (in press)
Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323–337
Politi V, Perini G, Trazzi S, Pliss A, Raska I, Earnshaw WC, Della Valle G (2002) CENP-C binds the alpha-satellite DNA in vivo at specific centromere domains. J Cell Sci 11:2317–2327
Pons J, Bruvo B, Petitpierre E, Plohl M, Ugarković D, Juan C (2004) Complex structural feature of satellite DNA sequences in the genus Pimelia (Coleoptera: Tenebrionidae): random differential amplification from a common “satellite DNA library”. Heredity 92:418–427
Renault S, Roulex-Bonnin F, Periquet G, Bigot Y (1999) Satellite DNA transcription in Diadromus pulchellus (Hymenoptera). Insect Biochem Mol Biol 29:103–111
Romanova LY, Deriagin GV, Mashkova TG, Tumeneva IG, Mushegian AR, Kisselev LL, Alexandrov IA (1996) Evidence for selection in evolution of alpha satellite DNA: the central role of CENP-B/pJα binding region. J Mol Biol 261:334–340
Rudd MA, Wray GA, Willard HF (2006) The evolutionary dynamics of α-satellite. Genome Res 16:88–96
Sanyal K, Baum M, Carbon J (2004) Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci USA 101:1134–11379
Schueler MG, Sullivan B (2006) Structural and functional dynamics of human centromeric heterochromatin. Annu Rev Genomics Hum Genet 7:301–313
Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF (2001) Genomic and genetic definition of a functional human centromere. Science 294:109–115
Schueler MG, Dunn JM, Bird CP, Ross MT, Viggiano L; NISC Comparative Sequencing Program, Rocchi M, Willard HF, Green ED (2005) Progressive proximal expansion of the primate X chromosome centromere. Proc Natl Acad Sci USA 102:10563–10568
Slamovits CH, Cook JA, Lessa EP, Rossi MS (2001) Recurrent amplifications and deletions of satellite DNA accompanied chromosomal diversification in South American tuco-tucos (genus Ctenomys, Rodentia: Octodontidae): a phylogenetic approach. Mol Biol Evol 18:1708–1719
Smith PG (1976) Evolution of repeated sequences by unequal crossover. Science 191:528–535
Stephan W (1986) Recombination and the evolution of satellite DNA. Genet Res 47:167–174
Stephan W (2007) Evolution of genome organization. In: Encyclopedia of Life Sciences (ELS). Wiley, Chichester
Sun X, Le HD, Janice M, Wahlstrom JM, Karpen GH (2003) Sequence analysis of a functional Drosophila centromere. Genome Res 13:182–194
Tal M, Shimron F, Yagil G (1994) Unwound regions in yeast centromere IV DNA. J Mol Biol 243:179–189
Talbert PB, Bryson TD, Henikoff S (2004) Adaptive evolution of centromere proteins in plants and animals. J Biol 3:18
Takasuka TE, Cioffi A, Stein A (2008) Sequence information encoded in DNA that may influence long-range chromatin structure correlates with human chromosome functions. PLoS ONE 3:e2643
Topp CN, Zhong CX, Dawe RK (2004) Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci USA 101:15986–15991
Ugarković Đ (2005) Functional elements residing within satellite DNAs. EMBO Rep 6:1035–1039
Ugarković Đ (2008a) Evolution of Alpha satellite DNA. In: Encyclopedia of Life Sciences (ELS). Wiley, Chichester
Ugarković Đ (2008b) Satellite DNA libraries and centromere evolution. Open Evol J 2:1–6
Ugarković Đ, Plohl M (2002) Variation in satellite DNA profiles – causes and effects. EMBO J 21:5955–5959
Ugarković Đ, Podnar M, Plohl M (1996a) Satellite DNA of the red flour beetle Tribolium castaneum-comparative study of satellites from the genus Tribolium. Mol Biol Evol 13:1059–1066
Ugarković Đ, Durajlija S, Plohl M (1996b) Evolution of Tribolium madens (Insecta, Coleoptera) satellite DNA through DNA inversion and insertion. J Mol Evol 42:350–358
Ugarković ĐL, Plohl M, Lucijanić-Justić V, Borštnik B (1992) Detection of satellite DNA in Palorus atzeburgii: Analysis of curvature profiles and comparison with Tenebrio molitor satellite DNA. Biochimie 74:1075–1082
Vagnarelli P, Ribeiro SA, Earnshaw WC (2008) Centromeres: old tales and new tools. FEBS Lett 582:1950–1959
Verdel A, Jia S, Gerber S, Suglyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi-mediated targeting of heterochromatin with the RITS complex. Science 303:672–676
Vershinin AV, Alkhimova EG, Heslop-Harrison JS (1996) Molecular diversification of tandemly organised sequences and heterochromatic chromosome regions in some Triticeae species. Chromosome Res 4:517–525
Vogt P (1990) Potential genetic functions of tandem repeated DNA sequence blocks in the human genome are based on a highly conserved “chromatin folding code”. Hum Genet 84:301–336
Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833–1837
Wang F, Koyama N, Nishida H, Haraguchi T, Reith W, Tsukamoto T (2006) The assembly and maintenance of heterochromatin initiated by transgene repeats are independent of the RNA interference pathway in mammalian cells. Mol Cell Biol 26:4028–4040
Westermann S, Drubin DG, Barnes G (2007) Structures and functions of yeast kinetochore complexes. Ann Rev Biochem 76:563–592
Win TZ, Stevenson AL, Wang SW (2006) Fission yeast Cid12 has dual functions in chromosome segregation and checkpoint control. Mol Cell Biol 26:4435–4447
Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, Choo KHA (2007) Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res 17:1146–1160
Yamagishi Y, Sakuno T, Shimura M, Watanabe Y (2008) Heterochromatin links to centromeric protection by recruiting shugoshin. Nature 455:251–256
Zhang Y, Huang YC, Zhang L, Li Y, Lu TT, Lu YQ, Feng Q, Zhao Q, Cheng ZK, Xue YB, Wing RA, Han B (2004) Structural features of the rice chromosome 4 centromere. Nucleic Acids Res 32:2023–2030
Zhu L, Chou SH, Reid BR (1996) A single G-to-C change causes human centromere TGGAA repeats to fold back into hairpins. Proc Natl Acad Sci USA 93:12159–12164
Acknowledgements
This work was supported by grant 00982604 from the Croatian Ministry of Science and EU FP6 Marie Curie Transfer of Knowledge Grant MTKD-CT-2006-042248. The author is grateful to Josip Brajković for the help with figures.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2009 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Ugarković, Đ. (2009). Centromere-Competent DNA: Structure and Evolution. In: Ugarkovic, D. (eds) Centromere. Progress in Molecular and Subcellular Biology, vol 48. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-00182-6_3
Download citation
DOI: https://doi.org/10.1007/978-3-642-00182-6_3
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-00181-9
Online ISBN: 978-3-642-00182-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)