Abstract
Medicine will experience many changes in the coming years because the so-called “medicine of the future” will be increasingly proactive, featuring four basic elements: predictive, personalized, preventive, and participatory. Drivers for these changes include the digitization of data in medicine and the availability of computational tools that deal with massive volumes of data. Thus, the need to apply machine-learning methods to medicine has increased dramatically in recent years while facing challenges related to an unprecedented large number of clinically relevant features and highly specific diagnostic tests. Advances regarding data-storage technology and the progress concerning genome studies have enabled collecting vast amounts of patient clinical details, thus permitting the extraction of valuable information. In consequence, big-data analytics is becoming a mandatory technology to be used in the clinical domain.
Machine learning and big-data analytics can be used in the field of cardiology, for example, for the prediction of individual risk factors for cardiovascular disease, for clinical decision support, and for practicing precision medicine using genomic information. Several projects employ machine-learning techniques to address the problem of classification and prediction of heart failure (HF) subtypes and unbiased clustering analysis using dense phenomapping to identify phenotypically distinct HF categories. In this chapter, these ideas are further presented, and a computerized model allowing the distinction between two major HF phenotypes on the basis of ventricular-volume data analysis is discussed in detail.
 Art work by Piet Michiels, Leuven, Belgium
Art work by Piet Michiels, Leuven, Belgium
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Machine learning
- Big-data analysis
- Cluster analysis
- Precision medicine
- Heart failure phenotyping
- Support vector machine
Introduction
In July 2016, the Food and Drug Administration approved the first monoclonal antibody that inactivates the components responsible for the degradation of low-density lipoprotein receptors in the liver, decreasing their blood levels to much lower numbers than those that can be achieved with statins. This development is relevant because it represents the first important step toward a new version, as an information science, of the field of medicine. Genomics pioneer L. Hood [1] calls this the “medicine of the future,” which is changing its way of working from reactive to proactive toward “4P medicine” (powerfully predictive, personalized, preventive [a shift of focus from illness to wellness], and participatory). Several things will eventually drive this change, and among the most important will be the digitization of medicine and the development of computational tools —that is, machine-learning tools and methods, including preprocessing techniques needed to eliminate noise and irrelevant variables from the data—with the ability to deal with big data. Because data dimensionality is continuously increasing, it will be necessary to incorporate big-data analytics in order to be able to deal with the billions of data points that are expected for each individual patient in the next decade. Predictive medicine must correlate this high number of dimensional data sets with individual genotypes and phenotypes.
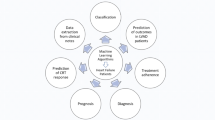
Thus, in this medicine of the future , machine learning [2] and big-data analytics [3,4,5,6,7] will be key disciplines. Data, often in large volumes, will be analyzed based on epidemiological variables, electronic health records (EHRs) , genomic databases, and so on. These data will allow for the practice of preventive and precision medicine and the avoidance of medical errors that might occur because of medical doctors’ distress due to their increasing and intensifying workloads, thereby increasing quality at affordable prices (see Fig. 37.1). For example, 10 years ago the expense of sequencing the genome of just one individual was approximately €200,000, whereas currently it is <€600.
Computerized and artificial intelligence (AI)–based methods for the analysis and interpretation of medical databases are not that new; these techniques found relatively early application in the medical sciences [8]. Machine learning and big-data analytics can be used in the field of cardiology in several ways, such as predicting an individual’s risk for cardiovascular disease, clinical decision support, precision medicine using genomic information, and so on. Some works that can be found in the literature using machine-learning techniques investigate the problem of classification and prediction of heart failure (HF) subtypes [9] and unbiased clustering analysis using dense phenomapping to identify phenotypically distinct categories of preserved ejection fraction (HFpEF). Alonso-Betanzos et al. [10] propose a computerized model that allows a clearer distinction between the two major phenotypes of patients with HF based on ventricular volume data analysis. In that work, a gray zone—which might correspond to a third separate phenotype—was identified, thus corroborating the capacity of machine-learning techniques to discover knowledge in medicine. Narula et al. [11] used an ensemble machine-learning model to aid in cardiac phenotypic recognition, specifically for the automated discrimination of hypertrophic cardiomyopathy from the physiological hypertrophy seen in athletes. The model used the previous feature of selection preprocessing (using the information gain filter [12]) step-over features of cardiac tissue deformation. Three different models integrated the ensemble (support vector machine [SVM] , random forest [RF] , and artificial neural network), and majority voting was used to reach a final decision.
However, studies using big-data analytics in the field of cardiology are not yet frequent in the literature. Some investigators described a framework aiming at setting an “initial but timely step toward a more intelligent and learning health care system that will require innovative bonds among patients, clinicians, data scientists, and health care systems” [13]. Motwani et al. [14] used machine learning on a data set from patients (10,030 patients during a 5-year follow-up period) undergoing coronary computed tomographic angiography. The aim of the study was to compare the results of cardiovascular outcome prediction using machine learning (including automated feature selection and an ensemble algorithm for learning) with those of traditional prognosis, which was limited regarding the use of clinical and imaging findings. The results showed considerable improvement in predicting all-cause mortality of those patients. In another study, the investigators studied the use of a Bayesian statistical model to address the limited predictive capacity of existing risk scores derived from multi-variate analyses [15]. The prognostic model showed superior prediction of acute, early, and late right-ventricular failure after left-ventricular (LV) assist device (LVAD) therapy compared with the currently available risk-prediction model. In conclusion, these models might facilitate clinical decision making while screening candidates for LVAD therapy. The trade-offs between data requirements and model utility were analyzed by Ng et al. [16] and Spertus et al. [17], who concluded that machine-learning techniques should be more frequently used in health care—in particular in cardiovascular risk estimations and mortality predictions—because they can greatly contribute to minimizing bias in hospital performance assessments. Despite these seminal works, and without regard to how the promising results have been reached, the use of big-data analytics in cardiovascular practice is still incipient, and it remains a long way from being a reality in daily practice.
Some areas in which impact is expected in a few years are the fields of cardiovascular epidemiology and cardiac imaging, among others. Using data from EHRs will not only obtain more accurate predictions, because it will permit balancing primary well-known risk factors with other secondary less-investigated ones, it also crosses those risk factors with other illnesses, such as cancer or cerebrovascular diseases. At the same time, more general health models could be obtained in this way. Machine learning with big data will also permit the selection of sub-populations in specific geographic areas, thus opening a door to the design of local health policies and adequate resource planning, which are lacking in many countries. Image analysis also soon will probably see changes in patient classification, diagnosis, and visualization because multi-modal big data are increasingly being generated from echocardiography studies, computerized tomography studies, magnetic resonance studies, and so on. Patients will be empowered by the generalized use of mobile devices and apps, thus allowing for extra-hospital management of cardiac conditions, such as cardiac insufficiency, auricular fibrillation, etc., and helping decrease the incidence of costly patient re-hospitalization.
Some of the difficulties encountered are data standardization between and within hospitals; the need to render data anonymous; and data heterogeneity, complexity, and disorganization, which in turn leads to the need for preprocessing techniques aiming at removing noise, discretizing and filtering data, removing irrelevant variables, and so on. In addition to technical difficulties, some other aspects to be considered are the security and privacy of data, which is of special importance in a medical context.
Big Data
Managing and using big data effectively is currently challenging, but in fact the existence of data is not new. Since ancient times, humans have tried to save data and information, but never has it been so easy, inexpensive, and quick to save, copy, share, and process data. In addition, we have evolved from saving simple scientific numbers at the origin of computation to the possibility of representing digitally almost anything, such as music, travel, or even the human body, among others. This growing digitalization process is possible thanks to the existence of myriad sensors that register events and activities, which permits the transformation from the physical to the digital. Because digital entities can be easily replicated, saved, transmitted, modified, sold, and so on, health sectors, for example, are being transformed into information and knowledge services.
However, sensors are not the only difference. Until some years ago, we were happily living with our database relational model. However, some companies (such as Google and Yahoo) found out that the database model limited the type and quantity of data that could be saved and analyzed and that this fact was contrary to their business. Thus, they decided to confront certain problems in a non-traditional way, which included the fact that all data could be considered important in some way. All data have value and saving and analyzing large volumes of data was a key point in their new business proposal. The problem? The value of big data is really discovered only after analyzing large volumes of them. Since then, analyzing big-data analytics has become a major driver of the economy.
As mentioned, the use of data is not new in the field of health care, where researchers have always been involved in collecting and analyzing data. However, the new digitization context, toward which we are currently moving, implies a volume, variety, and velocity of data production that pose new opportunities and demands in terms of both scale and complexity. Those challenges are the main impetus behind the development of the National Institutes of Health Big Data to Knowledge initiative [18] for addressing the opportunities and challenges presented by biomedical big data as well as the partnership between the National Cancer Institute and the United States Department of Energy to research years of cancer data and analyze them to develop new and more effective cancer treatments [19].
Several characteristics are required for data sets to be considered “big data” (see Fig. 37.2). Among the most important so-called five “Vs” of big data [20] are volume, velocity, variety, validity/veracity, and value.
-
The volume of data that must be processed by algorithms is substantially large (on the order of petabytes [i.e. 1015 bytes] and zettabytes [i.e. 1021 bytes] and continuously growing. Large volumes of data demand different data-storage and -processing tools and new characteristics of the data-preparation and -preprocessing steps (see section “Preprocessing methods for big data”).
-
Data might appear in some context at high velocities , in other words, important volumes of data manifesting in short time periods. Being able to analyze this data in real time might be crucial for some applications, but this requires a specific infrastructure to manage data streams. There are use cases, however, in which velocity is not a problem. For example, many more “tweets” are generated per minute than magnetic resonance imaging (MRI) scan images, and while reacting to a negative tweet might be relevant for a company, a strict real-time response is rarely required for MRI diagnostics.
-
Data currently come in different types (structured, semi-structured, and unstructured), and formats (text, images, audio, video, XML, etc.). This great variety increases complexity in data-saving and analytics solutions.
-
The variety of data types and formats, with large volumes at being generated at high velocities, constitutes the ideal situation to raise doubts about their degree of date quality and/or veracity or validity . Are the data correct? Are they of good-enough quality? Can I simultaneously use data that have different degrees of precision or temporal or spatial scale? Are these data relevant for my problem? Can they lead me to “actionable” information? Data preprocessing techniques are mostly unavoidable because they remove noise, invalid data, or redundant features, for example. However, these processes also pose a problem and a challenge. Regarding the problem, on one hand their use implies an extra effort that perhaps is not justified if the results are not really affected; in contrast, if the information to be obtained is sensible, the preprocessing step should not be avoided. Thus, depending on the application, veracity could be mandatory or secondary. Regarding the challenge, most preprocessing techniques (such as discretization or feature selection) were not designed for the use of big data, and the data suffer when scalability is needed.
-
Validity and veracity are critical determinants for users of big data because they affect the last “V”: value. Value implies that the knowledge and information derived from the data must be useful for the company or entity. To derive that knowledge, big-data analytics must be used, which makes data scientists essential. Although some aspects could be automatized, management of the entire process (from designing the appropriate infrastructure needed to adequately visualize the results) is complex and requires high-level human expertise.
Many issues require planning and careful execution for the use of big data. Among the most important, especially in the field of health care, are security and privacy. The privacy of those individuals whose data are being managed is crucial, and addressing issues of this type requires deep understanding of the nature of the data, the relevant norms and regulations, and the techniques that should be used, such as, for example, anonymization . A careless analysis might reveal private information, thus opening a possibly unnoticed gap in privacy. Big-data security issues should be also considered, mostly because the types of databases that should be used do not provide as robust a built-in security mechanism as do traditional relational database-management systems. Similar situations might appear in the data-analysis phase, in which data might be distributed among several nodes, which is a common situation. If, for example, we are trying to derive prognostic models using patient data from different hospitals in a region or country, the machine-learning algorithms being used might need to interchange or combine intermediate results, again opening the possibility for inadvertent breaches of privacy. Thus, privacy preservation should be a requirement for the employed algorithms. Because the analysis of big data is different from traditional data analysis, a systematic methodology [20, 21] is needed to organize the diverse associated activities (see Fig. 37.3), and these must be managed by specialized data scientists.
Data Identification
In the first step, the business case to be confronted should be clearly identified , with an adequate motivation for the analysis, together with the types of data and the analytics needed. The projects goals also should be established. An important outcome of this stage is definition of the tools and other economic or personnel resources needed.
In the data-identification stage , the sources of data to be employed and their quantity, quality, and format should be established. The data sources can be internal or external to the company. In the latter case, a list of data providers (this includes publicly available data sets) is needed. Finally, if personally identifiable information requires removal or masking, the involved requirements for anonymization and re-identification should be stated.
Data Collection
Data are collected from the data sources identified in the previous step during the acquisition phase. In addition, meta-data (such as data size, structure, date, time of creation, etc.) is added, if needed, to maintain the data during the rest of the methodology phases. Persistence of the data should be assured because fault-tolerance scenarios must be accounted for; therefore, data should be stored in a database.
Data Preprocessing
Preprocessing data implies several operations, such as removing noise, filtering data to remove some types possibly lacking value for posterior analysis, aggregating data that might be spread across several data sets, and removing irrelevant and redundant features in the data to simplify the data-analysis phase. Preprocessing is a fundamental step for assuring data quality and consistency, and it will be described in detail in section “Preprocessing methods for big data.”
Data Analysis
The data-analysis stage is devoted to discovering patterns, correlations, predictions, or anomalies in the data to give answers to the questions raised in the first stage, thus making it possible to derive business value from the data. Because large volumes of data should be analyzed, specialized software tools and applications for machine learning and statistics are needed. The algorithms to be employed should obviously be scalable, and the type of architecture and tools to be employed depend on the restrictions of the specific use case. Stream-processing architecture produces real-time data insights because it computes one data element at a time. The data analysis is performed in almost real time, and thus immediate action can be taken in response. An example of this is acting in response to a patient’s health status or the experience of a hospitalized patient. Tools are different for batch processing, which processes large volumes of data for which a quick response time is not critical; an example could be the elaboration of a monthly report on some activities of the hospital. Batch processing is more related to the volume and variety, whereas for stream processing velocity is most critical. Therefore, for some applications, batch-processed data might be outdated by the time it reaches health-care professionals. Of course, scenarios exist in which both types of data processing can be employed. For example, in marketing, batch data processing can be used to analyze the habits of consumers from historical data sets. Then health-care marketers can create tailored and targeted marketing campaigns that will ideally improve adherence and engagement from patients by establishing which communication channels will result in the best response rate from each group. From there onward, streaming processes can analyze which social media messages are most effective for each individual and take immediate action. Another example is the different use cases that can be derived from health services running on smartphones with sensors, which have become extremely popular, tracking regularly daily activities, such as sport, sleep, and diet habits across sport-oriented social networks. Because this is also an important phase, machine-learning methods will be described in more detail in section “Supervised and unsupervised machine-learning methods.”
Data Visualization
Finally, data visualization consists of a presentation of the output (from the previous phase) in a format that allows business users to understand the results obtained and thus be able to make decisions based on them. This might comprise tables, graphs, information “blocks,” and so on.
Choosing the appropriate infrastructure is crucial in big-data projects because companies build their competence around it. The selected infrastructure strongly affects big-data architecture for new products and services. Having the appropriate tools for storing, processing, and analyzing data is key. Among the most well-known tools are big-data databases (e.g., MongoDB, HBase, or Apache Cassandra); tools for transferring and aggregating data (e.g., Flume or Lucene); frameworks (e.g., Apache Hadoop [http://hadoop.apache.org/], Apache Spark [http://spark.apache.org/], or Apache Flink [https://flink.apache.org/]) with their corresponding machine-learning libraries (e.g., Mahout, MlLib, and Flink ML) or Spark components for graph analytics (e.g., GraphX and GraphFrames).
Preprocessing Methods for Big Data
As mentioned in the previous section, the advent of big data has brought an important number of challenges to the scientific community, which now must deal with unprecedented volumes in data and try to extract useful information from them. We continue to store data of all kinds, and usually a preprocessing step is necessary before applying an adequate machine-learning method (see section “Supervised and unsupervised machine-learning methods”). In fact, a typical scenario in health data is to be able to classify a patient as presenting with a particular condition or not (e.g., the risk of not catching a patient presenting with heart failure). From a machine-learning perspective, this is a classification task in which a learner or classifier must learn the characteristics of the data (i.e. variables about a given patient, such as age, sex, and the results of medical tests) and then provide a prediction. However, in many cases these variables are of different natures because attributes such as sex are discrete; other attributes, such as weight, have continuous values. Some classifiers can only deal with discrete data, so there exists an important preprocessing technique called “discretization .”
Another problem typically encountered by machine-learning researchers facing medical data are that it is likely that some variables are not relevant for the prediction task. For example, sex can be an important factor for determining the risk of presenting with heart failure but completely irrelevant for other conditions. To solve this problem, feature selection is usually applied as a preprocessing step to remove those unimportant variables from the task at hand. In this section we will focus on two popular preprocessing techniques: discretization and feature selection.
Discretization
Discretization is a preprocessing technique that consists of transforming the continuous variables of a data set into discrete variables. By applying this technique, quantitative data are transformed into qualitative data, thus procuring a nonoverlapping division of a continuous domain. Discretization also can be considered as a data-reduction mechanism because it reduces data from a large domain of numeric values to a subset of discrete values [22].
In some cases, discretization is a mandatory step because some machine-learning algorithms used afterward are not able to handle continuous variables. For instance, this is the case with the popular classifier Naïve Bayes [23] and also with feature-selection methods, such as the information gain filter [24]. Apart from this, discretization can have a beneficial impact on the performance of learning algorithms, for example, in terms of speed (especially important in the context of big data and real-time learning) and accuracy. The basic discretization process is formed by four steps, which are detailed here and also can be seen in Fig. 37.4.
-
Step 1: Sort the continuous values for an attribute (either in ascending or descending order). It is crucial to choose an efficient sorting algorithm to perform this step.
-
Step 2: After sorting, select the best cut point of the best pair of adjacent intervals in the attribute range to split or merge them in the next step. It is necessary to define an evaluation measure of function, which can be correlation, gain, improvement in performance, or any other benefit according to the class label.
-
Step 3: Split or merge intervals according to the operation method of each discretization algorithm. To split, the possible cut points are the different continuous values in each attribute. To merge, the discretization algorithm tries to find the best adjacent intervals in each iteration.
-
Step 4: Stop according to some criterion or otherwise return to step 2. Usually a trade-off between a low number of intervals, good comprehension, and consistency is assumed.
A broad suite of discretization algorithms can be found in the literature, and the selection of one or another depends on the type of the data. For a complete taxonomy about discretization methods, see Ramírez et al. [22]. In the following text, some of the most popular methods will be described (including those in the popular Weka tool [25]).
-
Equal width: This simple unsupervised discretization algorithm calculates the range of the variable and then divides it into equal parts. The resulting intervals will generally be unbalanced, with many items ending in a few of them and some much less populated. The split/merge step is disregarded in this simple method.
-
Equal frequency: This algorithm obtains intervals that contain a constant number of items. The basic version of this method aims to obtain a fixed number of intervals, although this is suboptimal for some classification algorithms. Therefore, a variation called Proportional k-Interval Discretization (PKID) [26] can be used instead. This algorithm adjusts the number of intervals according to the number of samples.
-
Minimum Descriptive Length (MDL): This popular method uses information-entropy minimization as a heuristic to calculate the most suitable cut points [27].
In a big-data scenario, the problem is that classical data-reduction methods were not designed to handle such a large amount of data, which makes their use difficult or even impossible in some cases. To solve this issue, in the past few years new implementations of the most popular methods have appeared that take advantage of distributed computational frameworks. For example, a distributed implementation of PKID is available in Spark MlLib. Moreover, a distributed implementation of MDL is available for Apache Spark [28], which leverages a computer cluster to speed up the sorting and cut point–evaluation steps involved in this method, thus enabling it to deal with large data sets.
Feature Selection
Analogous to the term “big data,” the term “big dimensionality” has been coined to refer to the unprecedented number of features arriving at levels that render existing machine-learning methods inadequate [29]. Thus, dimensionality-reduction techniques, such as feature selection, have become almost essential. Feature selection is the process of selecting relevant variables (features), by removing the irrelevant and/or redundant ones, with the aim of obtaining better and simpler models. Because the process does not transform the original features (unlike feature-extraction methods), it obtains models that might be easier to interpret for researchers or medical practitioners. Moreover, it presents other benefits, such as enhancing generalization by decreasing overfitting and the confers the requirement of shorter training times (a crucial point in real-time application).
Typically, feature-selection methods are classified into filters, wrappers, and embedded methods based on their relationship with the learning algorithm (see Fig. 37.5). The simplest model is the filter, which relies on the general characteristics of training data and performing the feature-selection process as a preprocessing step with independence of the induction algorithm. Wrappers involve a learning algorithm as a black box, and they use their prediction performance to assess the relative usefulness of subsets of variables. Finally, embedded methods perform feature selection in the process of training and are usually specific to given learning machines. They learn which features best contribute to the accuracy of the model while the model is being created. Because of this interaction with the learning algorithm, wrappers and embedded methods tend to give more accurate subsets of features, but they are usually specific for a particular classifier and are computationally expensive (especially wrappers). In contrast, filters are advantageous for their low computational cost and good generalization abilities [30]. Each of the three approaches is extensively used, although the filter model is more adequate for big-data settings.
Considering that several algorithms exist for each one of the previously commented approaches, there is a vast body of feature-selection methods. We describe some of the most popular ones here:
-
Correlation-based feature selection : This is a simple multi-variate filter algorithm that ranks feature subsets according to a correlation-based heuristic-evaluation function [31]. The bias of the evaluation function is toward subsets that contain features that are highly correlated with the class and not correlated with each other. Irrelevant features should be ignored because they will have low correlation with the class. Redundant features should be screened out because they will be highly correlated with one or more of the remaining features. The acceptance of a feature will depend on the extent to which it predicts classes in areas of the instance space not already predicted by other features.
-
Consistency-based : This filter [32] evaluates the worth of a subset of features by the level of consistency in the class values when the training instances are projected onto the subset of attributes.
-
Information gain : This filter [12] provides an ordered ranking of all features , and then a threshold is required.
-
ReliefF : This filter [33] is an extension of the original Relief algorithm. The original Relief works by randomly sampling an instance from the data and then locating its nearest neighbor from the same class and from the opposite class. The values of the attributes of the nearest neighbors are compared with the sampled instance and used to update relevance scores for each attribute. The rationale is that a useful attribute should differentiate between instances from different classes and have the same value for instances from the same class. ReliefF adds the ability of dealing with multi-class problems and is also more robust and capable of dealing with incomplete and noisy data. ReliefF is applicable in all situations; it has low bias; it includes interaction among features; and it may capture local dependencies missed by other methods.
-
Minimum redundancy maximum relevance : This filter [34] selects features that have the greatest relevance with the target class and that are also minimally redundant. In other words,” it selects features that are maximally dissimilar to each other. Both optimization criteria (maximum relevance and minimum redundancy) are based on mutual information.
-
Recursive Feature Elimination for SVMs : This embedded method [35] performs feature selection by iteratively training an SVM classifier with the current set of features and removing the least important feature indicated by the SVM.
Although feature selection is almost mandatory for machine-learning algorithms to be able to manage large dimensional data sets, most available methods were not developed considering this scenario, and their computational costs prevent their use in big-data settings. Recently, some approaches—such as employing graphical processing units (to implement faster versions [36] of well-known algorithms) or parallelization using MapReduce, Hadoop, or Apache Spark—have been developed to solve this problem.
Supervised and Unsupervised Machine-Learning Methods
As mentioned in the Introduction section, digitization seems to be an unstoppable process in the fields of medicine and health, among others. For the analysis of these ever-increasing amounts of data, thus being able to derive information and knowledge from them, it is unavoidable to use automated methods, which should be scalable to keep pace with the crescent input loads. Machine learning [37] is a sub-discipline of the field of AI that consists of a set of methods and algorithms that can learn from data and devise models for different processes, such as pattern recognition or prediction, for example. However, machine learning [38,39,40,41] is not a new field: It appeared due to the early interest of AI researchers in determining if computers could learn directly from data without being programmed to perform specific tasks. However, due to the appearance of big data, machine learning is currently a hot buzzword, and an area of very active research, which—together with other factors— have brought a “new spring to the step” of the field of AI . Although many machine-learning algorithms have been around for several years, the ability to automatically apply complex mathematical calculations to large quantities of data in competitive time is a recent development. Machine-learning algorithms learn a function f: X➔Y. This function belongs to a certain “family” [42] and maps the input domain of data X to a certain output domain Y (a prediction, for example). The five main types of problems machine learning can solve [43] include:
-
1.
Classification, where the algorithm must assign unseen inputs to a series of predefined classes
-
2.
Regression , where the focus is predicting a continuous output
-
3.
Clustering , where inputs must be labeled into unknown groups (unlike classification)
-
4.
Density estimation , where the goal is finding the distribution of a set of inputs
-
5.
Dimensionality reduction , where inputs are simplified by mapping them to lower dimensional spaces
These tasks can also be classified according to the nature of the available learning data, which is provided in the form of examples (xi, yi) € X Y. In this case, three basic forms of learning can be distinguished:
-
Supervised learning , where a set of known patterns are used for training, that is, the training data set is labeled, and thus yi is the corresponding ground truth for xi, and the aim of the process is to classify data based on that a priori knowledge. This previous knowledge of the data set’s instance classes (i.e. the value to be predicted) is used to learn predictive models from the data set of examples in order to classify unseen instances. One important aspect of supervised classification is the evaluation of algorithms by means of an evaluation function, which usually quantifies the generalization ability of the classifier. That is, the goal is to minimize the error or loss function, fˆ = Y × Y➔R, which quantifies the difference between the predicted output and the real ground-truth label for that sample. In real-world problems, the true classification error is unknown, and thus so is its underlying probability distribution. Therefore, it must be estimated from the data. Because the loss cannot be minimized directly on the test instances and their labels, because typically these are not available at training times, supervised algorithms aim to construct functions that generalize well to previously unseen data, not to those that perform optimally on the given training data set (thus overfitting the data). In training and evaluating the devised model, two sources of data are employed. The parameters of the model are set based on the train data only, and if the test data are generated from the same underlying process that has generated the training data, an unbiased estimate of the generalization performance can be obtained by measuring the test-data performance of the trained model. It is important to recall that the test performance should not be used to adjust the model parameters because in this case the measure of performance will lose its independence. In particular, the mean squared error (MSE) is the measure typically employed for evaluating estimations made by the algorithms. The MSE is the second-order moment of the error, and therefore it incorporates both the variance and the bias of the estimator. The most common supervised learning tasks are classification and regression.
As an example, we show the results of classification for the data set Heart Disease (Cleveland) from the University of California-Irvine learning repository (http://archive.ics.uci.edu/ml/). We used the well-known platform Weka [25] to apply the selected algorithms. The Heart Disease data set has 13 attributes and 5 different classes as output (with 160, 35, 54, 35, and 13 samples respectively), with a total of 297 samples, because 6 of the 303 total have missing data and thus were eliminated from this study (see Fig. 37.6).
Several different classification algorithms can be used, and they show the results employing the RF classifier [44] because it is one of the state-of-the-art and more accurate classifiers. For this data set, the number of data available is not large, and thus if we must divide the data set into training (usually 80%) and test (20%) of the data, the estimation of the true error will be not very accurate. In these cases, a cross-validation procedure is often used. Cross-validation consists of making k partitions (folds) of the data, using k-1 for training and the remaining one for testing, and repeating the procedure k times until all folds have been used for testing the model. Thus, we are evaluating k models, and by averaging the results we have an idea of the variance of the learning algorithm with the variations in the training data and thus can obtain a more real approach to the error of the model. Cross-validation is conventionally applied with k = 10, although if the number of samples in each fold is low (usually < 30 [because this allows for approximating the binomial distribution of the number of correctly classified samples in a fold by normal distribution]), other k are used (most commonly k = 5). An extreme value is k = n, in which all samples except one are used to train, and that one is also used to test. This method is called “leaving-one-out” [17]. Using 10-fold cross-validation, the results obtained are 57.6% accurate; the confusion matrix is listed in Table 37.1.
If the data set is converted to a binary one, then only the class absence of heart disease (class 1, 160 samples) and the presence of heart disease (classes 2 through 5 with 137 samples total) are taken into account; the accuracy increases to 83.5% using the same classifier and with the contingency table listed in Table 37.2.
Thus, it can be seen that multi-class classification is a more difficult problem for the algorithm because the number of samples available is not enough for a good generalization. Another problem mentioned, one that is quite common in medical data bases, is the existence of missing values that should be treated accordingly [45, 45]. In our case, because the missing data are only present in three samples, we opted for the simplest operation, that is, elimination. Another common problem in medical data sets [37] are incorrectness (the presence of noise, probably due to sensor errors), inexactness (the presence of redundant data, which can imply the need of more complex models that in fact would not be necessary), and sparseness (sometimes there might be few records available for certain studies).
-
Unsupervised learning , in which the training data set is unlabeled {xi} and the aim is to unravel the underlying similarities, obtains a plausible compact description of the data. An objective is used to quantify the accuracy of the description. In the case of unsupervised learning, the aim is to model the distribution p(x). The likelihood of the model to generate the data is a popular measure of the accuracy of the description.
The most common unsupervised learning task is clustering , in which the aim is the construction of a function (f), which partitions the training data set into k clusters. Several algorithms can be applied for clustering, but typically they work by assessing the similarity between instances by assigning similar samples to the same cluster and dissimilar ones to different cluster. Using a simple and well-known cluster algorithm, k-means, with the binary Heart Disease (Cleveland) data set and not supplying the information related to the class (because it is assumed that the data set is unlabeled in this case), the error of the algorithm is 20.2%; the results are listed in Table 37.3.
Other unsupervised tasks also exist—such as association rules (which build rules associating items that occur together with a certain frequency, discover patterns in the data, or can alternatively use correlation between real-valued variables—that are popular in machine learning. Then, to end we can say that machine learning is the task of building a model from data that generalizes a decision against a performance measure.
Most times, learning pipelines must include some kind of preprocessing operations, for example, noise must be eliminated, data discretized, and so on. Feature selection is also an important operation to consider because it can help with the generalization of machine-learning algorithms, thus improving their performance and perhaps the interpretability of the obtained results (see Fig. 37.7).
Example of feature-selection and -classification process on the Heart Disease (Cleveland) data set. At the bottom, we can see a part of the C4.5 decision tree built for predicting class 3 (presence of heart disease). The variables used are Thal (Thalassemia), ca 8 (number of major vessels colored by fluoroscopy), exang (Exercise-Induced Angina), slope (Slope of the Peak Exercise ST Segment), cp (Chest Pain Type), and thalach (Maximum Heart Rate Achieved)
A Case Study
In this section we present a case study in which we applied machine-learning methods to classify HF subtypes based on the work by Alonso-Betanzos et al. [10]. HF is a relatively common cardiac syndrome known for its severe sequelae, including death. The diagnosis is often only evident from the combination of symptoms (e.g., fatigue, dyspnea, etc.) and signs (e.g., ankle edema), plus clinical investigations—including the determination of LV size and chamber filling pressure—and information derived from specific biomarkers.
HF is manifested in at least two subtypes . The current paradigm distinguishes them by using metric EF and constraint for end-diastolic volume. Approximately half of all HF patients, often including women and elderly, exhibit HFpEF. Thus, as life expectancies continue to increase in western societies, the prevalence of HFpEF will continue to grow. However, compared with “classical” HF with decreased ejection fraction (HFrEF), only a limited spectrum of treatment modalities seems to be effective for improving the morbidity and mortality rates in patients with HFpEF.
Traditionally, EF has been widely applied to assess the severity of cardiac problems. In the particular case of HF, EF is one of the many indicators to characterize the various aspects of the syndrome [47]. Typically, a low EF value corresponds with serious cardiac problems and a poor prognosis. Calculation of EF is performed by taking the ratio of two LV volume determinations during a cardiac cycle, namely, at the completion of filling and again at maximal contraction. Advised cut-off levels to distinguish HFrEF from HFpEF are clearly formulated, but they vary between 40% and 50%, which defines a linear divider. In addition, some studies opt for eliminating HF patients from consideration if 40 < EF < 50% (“gray zone” [currently often referred to as the “mid-range” phenotype]). Thus, there is a need to develop documented classification guidelines, solve gray-zone ambiguity, and formulate crisp delineation of the transition between phenotypes.
Subgroups of HF patients are located in at least two distinct regions on the basis of their end-systolic volume index and end-diastolic volume index; therefore, they are uniquely located within the LV-volume domain. In this case study, we present the application of machine-learning techniques to explore a more rational foundation for classifying two phenotypes of HF .
In summary, this case study addresses several relevant issues regarding the classification of HF patients: How can machine-learning models assist clinicians in the classification of major HF subtypes, the consequences of varying the cut-off values, and describe implications for borderline patients (in the gray zone).
Results for Applying Machine-Learning Techniques
The first set of experiments performed consisted of using unsupervised ML methods for the following three data sets (for more details, check Alonso-Betanzos et al. [10]):
-
Data set 1: Data from real patients, a total of 48 instances where 35 belong to class HFpEF and 13 to class HFrEF.
-
Data set 2: Data simulated with Monte Carlo, a total of 63 instances where 34 belong to class HFpEF and 29 to class HFrEF.
-
Data set 3: Monte Carlo data generated as testing data, a total of 403 instances where 150 refer to class HFpEF; 137 belong to class HFrEF; and a third group (n = 116) still requires classification because on the basis of current guidelines they belong to neither HFpEF nor HFrEF. The third group is specifically introduced to challenge the universal validity of the current EF–EDVI paradigm, which favors a linear separator based on a fixed EF value.
Because we are using unsupervised algorithms, we focused on clustering , which consists of grouping a set of data in such a way that those belonging to the same group (called a “cluster”) are more similar (in one sense or another [which is defined by the type of algorithm and its parameters]) to each other than to those in other clusters. To perform an unsupervised separation of the two major phenotypes of HF patients , we evaluated several different clustering algorithms, using different approaches all implemented in the Weka software tool [25]: K-means, Expectation Minimization (EM), and Sequential Information Bottleneck (sIB). As can be seen in Fig. 37.8, for Data sets 1 and 2 (real patients and simulated Monte Carlo, including only the two major types of patient subgroups), only the sIB algorithm tried to separate the samples using a similar approach as the current clinical guidelines. However, it can also be seen that the patients reclassified in an alternative manner (see squares in Fig. 37.8) and are all located within a region, which in some other studies is neglected and referred to as the “gray zone.”
Results of clustering analysis . Squares represent instances that are incorrectly assigned to a cluster. EM expectation minimization, sIB sequential-information bottleneck. (Image reprinted with permission from Alonso-Betanzos et al. [10])
Then, we performed a supervised automatic classification of both major HF types using SVM PEGASOS, which implements a sequential minimal-optimization algorithm for training an SVM, also available in Weka [25]. The set of experiments carried out included different cut-off points (EF at 40%, 45%, 50%, and 55%) in the training data set (Data Set 1) to evaluate the consequences of adopting different criteria for defining major HF phenotypes and the ability of machine-learning methods to correctly classify the patients in each case. A summary of the results is listed in Table 37.4; more details can be found in Alonso-Betanzos et al. [10].
As can be seen, the results obtained after classifying the data with an SVM are quite satisfactory, with true-positive rates >0.90 in most of the cases. Moreover, we performed experiments to see how a machine-learning method would classify those patients belonging to the gray zone (i.e. the area where 40 < EF < 50%), which is not yet classified.
In Figs. 37.9 and 37.10, we see an example for cut-off 45%; the complete results can be checked in Alonso-Betanzos et al. [10]. In general, we can see that that the third group can largely be classified as HFpEF (although it varies when changing the cut-off). Interestingly, the separation does not follow the linear division as prescribed by the concept referring to a constant EF value for the cut-off. As seen, the points that are labeled differently seem to be located on the border between the main classes.
Real labels for the test set (n = 403 [including the third group, which is newly assigned to either of the existing phenotypes]) for the 45% cut-off of the patient data as a training set (image reprinted with permission from Alonso-Betanzos et al. [10])
Enlarged picture of the third group of data labels showing in detail that the algorithm applies a nonlinear division rather than a straight EF line (image reprinted with permission from Alonso-Betanzos et al. [10])
We can thus conclude that machine-learning models offer promise for making a computer-assisted distinction between the two major phenotypes of HF patients on the basis of ventricular-volume analysis. Moreover, selected machine-learning tools may assist during the classification of individual patients having measurements located in the (clinically often neglected) gray zone where 40 < EF < 50%.
Future Directions
The need to apply machine-learning methods to the field of medicine has increased dramatically in recent years to face challenges brought by the advent of big data, for which it is necessary to cope with an unprecedented large number of features and samples. The increasingly decreased cost of storage technology has enabled us to store all kind of information about patients, with the aim of extracting useful and valuable information. However, these data are messy when they come out of the electronic health record of a patient and, even when they were collected from a study, the data might be of very different natures, which complicates the task of developing machine-learning algorithms.
Several research lines are open in this area. First is the problem of data distribution and data privacy. In some cases, information about patients is distributed across geographical and organizational boundaries (i.e. different hospitals), and it is not legal or affordable to gather it in a single location. In this case, it is necessary to develop distributed approaches for existing machine-learning methods that preserve privacy. It can be the case of a vertical distribution (each party has partial information about all the patients) or a horizontal distribution (each party involved in data sharing has information about all the variables but for different sets of patients). Although some approaches already exist that try to deal with this issue, there is still room for more works solving a problem that is especially important in the medical field. Another open question is the necessity of real-time processing in computer-assisted methods for the analysis of medical data. If a practitioner must wait a long time to obtain a recommendation from a computerized system, it is likely that he or she will stop using it. To avoid this, it is crucial to process and analyze data in real time, which can be performed either by accelerating the processing of the data (with feature selection or discretization methods) or by using online approaches (which are still relatively scarce in the literature). Anonymization , a process in which data sets are purged of personally identifying information, is another important challenge that has constituted an open research question for years. Although some recent attempts have been made to be able to use well-known privacy models, such as k-anonymity, in big data [48] and others, such as ε-differential privacy [49], there is still a long way to go because there are in fact very sophisticated methods [50] that can work backwards and re-identify individuals. Finally, another challenge is the visualization and interpretation of results. In recent years, several dimensionality-reduction techniques have been developed, aiming at a better visualization of the data. However, some of them have the limitation that the features being visualized are transformations of the original features, which greatly complicates the task of interpretation usually required by practitioners.
In conclusion, machine-learning models are much needed to help in clinical medicine . However, this enthusiasm does not generally match the level of actual activity in the field. It is very well to have machine-learning algorithms that are good at predicting and may help in clinical routine, but it is more complicated to employ them in the real world. We must make sure that they can be applied in a safe, responsible, and ethical way and—most of all—that people would accept being diagnosed by a machine rather than a person.
References
Hood L. A vision for personalized medicine. MIT Technology Review. Available at: https://www.technologyreview.com/s/417929/a-vision-forpersonalized-medicine/. Accessed 6 Apr 2017.
Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–30.
Krittanawong C, et al. Future physicians in the era of precision cardiovascular medicine. Circulation. 2017;136:1572–4.
Shah SJ, Katz DH, Selvaraj S, Burke MA, Yancy CW, Gheorghiade M, Bonow RO, Huang CC, Deo RC. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation. 2015;131:269–79.
Zhang X, Ambale-Venkatesh B, Bluemke DA, Cowan BR, Finn JP, Kadish AH, Lee DC, Lima JA, Hundley WG, Suinesiaputra A, Young AA, Medrano-Gracia P. Information maximizing component analysis of left ventricular remodeling due to myocardial infarction. J Transl Med. 2015 Nov 3;13(1):343. https://doi.org/10.1186/s12967-015-0709-4.
Rumsfeld JS, Joynt KE, Maddox TM. Big data analytics to improve cardiovascular care: promise and challenges. Nat Rev Cardiol. 2016 Jun;13(6):350–9.
Tripoliti EE, Papadopoulos TG, Karanasiou GS, Naka KK, Fotiadis DI. Heart failure: diagnosis, severity estimation and prediction of adverse events through machine learning techniques. Comput Struct Biotechnol. 2017;15:26–47.
Kerkhof PLM, Alonso-Betanzos A, Moret-Bonillo V. Medical expert systems. In: Wiley encyclopedia of electrical and electronics engineering. Wiley; 2017.
Austin PC, Tu JV, Ho JE, Levy D, Lee DS. Using methods from the data mining and machine learning literature for disease classification and prediction: a case study examining classification of heart failure subtypes. J Clin Epidemiol. 2013;66:398–407.
Alonso-Betanzos A, Bolón-Canedo V, Heyndrickx GR, Kerkhof PLM. Exploring guidelines for classification of major heart failure subtypes by using machine learning. Clin Med Insights Cardiol. 2015;9(Suppl 1):57–71. https://doi.org/10.4137/CMC.S18744.
Narula S, Shameer K, Omar AMS, Dudley JT, Sengupta PP. Machine-learning algorithms to Automate morphological and functional assessments in 2D echocardiography. J Am Coll Cardiol. 2016 Nov;68(21):2287–95. https://doi.org/10.1016/j.jacc.2016.08.062.
Quinlan JR. Induction of decision trees. Mach Learn. 1986;1(1):81–106.
Ahmad T, Testani JM, Desai NR. Can big data simplify the complexity of modern medicine? Prediction of right ventricular failure after left ventricular assist device support as a test case. JACC Heart Failure. 2016;4(9):722–4.
Motwani M, Dey D, Berman DS, Germano G, Achenbach S, Al-Mallah MH, Andreini D, Budoff MJ, Cademartiri F, Callister TQ, Chang HJ, Chinnaiyan K, Chow BJ, Cury RC, Delago A, Gomez M, Gransar H, Hadamitzky M, Hausleiter J, Hindoyan N, Feuchtner G, Kaufmann PA, Kim YJ, Leipsic J, Lin FY, Maffei E, Marques H, Pontone G, Raff G, Rubinshtein R, Shaw LJ, Stehli J, Villines TC, Dunning A, Min JK, Slomka PJ. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur Heart J. 2017;38(7):500–7. https://doi.org/10.1093/eurheartj/ehw188.
Loghmanpour NA, Kormos RL, Kanwar MK, Teuteberg JJ, Murali S, Antaki JF. A Bayesian model to predict right ventricular failure following left ventricular assist device therapy. JACC Heart Failure. 2016;4(9):711–21.
Ng K, Steinhubl SR, de Filippi C, Dey S, Stewart WF. Early detection of heart failure using electronic health records: practical implications for time before diagnosis, data diversity, data quantity, and data density. Circ Cardiovasc Qual Outcomes. 2016;9:649–58.
Spertus JV, Normand ST, Wolf R, Cioffi M, Lovett A, Rose S. Assessing hospital performance after percutaneous coronary intervention using big data. Circ Cardiovasc Qual Outcomes. 2016;9:659–69.
Bourne PE, Bonazzi V, Dunn M, Green ED, Guyer M, Komatsoulis G, Larkin J, Russell B. The NIH big data to knowledge (BD2K) initiative. J Am Med Inform Assoc. 2015;22(6):1114. https://doi.org/10.1093/jamia/ocv136.
Shein E. Combating Cancer with data. Supercomputers will shift massive amounts of data in search of therapies that work. Commun ACM. 2017;60(5):10–2.
Erl T, Khattak W, Buhler P. Big data fundamentals. Concepts, drivers & techniques. Boston: Prentice-Hall; 2016.
Ankam V. Big data analytics. Birmingham: Packt Publishing; 2016.
Ramírez-Gallego S, García S, Mouriño-Talín H, Martínez-Rego D, Bolón-Canedo V, Alonso-Betanzos S, Benítez JM, Herrera F. Data discretization: taxonomy and big data challenge. Wiley Interdiscip Rev Data Min Knowl Disc. 2016;6(1):5–21.
Yang Y, Webb GI. Discretization for naive-Bayes learning: managing discretization bias and variance. Mach Learn. 2009;74(1):39–74.
Quinlan JR. Induction of decision trees. Mach Learn. 1986;1(1):81–106.
Frank E, Hall MA, Witten IH. The WEKA Workbench. Online appendix for “Data mining: practical machine learning tools and techniques”. 4th ed. Cambridge, MA: Morgan Kaufmann; 2016.
Yang Y, Webb GI. Proportional k-interval discretization for naive-Bayes classifiers. In: European conference on machine learning. Berlin/Heidelberg: Springer; 2001. p. 564–575.
Irani KB. Multi-interval discretization of continuous-valued attributes for classification learning. 1993.
Ramírez-Gallego S, García S, Mourino-Talin H, Martínez-Rego D, Bolón-Canedo V, Alonso-Betanzos A, Herrera F. Distributed entropy minimization discretizer for big data analysis under apache spark. In Trustcom/BigDataSE/ISPA, 2015 IEEE. Vol. 2. IEEE; 2015, August. p. 33–40.
Zhai Y, Ong YS, Tsang IW. The emerging “Big Dimensionality”. IEEE Comput Intell Mag. 2014;9(3):14–26.
Bolón-Canedo V, Sanchez-Marono N, Alonso-Betanzos A. Feature selection for high-dimensional data. Cham: Springer; 2015.
Hall MA. Correlation-based feature selection for machine learning. Doctoral dissertation, The University of Waikato. 1999.
Dash M, Liu H. Consistency-based search in feature selection. Artif Intell. 2003;151(1–2):155–76.
Kononenko I. Estimating attributes: analysis and extensions of RELIEF. In European conference on machine learning. Berlin/Heidelberg: Springer; 1994, April. p. 171–82.
Peng H, Long F, Ding C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell. 2005;27(8):1226–38.
Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learn. 2002;46(1):389–422.
Ramírez-Gallego S, Lastra I, Martínez-Rego D, Bolón-Canedo V, Benítez JM, Herrera F, Alonso-Betanzos A. Fast-mRMR: fast minimum redundancy maximum relevance algorithm for high-dimensional big data. Int J Intell Syst. 2016;0:1–19.
Bolón-Canedo V, Remeseiro B, Alonso-Betanzos A, Campilho A. Machine learning for medical applications. In: Proceedings of the European Symposium on Artificial Neural Networks (ESANN). 2016. p. 225–34.
Flach P. Machine learning. The art and science of algorithms that make sense of data. Cambridge: Cambridge University Press; 2012.
Murphy KP. Machine learning. A probabilistic perspective. Cambridge, MA: MIT Press; 2012.
Shalev-Schwartz S, Ben-David S. Understanding machine learning. Cambridge: Cambridge University Press; 2014.
Barber D. Bayesian reasoning and machine learning. Cambridge: Cambridge University Press; 2012.
Domingos P. The master algorithm. How the quest for the ultimate learning machine will remake our world. New York: Basic Books; 2015.
Bishop CM. Pattern recognition and machine learning. New York: Springer; 2006.
Fernández-Delgado M, Cernadas E, Barro S, Amorim D. Do we need hundreds of classifiers to solve real worls classification problems? J Mach Learn Res. 2014;15:3133–81.
Little RJA, Rubin DB. Statistical analysis with missing data. Chicester: Wiley; 2002.
Horton NJ, Kleiman KP. Much ado about nothing: a comparison of missing data methods and software to fit incomplete data regression model. The American Statiscian. 2007;61(1):79–90.
Kerkhof PL. Characterizing heart failure in the ventricular volume domain. Clin Med Insights Cardiol. 2015;2015(Suppl. 1):11.
Domingo-Ferrer J, Soria-Comas J. Anonymization in the time of big data. In: Domingo-Ferrer J, Pejić-Bach M, editors. Privacy in statistical databases. PSD 2016, Lecture notes in computer science, vol. 9867. Cham: Springer; 2016.
Domingo-Ferrer J, Soria-Comas J. From t-closeness to differential privacy and vice versa in data anonymization. Knowl-Based Syst. 2015;74:151–8. https://doi.org/10.1016/j.knosys.2014.11.011.
Sweeney L, Abu A, Winn J. Identifying participants in the Personal Genome Project by Name. Harvard University. Data Privacy Lab. White Paper 1021-1. April 24, 2013.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Alonso-Betanzos, A., Bolón-Canedo, V. (2018). Big-Data Analysis, Cluster Analysis, and Machine-Learning Approaches. In: Kerkhof, P., Miller, V. (eds) Sex-Specific Analysis of Cardiovascular Function. Advances in Experimental Medicine and Biology, vol 1065. Springer, Cham. https://doi.org/10.1007/978-3-319-77932-4_37
Download citation
DOI: https://doi.org/10.1007/978-3-319-77932-4_37
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-77931-7
Online ISBN: 978-3-319-77932-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)