Abstract
In this paper, we present a clinically pediatric disease prediction system based on a new efficient hybrid knowledge graph. Firstly, we automatically extract a set of triples by modeling and analyzing 1454 clinically pediatric cases, building a weighted knowledge graph based Naïve Bayes. Secondly, to extract new prediction opportunities from heterogeneous data sources, we model and analyze both classically professional pediatrics textbooks and clinical experiences of pediatric doctors respectively in order to derive prediction rules. Thirdly, we mix up those rules with the weighted knowledge graph we built to propose a new hybrid knowledge graph which can carry on both the Bayesian reasoning and the logic calculation at the same time. Fourthly, in term of that hybrid knowledge graph, we further design a new multi-label classifier based on the well-known Bayesian Ranking for the disease prediction. Finally, we implement such a hybrid knowledge graph based disease pediatric prediction system (HKDP) which uses the descriptions of the patients’ symptoms as the inputs so as to return the predicted candidates of diseases for a child. In our experiments, the comparisons with classical prediction methods prove the validity and advantage of our system, especially guaranteeing good balance between the interpretability and precision of predictions in HKDP.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Various computational models have been used to predict disease based on clinical data [1, 2]. Expert systems based on logic and rules were developed in last century [3]. Recent developments in statistical models provide new opportunities for implementing intelligent disease prediction [4]. There are still many challenges in realizing a practical predictive model. One of key problems is how to build a predictive model from heterogeneous data sources. In particular, human doctors make decisions on multiple possible diseases based on both clinical experiences and knowledge learned from text books, while most of machine learning models focus on the clinical data which contains just one type of diseases. There is still, to the best of the author’s knowledge, few works to build a comprehensive model over heterogeneous data sources for predicting multiple diseases.
The work presented in this paper provides a systematic solution towards this issue. Specifically, we proposed a Hybrid Knowledge graph model based pediatric Disease Prediction system (HKDP) to integrate a Bayesian statistic model with knowledge pieces extracted from clinical data, text book and expert experiences. In the system, a multi-label prediction classifier is designed to predict diseases based on clinical symptom descriptive texts. We compare the proposed predictive models with other classical methods. The results show that the precision rate, recall rate and F-score are all improved.
The organization of this paper is as follows: Sect. 2 provides an overview of related work on disease perdition models. Section 3 describes the whole framework of the proposed system and the hybrid knowledge graph construction. Section 4 focuses on the disease diagnosis model. Section 5 presents the experimental evaluation. Section 6 gives conclusion and future works.
2 Related Work
Recently more and more computational models have been applied in intelligent tasks such as question-answer [5] and topic discovering from e-health data [6]. Disease prediction, as a classical problem of Clinical Decision Support System (CDSS) research [3], is still receiving much more attentions. Current major researches on disease prediction focus on two trends: Bayesian models and Artificial Neural Network (ANN) models.
An early Bayesian model in [7] was investigated to predict the on-set of Alzheimer’s disease. [8] has done some research on the suitability of the Gaussian Bayes classifier in determining DR severity level. The Bayesian model was also used in the prediction of heart disease [9, 10]. In [11, 12], two similar heart disease prediction systems based on a naïve Bayes model were proposed respectively, both of which predict the likelihood of patients getting a heart disease, whereas the latter also used a Jelinek-mercer smoothing technique to improve the prediction. In order to compensate for the deficiencies of Bayesian models in heart disease prediction, Nahar has proposed two different improvement methods. One was a feature extraction method based on medical knowledge, which had shown promise in heart disease diagnostics [13]; another was using several rule mining algorithms (Apriori, Predictive Apriori and Tertius) to find association rules in heart disease data which then be applied to a naïve Bayes classifier and had made a favor to identify the key factors behind the disease [14].
Neural network models have been extensively applied in intelligent medication systems. The work can be traced back to 1990s, when artificial neural network began to apply to clinical medicine [15]. In [16], Dawson applied the Bayesian model and Recurrent Neural Networks (RNN) to medical image analysis. Inouye et al. [17] presented a predictive model using admission characteristics which classify delirium in order to help discover potential disease. Recently, significant achievements in deep neural network models provide new opportunities in making more intelligent prediction models based on big data. For example, a diagnostic model based on the Long Short Term Memory (LSTM) model was proposed which established the patient characteristics by identifying the time series and then used the RNN to build the predictive model [18]. This model has a good effect in diagnosing diabetes. On the same time, Google researchers proposed an unsupervised deep feature learning method to extract feature information from Electronic Medical Records (EHR) data and established a better model to predict latent possible diseases [19]. Neural network models are black-box models and are difficult to explain. In order to solve this problem, Zhihua Zhou proposed a C4.5 rule model using artificial neural network to do data preprocessing, which has strong interpretability due to the advantages of rule induction [20].
Most previous work are based on EHR. There is still few work on incorporating heterogeneous data and knowledge that are from both EHR and text books. Moreover, most models are single-disease model, which is not suitable for a real clinical application that will face multiple possible diseases. In this paper, we provide a hybrid knowledge graph model to address these two problems.
3 System Architecture
Figure 1 shows the architecture diagram of the proposed system which consists of following models: The human - computer interaction module receives input from users and displays the returned results. The inquiry processing module is built to process the user input and apply the natural language processing method to identify the relevant feature information from the text of user input. The answers generation module generates the output sentences according to the user input and the results from the intelligent reasoning model. The intelligent reasoning module conduct reasons over the hybrid knowledge graph according to the information from heterogeneous data sources in expertise knowledge, case base and book knowledge and the module will provide disease prediction results or prompt for user to continue providing more symptoms. The symptom prompt module use a Latent Dirichlet Allocation (LDA) [21] to analyze the clinical case feature graph and locates the top-k symptoms as the recommendation for users to evaluate. The system also recommends more symptoms with the largest weights according to the previous inputs of users. Differential diagnosis module combines symptom information entered by the user with the somatoscopy information and uses the diagnostic criteria of the top-k most matching diseases to build a final differential diagnosis based on a multiple labeling prediction model that is built based on the hybrid knowledge graph. Differential diagnosis module is the core module to implement the prediction function in the system. The core methods will be introduced in next sections.
The prediction is based on knowledge extracted from different sources including: Expert experience library mainly comes from the oral dictation of doctors; Semantic Knowledge Base containing common symptoms and signs in medicine, and syntax pattern expression rules that are used to parse the natural language text. Book Knowledge Base contains a large number of entity-relationship information that extracted from a college text book in Pediatrics. Case database contains 1454 raw clinically pediatric cases that come from a hospital. We build a hybrid knowledge graph to integrate knowledge extracted from these heterogeneous data sources for further reasoning.
3.1 Structure of Hybrid Knowledge Graph
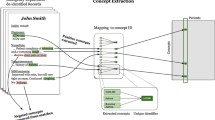
We design a hybrid knowledge graph structure to hold knowledge information from heterogeneous sources. More specifically, we use the hybrid knowledge graph to integrate a statistical model, a rule based model, and a semantic pattern model in one structure so that it can support future extension. The hybrid knowledge graph is a labelled graph consisting of following types of nodes: disease types, symptom types, syntax objects and computational nodes; Links are of types: Bayesian priori-probability, sufficient condition, existence positioning relation and computer input/output. Disease types, symptoms and syntax objects are linked by those edges to represent reasoning semantics and natural language parsing semantics. Computational nodes in the graph are processing algorithms that take inputting nodes as arguments and take outputs also as nodes in the graph. Figure 2 shows a basic structure of a hybrid knowledge graph.
For example, the text parsing node in Fig. 2 takes a set of semantic parsing links as its input and extracts semantics information from natural language of the text book and user input texts. The output of the text parsing node contains multiple entity-relationships that will be integrated into the hybrid knowledge graph. When processing a reasoning task, the text parsing node is also invoked to parse relevant texts into semantic structures for further usage. A run-time execution engine is implemented in the intelligent reasoning module to hold the hybrid knowledge graph and support the running. In this way, we can dynamically update the hybrid knowledge graph to extend the system function.
3.2 Automatic Construction of Hybrid Knowledge Graph
During the doctor’s diagnosis, different symptoms provide different levels of information. For example, “cough” will provide more information than “having a good spirit” for a patient with respiratory infection. Therefore, to simulate a real doctor’s diagnosis process in our system, we assign different weights to each edge in the graph, and the weights represent the diagnostic information that the symptoms can provide in this disease. Finally, we use Eq. 1 to assign different weights to each triple in the knowledge graph for follow-up reasoning.
In it, \( t_{ij} \) is the number of occurrences of the triple object \( t_{i} \) in all cases of disease \( d_{j} \), \( \sum\nolimits_{k} {t_{kj} } \) indicates the total number of triples in all cases of disease \( d_{j} \), and \( \left| D \right| \) indicates the total number of diseases. \( \left| {\left\{ {m:t_{i} \in d_{m} } \right\}} \right| \) indicates the number of diseases with the triple object \( t_{i} \).
The automatic construction of knowledge graphs focuses on the automatic recognition of symptoms in clinical cases. In this study, we mainly use a domain dictionary and a regular expression matching method. The domain dictionary is constructed by hand and contains common symptoms in relevant medical domains. Subsequently, regular expressions are constructed by this domain dictionary, which is used to identify the symptoms of each clinically pediatric case. Besides, the extraction method based on the domain dictionary and regular expressions are also used to find rules from expert experiences and to parse the text book, and then we mix up all results into the hybrid knowledge graph. In summary, the construction of the disease knowledge graph can be described by the following steps.
-
1.
Use the constructed regular expression to extract the symptoms in the cases.
-
2.
Construct triples with the symptoms.
-
3.
Represent the disease as a set of triples and calculate the number of occurrences of different diseases.
-
4.
Construct graph for each disease, calculate the prior probabilities for symptoms and add the weights to the graph.
-
5.
Find rules from expert experience and parse the text book.
-
6.
Mix up the graph, rules and the knowledge of text book into a hybrid knowledge graph and add edges.
4 Disease Diagnosis Model
Based on the detailed analysis of 1454 clinically pediatric cases, we found that doctors always consider more than one type of diseases when making a diagnose over a case, and rank the diseases according to the likelihood they measure. With this in mind, we model the problem of disease diagnosis as a multi-label classification problem considering labels order. In this section, we propose a hierarchical disease diagnosis model based on a naive Bayesian model, rules as well as book knowledge which have been formalized in the hybrid knowledge graph. Figure 3 shows the block diagram of this hierarchical model. When diagnosing a clinical case, the multi-label classification based on naïve Bayes is used firstly, which predict several diseases with probability, then we use the rule-based and the book-knowledge-based reasoning to filter and fill up possible results.
4.1 Naïve Bayes Based Multi-label Classifier
Naive Bayes Classifier
Naïve Bayes is a classical statistical classifier which assumes no dependency between attributes. It works as follows:
Let the input space \( {\fancyscript{x}} \subseteq {R^n} \) be a set of n-dimensional vectors, and the output space \( {\fancyscript{y}} = \left\{ c_{1},c_{2}, \ldots,c_{i} \right\} \) be a set of class labels. The input is a feature vector \( x \in {\fancyscript{x}} \), and the output is a class label \( y \in \fancyscript{y} \). X is a random variable defined in the input space and Y is a random variable defined in the output space. \( P(X,Y) \) is the joint probability distribution of X and Y. The training dataset \( T = \left\{ {(x_{1} ,y_{1} ),(x_{2} ,y_{2} ), \ldots ,(x_{N} ,y_{N} )} \right\} \) is generated by an independent identical distribution \( P(X,Y) \). A Naïve Bayesian model learns the joint probability distribution \( P(X,Y) \) by training over the known dataset. Given the input x, the posterior probability distribution is calculated by the learned model, and the label with the largest posterior probability is regarded as the outputs.
Note that the denominator in the above equation is the same for all \( c_{i} \), so the final formula is
where \( \omega_{ij} \) is the weight of tripe in disease \( c_{i} \) in the hybrid knowledge graph.
Multi-label Classifier
The classic Naïve Bayes classification algorithm is effective for single-label samples, however, in our clinical cases, one sample always contains multiple class labels, which makes it tricky for us to directly use. To solve this problem, we adopted a one-vs-all strategy [22] which learns \( \left| L \right| \) naïve Bayes binary classifiers \( H_{l} :X \to \left\{ {l,\neg l} \right\} \), one for each different disease label \( l \) in L, where L is the number of labels of diseases. The original training data set is transformed into \( \left| L \right| \) data sets \( D_{l} \) that contain all examples of the original training data set, labelled as \( l \) if the labels of the original example contained \( l \) and as \( \neg l \) otherwise. For the classification of a new instance \( x \) this method outputs as a set of labels the union of the labels that are output by the \( \left| L \right| \) Bayes classifier:
Next, we focus on the determination of the order and number of labels. Here we proposed a mechanism called Average Interval Filtering for dynamically adjusting the number of predicted labels. We rank the labels reversely by their posterior probability, and then extract the top \( k \) labels, calculate the average interval between them.
Lastly, we traverse the ranked labels from the second label to the kth label one by one in a way that the current label will be added into the final labels set if the absolute value of the posterior probability of current label and its previous one is smaller than the average interval otherwise the traversing is stopped. Due to the fact that a sample must have at least one label, the label with the largest posterior probability will be added into the labels set firstly with no doubt, Table 1 shows the pseudo code.
4.2 Rule Based Reasoning
In the above work, we have formalized the experience of domain experts into the form of \( \left\{ {IF \ldots THEN \ldots } \right\} \) rules in the hybrid knowledge graph. There are two different kinds of rules in the hybrid knowledge graph, namely, the sufficient condition rule and the necessary condition rule. The sufficient condition rules can be used straightforwardly with the form of \( \left\{ {IF\,s_{1} ,s_{2} \ldots THEN\,d_{1} ,d_{2} \ldots } \right\} \). With the symptoms that we extract, we try to match them using the prefixes in the sufficient condition rules. If the match is successful, the suffixes of the sufficient condition rules which are also the disease labels will be directly added into the set of sample labels. The difference we make use of the necessary condition rules with the form of \( \left\{ {IF\,d_{1} \,THEN\,s_{1} ,s_{2} \ldots } \right\} \) is that the labels we predicted from the above classifier will be re-filtered by the necessary condition rules. Given the label \( l \) in the predicted labels set, we will check whether the symptoms of patients meet the suffixes of the necessary condition rule. If not, the label \( l \) will be eliminated from the predicted labels set.
4.3 Medical Knowledge Driven Approach
In order to further improve the accuracy of the predicted results, we finally use the knowledge of medical books driven approach to identify the disease label in the predicted labels set. The knowledge of medical books has been formalized to \( {\text{XML}} \) format, in which the \( \left\langle {{\text{differential}}\,{\text{diagnosis}}} \right\rangle \) tag records the conditions for the final diagnosis of a disease. Therefore, we will compare the symptoms that we extract from clinical case with the conditions for diagnosis of each disease label in predicted labels set, and the disease label will be one of the eventually outputs if the difference is within an acceptable range, otherwise the disease label is removed.
5 Evaluation
In this section, we introduce the evaluation metrics used in the system. Compared with the traditional single-label classification, multi-label classification usually requires more complex evaluation metrics. In our system, we use precision, recall and F-score [22]. Let D is a data set containing D multi-label samples which can be expressed as \( (x_{i} ,y_{i} ),i = 1 \ldots \left| D \right| \). L is the set of disease tags. \( H \) is a multi-label classifier, \( Z_{i} = H(x_{i} ) \) is the set of labels predicted by sample \( x_{i} \).
The precision is the ratio of all correct results in the returned results with all returned results.
The recall is expressed as the ratio of all correct results in the returned results with all the original correct results.
Accuracy and recall have different emphases, and F-score can be used to balance them. F-value is defined as follows.
5.1 Experiment Results
The dataset contains 1454 clinically pediatric cases in which each sample contains several disease labels and the total number of types of diseases is 71. Because of the imbalance distribution of disease labels in the raw dataset, we totally conducted 5 experiments for each test model and each time we randomly selected 80% samples for training and prediction. Here, we mainly do two groups of contrast experiments. First, we compared our model with the traditional Bayes based multi-label classification model which uses the TF-IDF method for feature extraction. Table 2 presents the values of the evaluation parameters (precision, recall, F-score) of the predictive models. We can see that the average of precision value, recall value and F-score of HKDP are all about 12% larger than TFIDF-NB. Figure 4 illustrates the trends of precision value, recall value and F-score of HKDP and TFIDF-NB in 5 experiments.
Second, we applied the method of feature extraction we used for constructing the hybrid knowledge graph automatically to the classical classifier including SVM, KNN and Logistic Regression. As we can see in Table 3, HKDP also performs better than all other classifiers. The reason is that the book knowledge and expert rules play a significant role in identifying and replenishing the diseases. Besides, the trend graph in Fig. 5 shows that the precision rate, recall rate and F-score of HKDP fluctuate within a range of only 1% in 5 randomized experiments, which indicates that the HKDP also has good stability and robustness.
6 Conclusion and Future Work
Disease prediction is a classic problem in clinical decision support systems (CDSS). One of the new problems is how to improve the accuracy of prediction with multiple heterogeneous data sources. In this paper, we presented a hybrid knowledge graph, which is used to integrate the clinically pediatric cases, classically professional pediatrics textbooks as well as clinical experiences of pediatric doctors, and the automatic construction method of this hybrid knowledge graph. Based on this graph, we built a disease prediction system which provides a multi-label classifier with label filtering. The system parses the descriptive text of patent’s symptoms and uses the knowledge in hybrid knowledge graph for intelligent reasoning and returns the predicted diseases results for patients finally. We conducted experiments on the 1454 clinical cases and the results show that HKDP has better prediction precision, recall and F-score than SVM, KNN, and LR, and has good interpretability at the same time.
In the future work, we will further expand the hybrid knowledge graph, introducing and studying the cognitive simulation model, so that the knowledge graph can simulate the thinking and evaluation process of medical diagnosis of doctors, and thus has better extensibility and self-explanation. Furthermore, we will apply our methods to the Traditional Chinese Medicine(TCM), in order to build a hybrid knowledge graph over TCM data for assisting diagnosis making of TCM.
References
Alshayea, Q.: Artificial neural networks in medical diagnosis. Int. J. Comput. Sci. Issues 96, 197–228 (2011)
Lin, D., Vasilakos, A.V., Tang, Y., Yao, Y.: Neural networks for computer-aided diagnosis in medicine: a review. Neurocomputing 216, 700–708 (2016)
Shortliffe, E.H.: MYCIN: Computer-Based Medical Consultations. Elsevier, New York (1976)
Prema, S., Umamaheswari, P.: Multitude classifier using rough set jelinek mercer Naïve Bayes for disease diagnosis. Circ. Syst. 07, 701–708 (2016)
Zhang, Y., Zhang, Y., Yin, Y., Xu, J., Xing, C., Chen, H.: Chronic disease related entity extraction in online chinese question and answer services. In: Zheng, X., Zeng, D.D., Chen, H., Leischow, Scott J. (eds.) ICSH 2015. LNCS, vol. 9545, pp. 55–67. Springer, Cham (2016). doi:10.1007/978-3-319-29175-8_6
Chen, X., Zhang, Y., Xu, J., Xing, C., Chen, H.: Deep learning based topic identification and categorization: mining diabetes-related topics on Chinese health websites. In: Navathe, Shamkant B., Wu, W., Shekhar, S., Du, X., Wang, X.S., Xiong, H. (eds.) DASFAA 2016. LNCS, vol. 9642, pp. 481–500. Springer, Cham (2016). doi:10.1007/978-3-319-32025-0_30
Prince, M.J.: Predicting the onset of Alzheimer’s disease using Bayes’ theorem. Am. J. Epidemiol. 143, 301–308 (1996)
Hani, A.F.M., Nugroho, H.A., Nugroho, H.: Gaussian Bayes classifier for medical diagnosis and grading: Application to diabetic retinopathy. In: Biomedical Engineering and Sciences, pp. 52–56 (2010)
Subbalakshmi, G., Ramesh, K., Rao, M.C.: Decision support in heart disease prediction system using Naive Bayes. Indian J. Comput. Sci. Eng. 2, 170–176 (2011)
Dangare, C.S., Apte, S.S.: Improved study of heart disease prediction system using data mining classification techniques. Int. J. Comput. Appl. 47, 44–48 (2012)
Patil, A.P., Bhosale, A.P., Ambre, G.: Intelligent Heart Disease Prediction System using Naive Bayes Classifier. Int. J. Adv. Innov. Res. (2013)
Patil, M.R.R.: Heart disease prediction system using Naive Bayes and Jelinek-mercer smoothing. Int. J. Adv. Res. Comput. Commun. Eng. 3, 6787–6792 (2014)
Nahar, J., Imam, T., Tickle, K.S., Chen, Y.P.P.: Computational intelligence for heart disease diagnosis: a medical knowledge driven approach. Expert Syst. Appl. 40, 96–104 (2013)
Nahar, J., Imam, T., Tickle, K.S., Chen, Y.P.P.: Association rule mining to detect factors which contribute to heart disease in males and females. Expert Syst. Appl. 40, 1086–1093 (2013)
Baxt, W.G.: Application of artificial neural networks to clinical medicine. Lancet 346, 1135–1138 (1995)
Dawson, A., Austin Jr., R., Weinberg, D.: Nuclear grading of breast carcinoma by image analysis. Classification by multivariate and neural network analysis. Am. J. Clin. Pathol. 95, S29–S37 (1991)
Inouye, S.K., Viscoli, C.M., Horwitz, R.I., Hurst, L.D., Tinetti, M.E.: A predictive model for delirium in hospitalized elderly medical patients based on admission characteristics. Ann. Internal Med. 119, 474–481 (1993)
Lipton, Z.C., Kale, D.C., Elkan, C., Wetzell, R.: Learning to diagnose with LSTM recurrent neural networks. Comput. Sci. (2015)
Miotto, R., Li, L., Kidd, B.A., Dudley, J.T.: Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci. Rep. 6 (2016)
Zhou, Z.-H., Jiang, Y.: Medical diagnosis with C4. 5 rule preceded by artificial neural network ensemble. IEEE Trans. Inf Technol. Biomed. 7, 37–42 (2003)
Blei, D.M., Ng, A.Y., Jordan, M.I.: Latent dirichlet allocation. J. Mach. Learn. Res. 3, 993–1022 (2003)
Tsoumakas, G., Katakis, I.: Multi-label Classification: An Overview. Department of Informatics, Aristotle University of Thessaloniki, Greece (2006)
Acknowledgments
This work was supported by the Networked Operating System for Cloud Computing (Grant No. 2016YFB1000505) and the Special Program for Applied Research on Super Computation of the NSFC-Guangdong Joint Fund (the second phase). Thanks Zongbo Zhang, Han Li and Dongxue Huo for some preliminary work.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Liu, P. et al. (2017). HKDP: A Hybrid Knowledge Graph Based Pediatric Disease Prediction System. In: Xing, C., Zhang, Y., Liang, Y. (eds) Smart Health. ICSH 2016. Lecture Notes in Computer Science(), vol 10219. Springer, Cham. https://doi.org/10.1007/978-3-319-59858-1_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-59858-1_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-59857-4
Online ISBN: 978-3-319-59858-1
eBook Packages: Computer ScienceComputer Science (R0)