Abstract
Ectomycorrhizal fungal assemblages in tropical regions have remained relatively understudied compared to temperate ecosystems. Sea grape (Coccoloba uvifera) from the Polygonaceae family is a common ectomycorrhizal tree species inhabiting coastal habitats in neotropical forests. We sampled roots of C. uvifera in six locations around the Caribbean basin to characterize the associated ectomycorrhizal fungal assemblages at the regional scale. In accordance with earlier reports, sea grape ectomycorrhizal fungal communities were species-poor, implying ecological host specificity. Species of the /tomentella-thelephora lineage were the most species-rich group inhabiting roots of C. uvifera. Inclusive phylogenetic analyses of Thelephoraceae from Northeast, Central and South America indicated that the mycobionts of C. uvifera tend to cluster with taxa associated with North and Central American Pinus and Fagaceae rather than with South American taxa. This suggests that C. uvifera may have received multiple EcM symbionts from host switching events due to a common habitat in maritime sand dunes or more widely shared historical habitat in North America and the Caribbean Islands. Considering the absence of shared fungal taxa with other Coccoloba species from inland rain forest habitats, we hypothesize that distinct ectomycorrhizal associations of sea grape are primarily driven by stressful environmental conditions in maritime sand dunes.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
16.1 Introduction
A range of biotic, abiotic and historical variables shape the structure and species richness of ectomycorrhizal (EcM) fungal communities. Host and fungal compatibility (i.e. host preference or specificity) that may vary widely across host taxa, has been increasingly shown to influence the structure and richness of EcM fungal assemblages at various taxonomic levels of plants (Ishida et al. 2007; Morris et al. 2008; Tedersoo et al. 2010b, 2013; Bahram et al. 2012; Põlme et al. 2013). To date, the influence of environment on EcM fungal richness and composition has received relatively limited attention and the biodiversity of several tropical plant groups has remained unknown. Unlike in most other organisms and fungi overall, EcM fungi exhibit greater diversity in temperate compared with tropical and arctic ecosystems (Tedresoo et al. 2012, 2014; Chap. 18). This has been ascribed to historical factors (the lack of earliest evolving Pinaceae hosts in lowland tropical habitats), rapid turnover of organic matter in tropical soils and low relative abundance of hosts.
The genus Coccoloba (Polygonaceae) comprises ca. 170 species of shrubs and trees with neotropical distribution (Howard 1960; Chap. 19). Coccoloba spp. are probably of South American origin, with greatest richness in northern Amazonia and the Atlantic rain forest (Howard 1961; Chap. 20). EcM associations were first described from sea grape (Coccoloba uvifera (L.) L.) in Cuba (Kreisel 1970) and thereafter in South American rain forest species (Moyersoen 1993; Bereau et al. 1997). The associated EcM fungi are poorly known, because host plants have remained unsettled in most South American mycological studies (Pegler 1983; Singer et al. 1983; Roy et al. 2016; but see Tedersoo et al. 2010b).
Sea grape is a tropical tree species with a native distribution from southern Florida to eastern Mexico to northeastern Brazil. Sea grape grows in immediate proximity of seashores, representing one of the first colonizers of sandy and rocky coastal areas. These habitats are characterized by high salinity, steady wind, seasonal drought and low soil nutrient availability. EcM symbiosis could potentially enhance plant tolerance of those harsh conditions, particularly of high salinity (Bandou et al. 2006). Séne et al. (2015) conducted a profound study sampling sporocarps and EcM root tips in sea grape communities and reported very limited EcM fungal diversity in Guadeloupe Island. The authors postulated four nonexclusive hypotheses to explain the low EcM fungal diversity: (1) isolation from mainland, (2) overall lower EcM fungal diversity at lower latitudes, (3) environmental filtering due to stressful conditions, (4) recent origin of EcM symbiosis within the Polygonaceae. In spite of profound local sampling effort in a small volcanic island, their study cannot be used to generalize about sea grape EcM communities over a wider geographic context.
In order to test the above hypotheses in a wider geographical and historical context, we sampled six additional sea grape communities around the Caribbean basin and compared these in the biogeographic perspective. We further addressed the potential specificity and origin of EcM fungi of C. uvifera by comparing the associated Tomentella species—the dominant group of mycobionts—to these from other hosts in North and South America.
16.2 Approaches
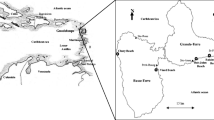
We performed root sampling in six study sites in the following locations: USA: Miami (June 2013; 26.0408°N; −80.1145°E), Mexico: Celestún (October 2015; 20.9336°N; −90.3748°E), Costa Rica: Cahuita (June 2013; 9.8590°N; −82.9458°E), Cuba: Cayo Santa Maria (December 2008; 22.6581°N; −79.0413°E), French Guiana: Montabo (November 2013; 4.9436°N; −52.2973°E) and Colombia: Los Naranjos (November 2014; 11.2973°N; −73.8946°E). From each site, roots from ten sea grape individuals were collected (except for the Cuba plot where 14 samples were collected). Randomly selected samples (15 × 15 cm to 10 cm depth) were situated at least 10 m apart.
Roots were cleaned from adhering soil in tap water and morphotyped under a stereomicroscope. EcM morphotypes were distinguished based on colour and roughness of mantle, presence of emanating hyphae and rhizomorphs. At least two EcM root tips from each morphotype per soil sample were stored in CTAB buffer (1% cetyltrimethylammonium bromide, 100 mM Tris–HCL (pH 8.0), 1.4 M NaCl, 20 mM ethylenediaminetetraacetic acid) for molecular analyses.
DNA was extracted from EcM root tips using Thermo Scientific Phire Plant Direct PCR Kit (Thermo Scentific, Waltham, MA, USA) according to the manufacturer’s instructions. In the course of the study, PCR was performed by use of 5× HOT FIREPol Blend Master Mix Ready to Load (Solis BioDyne, Tartu, Estonia). In EcM root tips, fungal rDNA Internal Transcribed Spacer (ITS) region was amplified with a forward primer ITSOF-T (5′-acttggtcatttagaggaagt-3′) in combination with reverse primers LB-W (5′-cttttcatctttccctcacgg-3′) or TW13 (5′-ggtccgtgtttcaagacg-3′). In case of PCR failure, we combined ITSOF-T with universal primers ITS4 (5′-tcctccgcttattgatatgc-3′), or basidiomycete-specific primer ITS4B (5′-caggagacttgtacacggtccag-3′) and LR0B (5′-acccgctgaacttaagc-3′) in order to amplify a shorter fragment of fungal DNA. To improve sequence quality, some root tip extracts were re-amplified with taxon-specific primers (Tedersoo et al. 2008). PCR and sequencing were run following Põlme et al. (2013). Sequences were assembled, checked, trimmed and manually corrected in Sequencher 5.1 software (GeneCodes Corp., Ann Arbor, MI, USA).
Sequences were confirmed to belong to EcM fungal lineages (cf. Tedersoo et al. 2010a; Chap. 6) by use of BLASTn searches against the International Sequence Databases (INSD) or UNITE (Abarenkov et al. 2010a). Sequences were partitioned into operational taxonomic units (OTUs), defined as a group of sequences sharing at least 97% pairwise similarity. Sequences with sufficient length and quality were assigned to UNITE species hypothesis (SHs; Kõljalg et al. 2013) with 3% dissimilarity threshold. We also included the recently published data of Séne et al. (2015) from Guadeluope. This study covered four study plots and had a much higher per-site sampling effort.
Using the PlutoF web platform (Abarenkov et al. 2010b), we downloaded all Tomentella sequences originating from South, Central and Northeast America. Identical sequences from the same sampling plots and hosts were removed prior to phylogeny construction. All sequences were aligned using MAFFT software (Katoh and Standley 2013). The alignment was manually adjusted in AliView software (Larsson 2014) and Maximum likelihood analysis was performed in FastTree 2.1 (Price et al. 2010) using default settings, with Odontia fibrosa (UDB000284) as an outgroup (Tedersoo et al. 2015).
16.3 Fungal Diversity
Out of 147 EcM root tips subjected to molecular analyses, 131 (89%) yielded good quality sequences. These sequences (including material collected by Séne et al. 2015 from Guadeloupe) were clustered into 42 OTUs (Table 16.1). Altogether 33 of OTUs were accommodated to existing SHs at the 97% sequence similarity cutoff level. Of these SHs, 26 (78.8%) were exclusively associated with sea grape. Only five fungal OTUs found from newly sampled sites overlapped with the Guadeloupe study, whereas eight OTUs remained exclusive to Guadeloupe.
The /tomentella-thelephora was by far most taxon-rich phylogenetic lineage of EcM fungi comprising 21 OTUs. Other sea grape-associating lineages were represented with the following number of OTUs: /boletus (four), /inocybe (four), /clavulina (three), /pisolithus-scleroderma (two), /sebacina (two), /paxillus-gyrodon (two), /russula-lactarius (one), /cantharellus (one), /serendipita (one; questionable mycorrhizal status) and /cenococcum (one). In spite of taxonomical richness of the /tomentella-thelephora lineage, Scleroderma bermudense (SH003700.07FU) and Scleroderma sp. (SH003702.07FU) were the most common individual taxa that were present in six and four sites out of seven, respectively.
Florida constituted the most OTU-rich site harbouring 13 EcM fungal taxa, followed by Cuba (11 OTUs), Colombia (10) and Costa Rica (9). Interestingly, Mexico and French Guiana sites harboured only two and one EcM fungal OTU, respectively (Fig. 16.1). In comparison, altogether 14 EcM OTUs were identified from root tips from four plots in Guadeloupe, with 4–9 EcM fungal OTUs per plot (Séne et al. 2015), but this can be ascribed to more extensive sampling effort on a local scale. None of the sea grape-associated OTUs overlapped with EcM fungal taxa associated with the five sampled Coccoloba spp. in Ecuador (Tedersoo et al. 2010b).
Map of study sites indicating number of fungal OTUs in each sampling area. Shared OTUs between areas are shown with arrows. Note that inland Coccoloba species from Ecuador (Tedersoo et al. 2010b) do not share any fungal OTUs with Coccoloba uvifera communities from the coastal areas
The overall OTU richness detected over a broader geographic scale increased nearly three-fold compared to the single study conducted in Guadeloupe (Séne et al. 2015). In addition to formerly reported EcM lineages, we found /sebacina, /clavulina and /boletus lineages from our sampling sites, which were not found in Guadeloupe. Interestingly, several EcM fungal lineages such as /paxillus-gyrodon, /serendipita, /russula-lactarius and /cantharellus were not found outside Guadeloupe. The two Guadeloupean species of Melanogaster belonging to the /paxillus-gyrodon lineage are remarkable, because this genus and the entire EcM lineage is not known to associate with hosts of tropical origin. Species of Cantharellus fruited in abundance in another C. uvifera stand ca. 3 km distant from the Colombian site, but ITS sequencing of these fruit-bodies failed, indicating either primer bias or extensive length of the ITS region (Tedersoo et al. 2016). It is likely that additional sampling effort would have revealed these taxa from the mainland as well and that the true EcM fungal species richness associated with sea grape is considerably higher than the currently reported 42 fungal OTUs (Fig. 16.2). However, the coarse structure of EcM fungal communities was relatively similar to that previously reported in Guadeloupe i.e. /tomentella-therephora being most taxon rich lineage and Scleroderma bermudense being the most abundant fungal taxon. Sites in French Guiana and Mexico were extremely species-poor, comprising only one and two fungal OTUs respectively. In French Guiana, S. bermudense colonized sea grape root systems in all samples. The French Guiana site was characterized by intense anthropogenic disturbance in addition to a small host tree population. However, the Florida site with the highest OTU richness was also characterized by substantial anthropogenic impact but with considerably larger host population.
Rarefied OTU accumulation curve of Coccoloba uvifera associated EcM fungi found in Caribbean basin. Closed circles and open circles represent the rarefied curve and its 95% confidence intervals, respectively. Triangles and squares represent the values of Chao2 and Jacknife2 minimum richness estimators, respectively. The values were calculated based on 999 permutations using EstimateS 9 (Colwell 2013)
16.4 Environmental Filtering and Host Specificity
Séne et al. (2015) proposed that the impoverished EcM fungal richness detected in Guadeloupe could partly result due to a founder effect and isolation from mainland. Although our sampling intensity is too low for comprehensive statistical comparison, the mainland and Guadeloupe sea grape communities harboured comparable EcM fungal richness, largely refuting this hypothesis. In spite of phylogenetic and geographical proximity, the absence of shared OTUs with inland Coccoloba species from neotropical forest, which harboured higher EcM diversity (Tedersoo et al. 2010b), supports the hypothesis of environmental filtering, also proposed by Séne et al. (2015) as an alternative. This explanation coincides well with the fact that vast majority of the SHs detected were exclusively associated with the sea grape. Similarly, Pisonia grandis (Nyctaginaceae) inhabiting small and often guano-rich Indian Ocean and Pacific islands harbours species poor and highly specific EcM assemblage (Suvi et al. 2010; Hayward and Horton 2012). Therefore, putative host specificity in such stressful habitats is most likely bounded with environmental filtering (but see Hayward and Horton 2012). The strong intrageneric ecological specificity in EcM fungi associated with Coccoloba contrast to the largely genus-level specific EcM fungi of Alnus (Põlme et al. 2013). Interestingly, environmental filtering is likely to have an important role in driving specificity of EcM interactions in both cases (Huggins et al. 2014). Nevertheless, we are unable to confidently disentangle the cause and consequence between the ecological host specificity and environmental filtering, because genetic and physiological compatibility between host and symbiont is likely to evolve mutually in extreme habitats over extended periods of time. Séne et al. (2015) pointed out that EcM origin in Coccoloba is relatively recent and this also holds true for Pisonia (Chap. 19), possibly explaining low EcM diversity in both groups. The fact that numerous EcM host taxa, with much broader range of mycobionts, have diverged in a comparable time frame (Chap. 19), makes this hypothesis disputable. Taken together, the hypothesis of strong environmental filtering seems the most plausible explanation for the low fungal diversity in C. uvifera.
16.5 Biogeography of Thelephoraceae
Using the PlutoF workbench, we were able to recover 832 sequences belonging to the /tomentella-thelephora lineage originating from eastern North America, northern South America and Central America. After removal of redundant sequences originating from the same host and study site, 525 sequences were subjected to a phylogenetic analysis (Fig. 16.3). In the large Thelephoraceae phylogram, sea grape-associated sequences clustered together more commonly with sequences originating from Central America and North America rather than with those from South America. This conflicts with the putative South American origin of the genus (Raven and Axelrod 1974; Chap. 20) and previously established patterns in Russulaceae (De Crop et al. 2017) and Sclerodermataceae (Wilson et al. 2012) putatively associating with Coccoloba spp. In terms of host identity, Thelephoraceae sequences associated with C. uvifera most often clustered together with sequences from Pinus spp. and to a lesser extent with those from Quercus spp. This suggests potential host shifts from phylogenetically distant host taxa. Previous studies focusing on the mycobionts of introduced plants support that host shifts for EcM fungi may occur between distantly related taxa such as Fagales and Pinales in a very short time frame (Bahram et al. 2013). Currently, the distribution of Coccoloba spp. overlaps with that of Pinus spp. and Fagales from Central Mexico to Nicaragua and in Cuba (Chap. 20). Furthermore, the fossil record indicates much greater overlap among the geographic range of Pinales, Fagales and Coccoloba during the Oligocene and Eocene (Gray 1960; Graham and Jarzen 1969), when Coccoloba spp. were distributed up to Central USA in the north, whereas Fagales spp. were more widely distributed in the Caribbean islands. In particular, C. uvifera shares its habitat with Pinus spp. in coastal sand dunes, although these species do not co-exist in present-day communities, except perhaps in the Bahamas. Nevertheless, the results of phylogenetic analysis should be interpreted with caution, because South America remains relatively under sampled compared to Central America and especially North America, which may slightly overestimate the links between Pinaceae and C. uvifera and particularly the genus Coccoloba in general.
Maximum likelihood phylogram of Thelephoraceae sequences from South, Central and Northeast America. Clades that do not contain Coccoloba associated Thelephoraceae sequences have been collapsed. Clades highlighted in purple, red and green represent Northeast, Central and South American groups, respectively. Blue, yellow and orange represent the mixture of North and Central American groups, North and South American groups, and Central and South American groups, respectively. Transparent clades represent a mixture of all three regions. Bold names indicate Thelephoraceae sequences associating with various Coccoloba species. The sequence name includes accession number, country, associating host species and species hypothesis (if available)
We also tested, whether the habitats historically influenced by Pinaceae and Fagales (Florida, Costa Rica, Mexico, Cuba) harbour more OTUs of Thelephoraceae shared with northern hosts than the sites far from natural Pinaceae and Fagales habitats (Guadeloupe, Colombia, French Guiana). Contrary to our expectations, there were no differences between the sites with shared and unshared habitats (n = 34; χ 2 = 0.123; P = 0.726). The wide distribution of EcM fungi of potentially northern temperate origin indicates their effective dispersal and good adaptation to tropical climate.
16.6 Conclusions
Our regional-scale study provides strong evidence that sea grape harbors distinct and relatively species-poor EcM fungal communities throughout its range. These associated fungi are highly distinct from the mycobionts of other Coccoloba spp., suggesting that strong environmental filtering due to salinity and perhaps high pH may cause the high observed ecological specificity. Close affinities of C. uvifera-associated Tomentella spp. with those from the phylogenetically distant Pinaceae and Fagaceae hosts supports multiple host shifts and/or broadening of host range due to more commonly shared habitats in the past. Future phylogeographic studies including more samples from South America are needed to enlighten the evolutionary history of EcM symbiosis in Central and South America.
References
Abarenkov K, Nilsson RH, Larsson K-H, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Ursing BM, Vrålstad T, Liimatainen K, Peintner U, Kõljalg U (2010a) The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol 186:281–285
Abarenkov K, Tedersoo L, Nilsson RH, Vellak K, Saar I, Veldre V, Parmasto E, Prous M, Aan A, Ots M, Kurina O, Ostonen I, Jõgeva J, Halapuu S, Põldmaa K, Toots M, Truu J, Larsson K-H, Kõjlag U (2010b) PlutoF—a web based workbench for ecological and taxonomic research, with an online implementation for fungal ITS sequences. Evol Bioinforma 6:189–196
Bahram M, Põlme S, Kõljalg U, Zarre S, Tedersoo L (2012) Regional and local patterns of ectomycorrhizal fungal diversity and community structure along an altitudinal gradient in the Hyrcanian forests of northern Iran. New Phytol 193:465–473
Bahram M, Kõljalg U, Kohout P, Mirshahvaladi S, Tedersoo L (2013) Ectomycorrhizal fungi of exotic pine plantations in relation to native host trees in Iran: evidence of host range expansion by local symbionts to distantly related host taxa. Mycorrhiza 23:11–19
Bandou E, Lebailly F, Muller F, Dulormne M, Toribio A, Chabrol J, Courtecuisse R, Plenchette C, Prin Y, Duponnois R, Thiao M (2006) The ectomycorrhizal fungus Scleroderma bermudense alleviates salt stress in seagrape (Coccoloba uvifera L.) seedlings. Mycorrhiza 16:559–565
Bereau M, Gazel M, Garbaye J (1997) Les symbioses mycorhiziennes des arbres de la foret tropicale humide de Guyane francaise. Can J Bot 75:711–716
Colwell RK (2013) EstimateS: statistical estimation of species richness and shared species from samples. Version 9. User’s guide and application published at: http://purl.oclc.org/estimates
De Crop E, Nuytinck J, Van de Putte K, Wisitrassameewong K, Hackel J, Stubbe D, Hyde KD, Roy M, Halling RE, Moreau PA, Eberhardt U (2017) A multi-gene phylogeny of Lactifluus (Basidiomycota, Russulales) translated into a new infrageneric classification of the genus. Persoonia 38:58–80
Graham A, Jarzen DM (1969) Studies in neotropical paleobotany. I. The Oligocene communities of Puerto Rico. Ann Mo Bot Gard 56:308–357
Gray J (1960) Temperate pollen genera in the Eocene (Claiborne) Flora, Alabama. Science 132:808–810
Hayward JA, Horton TR (2012) Edaphic factors do not govern the ectomycorrhizal specificity of Pisonia grandis (Nyctaginaceae). Mycorrhiza 22:647–652
Howard RA (1960) Studies in the genus Coccoloba IX, a critique of the South American species. J Arn Arbor 41:357–390
Howard RA (1961) Studies in the genus Coccoloba, X. New species and a summary distribution in South America. J Arn Arbor 42:87–95
Huggins JA, Talbot J, Gardes M, Kennedy PG (2014) Unlocking environmental keys to host specificity: differential tolerance of acidity and nitrate by Alnus-associated ectomycorrhizal fungi. Fungal Ecol 12:52–61
Ishida TA, Nara K, Hogetsu T (2007) Host effects on ectomycorrhizal fungal communities: insight from eight host species in mixed conifer–broadleaf forests. New Phytol 174:430–440
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AF, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson L, Lindahl BD, Lücking R, Martin MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KB, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson K-H (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277
Kreisel H (1970) El papel de los hongos en la veetacion forestal de Cuba. Bol Soc Mex Mic 4:39–43
Larsson A (2014) AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278
Morris MH, Smith ME, Rizzo DM, Rejmánek M, Bledsoe CS (2008) Contrasting ectomycorrhizal fungal communities on the roots of co-occurring oaks (Quercus spp.) in a California woodland. New Phytol 178:167–176
Moyersoen B (1993) Ectomicorrizas i micorrizas vesiculo-arbusculares en Caatinga Amazonica del Sur de Venezuela. Sci Guaianae 3:1–82
Pegler DN (1983) Agaric Flora of the lesser Antilles. Her Majesty’s stationary Office, London (Kew Bull. Additional Ser. IX)
Põlme S, Bahram M, Yamanaka T, Nara K, Dai YC, Grebenc T, Kraigher H, Toivonen M, Wang PH, Matsuda Y, Naadel T (2013) Biogeography of ectomycorrhizal fungi associated with alders (Alnus spp.) in relation to biotic and abiotic variables at the global scale. New Phytol 198:1239–1249
Price MN, Dehal PS, Arkin AP (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490
Raven PH, Axelrod DI (1974) Angiosperm biogeography and past continental movements. Ann Mo Bot Gard 61:539–673
Roy M, Schimann H, Braga-Neto R, Da Silva RA, Duque J, Frame D, Wartchow F, Neves MA (2016) Diversity and distribution of ectomycorrhizal fungi from Amazonian lowland white-sand forests in Brazil and French Guiana. Biotropica 48:90–100
Séne S, Avril R, Chaintreuil C, Geoffroy A, Ndiaye C, Diédhiou AG, Sadio O, Courtecuisse R, Sylla SN, Selosse MA, Bâ A (2015) Ectomycorrhizal fungal communities of Coccoloba uvifera (L.) L. mature trees and seedlings in the neotropical coastal forests of Guadeloupe (Lesser Antilles). Mycorrhiza 25:547–559
Singer R, Araujo I, Ivory MH (1983) The ectotrophically mycorrhizal fungi of the neotropical lowlands, especially Central Amazonia. Nova Hedw 77:1–352
Suvi T, Tedersoo L, Abarenkov K, Beaver K, Gerlach J, Koljalg U (2010) Mycorrhizal symbionts of Pisonia grandis and P. sechellarum in Seychelles: identification of mycorrhizal fungi and description of new Tomentella species. Mycologia 102:522–533
Tedersoo L, Jairus T, Horton BM, Abarenkov K, Suvi T, Saar I, Kõljalg U (2008) Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytol 180:479–490
Tedersoo L, May TW, Smith ME (2010a) Ectomycorrhizal lifestyle in fungi: global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 20:217–263
Tedersoo L, Sadam A, Zambrano M, Valencia R, Bahram M (2010b) Low diversity and high host preference of ectomycorrhizal fungi in Western Amazonia, a neotropical biodiversity hotspot. ISME J 4:465–471
Tedersoo L, Bahram M, Toots M, Diedhiou AG, Henkel TW, Kjøller R, Morris MH, Nara K, Nouhra E, Peay GK, Polme S, Ryberg M, Smith ME, Kõljalg U (2012) Towards global patterns in the diversity and community structure of ectomycorrhizal fungi. Mol Ecol 21:4160–4170
Tedersoo L, Mett M, Ishida TA, Bahram M (2013) Phylogenetic relationships among host plants explain differences in fungal species richness and community composition in ectomycorrhizal symbiosis. New Phytol 199:822–831
Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Villarreal-Ruiz L, Vasco-Palacios A, Quang Thu P, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Ratkowsky D, Pritsch K, Riit T, Põldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Pärtel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo L, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, De Kesel A, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K (2014) Global diversity and geography of soil fungi. Science 346:1078
Tedersoo L, Harend H, Buegger F, Pritsch K, Saar I, Kõljalg U (2015) Stable isotope analysis, field observations and synthesis experiments suggest that Odontia is a non-mycorrhizal sister genus of Tomentella and Thelephora. Fungal Ecol 11:80–90
Tedersoo L, Liiv I, Kivistik PA, Anslan S, Kõljalg U, Bahram M (2016) Genomics and metagenomics technologies to recover ribosomal DNA and single-copy genes from old fruitbody and ectomycorrhiza specimens. MycoKeys 13:1–20
Wilson AW, Binder M, Hibbett DS (2012) Diversity and evolution of ectomycorrhizal host associations in the Sclerodermatineae (Boletales, Basidiomycota). New Phytol 194:1079–1095
Acknowledgements
We thank two reviewers, E. Otsing and I. Hiiesalu for critical comments on the manuscript. J. A. Manjarrez and A. Arguelles helped with field sampling in Mexico. This study received funding from the Estonian Science Foundation (grants PUT1399 and PUT1317).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Põlme, S., Bahram, M., Kõljalg, U., Tedersoo, L. (2017). Biogeography and Specificity of Ectomycorrhizal Fungi of Coccoloba uvifera . In: Tedersoo, L. (eds) Biogeography of Mycorrhizal Symbiosis. Ecological Studies, vol 230. Springer, Cham. https://doi.org/10.1007/978-3-319-56363-3_16
Download citation
DOI: https://doi.org/10.1007/978-3-319-56363-3_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-56362-6
Online ISBN: 978-3-319-56363-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)