Abstract
The dynamics of a cell is always changing. Cells move, divide, communicate, adapt, and are always reacting to their surroundings non-synchronously. Currently, single-cell metabolomics has become the leading field in understanding the phenotypical variations between them, but sample volumes, low analyte concentrations, and validating gentle sample techniques have proven great barriers toward achieving accurate and complete metabolomics profiling. Certainly, advanced technologies such as nanodevices and microfluidic arrays are making great progress, and analytical techniques, such as matrix-assisted laser desorption ionization (MALDI), are gaining popularity with high-throughput methodology. Nevertheless, live single-cell mass spectrometry (LCSMS) values the sample quality and precision, turning once theoretical speculation into present-day applications in a variety of fields, including those of medicine, pharmaceutical, and agricultural industries. While there is still room for much improvement, it is clear that the metabolomics field is progressing toward analysis and discoveries at the single-cell level.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Single cell
- Metabolomics
- Mass spectrometry
- Nanodevices
- Matrix-assisted laser desorption ionization
- Live single-cell mass spectrometry
- Circulating tumor cell
- Drug discovery
1 Introduction
Metabolomics, the study of the complete complement of all small molecules (<1500 Da) found in a specific cell, organ, or organism [1], is considered the most recent – and arguably the end point – of the omics cascade [2]. In fact, it has been mentioned by Patti et al. to be the “apogee of the omics trilogy” [3], because unlike genomics and proteomics, metabolomics depicts real-time biochemical activity and therefore is the key in phenotype association and offers a more reliable depiction of the dynamics of the sample in question. As science progresses and technology advances, it has now become clear that tissue-scale metabolomics, or even multicell metabolomics, yields averaged data that can oftentimes be misleading in making assumptions relating to a cell’s condition. Cellular heterogeneity is dynamic and may result from a plethora of factors, including genetic, epigenetic, or phenotypic differences; morphological, biochemical, or functional changes; positional, exogenous, or endogenous mutations; and physical, chemical, or biological effects from the environment [4]. Even cells with identical genotypes can display phenotypical differences [5].
In addition to the above possible causes, stochasticity – induced phenotypic heterogeneity through gene and protein expression – was only recently added as another factor contributing to cell deviation [6, 7]. They explained that due to the low copy number of DNA and mRNA, “noise,” or random irregularities in the rate of their respective reactions, occurs in gene or protein expression, which then causes variable molecule concentrations from cell to cell.
Alas, it is near impossible to find two cells with the same metabolome, even if they originate from the same precursor cell. Furthermore, metabolites and other small molecules cannot be amplified like DNA, which is why most metabolomic and proteomic studies are comprised of large cell number homogenization. The ability to detect, identify, and quantify metabolites within a single cell will open new doors to understanding the reason behind cell-to-cell heterogeneity, even within a seemingly homogeneous population.
This chapter discusses the importance of single-cell metabolomics and significant points to consider when sampling a single cell. It also highlights the foremost approaches in sampling and analysis and gives a perspective into the future for single-cell analysis.
1.1 Why Single-Cell Metabolomics?
In life sciences, the cell is regarded as the minimal functional unit, and its analysis has been undoubtedly crucial. Single-cell analysis represents qualitatively and exhaustively analyzing a wide range of molecular information carried on numerous biomolecules at the single-cell level. It can give insight into unknown processes such as cellular evolution, adaptation, and communication.
Single-cell studies are theoretically the only types of analyses that can give a depiction of real-time biochemical reactions that oftentimes only take seconds or minutes to occur. The realization of how rapid the kinetics actually is within a cell can only move science toward the direction of single-cell metabolomics.
Not only that, but cells behave non-synchronously and therefore must be studied independently when examining topics such as the cell cycle in order to obtain an accurate metabolic profile. Concepts such as stochasticity and heterogeneity would be lost in translation when analyzing cell populations due to averaging. Therefore, in cases of phenotypic studies, single-cell measurements are essential to produce unbiased metabolic models.
During the past few years, single-cell technologies have undergone rapid development and reached a critical point where they have become a valuable tool for systematic characterization of cellular heterogeneity, which in turn has important implications in a wide range of biomedical issues such as gene regulation, cell lineage differentiation, signaling response, and disease characterization. The medical field has evolved significantly, which has led toward a paradigm shift in medicine: moving the observation from patient and organ toward a more in-depth observation – single cells within the organ. This advancement has been enabled to some extent by engineering sciences, among which microsystem technologies were a major driving force.
Perhaps the best example for the need of single-cell metabolomics is cancer. It only takes one abnormal cell in the whole body that contains 30 trillion cells to cause cancer [8]. Single-cell analysis has the potential to help in early detection of medical conditions involving modifications in the cellular functions such as cancer genesis and progression. There is also a need to differentiate between cells with different metabolomes within the same cancer in order to evaluate phenotype heterogeneity, and in effect, prevent drug resistance or discover new, more effective therapeutics.
2 Sample Considerations
2.1 The Size of the Cell Matters
The term “single cell” has no limitation on the actual size of the analyzed cell, and cells come in a wide spectrum in terms of size. To date, the largest single-cell organism documented is Caulerpa taxifolia, a member of the green algae family [9]. It ranges in size, but can grow up to several meters in length. Another much more common single cell is an egg. While the largest egg is that of an ostrich (measuring an average of 16 cm long and 13 cm in diameter and weighing 1.5 kg), chicken eggs (which are 24 times smaller than the ostrich egg on average) are of much more common use in science [10]. Mattsson et al. completed metabolic profiling of chicken embryos after exposure to perfluoroalkyl acids, a group of ecologically detrimental organic chemicals, by drilling holes in each “single cell” and injecting with the said substance [11]. Moving along the size spectrum, embryos of Xenopus laevis, or the South African clawed frog, have been studied to further understand embryonic development through single-cell metabolomics [12, 13]. The group of Sweedler has employed several techniques for extensive analysis of single neurons of the sea slug Aplysia californica, a cell that can grow up to 500 μm [14–19]. Concerning agricultural chemistry, large plant cells have often been the target of single-cell analysis due to their large size, including Allium cepa, commonly known as onion [20].
On the other side of the size spectrum, single-cell studies have been shifting toward smaller and smaller cells. The team of Masujima coined the term “live single-cell mass spectrometry” and have analyzed single plants cells – which can range from 10 to 100 μm – from the leaf, stem, and petal of Pelargonium zonale [21] and the leaf, stem, and root of Raphanus sativus [22]. The same group has done numerous studies on mammalian cells such as mouse embryonic fibroblasts Swiss 3 T3 [23], rat basophilic leukemia cell RBL-2H3 [24, 25], and hepatocellular carcinoma HepG2 [26], all ranging from 10 to 20 μm in size. Their most recent studies have been on single blood cells in relation to clinical studies, having successfully analyzed white blood cells and circulating tumor cells (CTC), ranging in size from 12 to 15 μm, and analyzed red blood cells, with an even smaller size of 6–8 μm [27]. Figure 13.1 shows actual size ratios of some of the most common cells used in single-cell analysis. The human oocyte is the largest single cell in the human body and is added as a point of comparison.
It seems that this may be the limit for true single-cell analysis. Ibáñez et al. recently published a study on single-cell yeast metabolomics [28], but their method involved using microarrays for mass spectrometry (MAMS) platform, in which each hydrophilic reservoir holds anywhere between one and 15 cells. In regard to bacteria, groups such as that of Tanaguchi have been successful in proteomics and transcriptomics of Escherichia coli [29–31], but no papers have been reported on single-cell bacteria metabolomics, revealing that the current instrumentation has reached a plateau in sensitivity. As previously stated, metabolites, in contrast to other biomolecules, cannot be amplified [32], and fluorescent labeling would negatively impact metabolic pathways within the cells, making sensitivity in smaller cells the major limiting factor in analysis.
2.2 The Condition of the Cell
Numerous techniques have been established for the isolation and analysis of a single cell, but one must ask themselves: what condition is the cell in? Even if the analysis is successful, will this be an accurate depiction of the metabolic profile? “A major issue is a suitable sample preparation that does not upset the metabolism of the cells to be investigated. One way to cope with this problem is to keep the cell in a native environment as long as possible” [33].
Cell sampling is arguably the most critical step for single-cell analysis. Protocols used for metabolomic studies on a cell population are deemed useless on the single-cell level. For a single cell, many methods have been established, and many separation kits have been developed [34]; but these processes are often long and contain many steps, causing a disturbance in the natural microenvironment around the cell and increasing the possibility of metabolite distortion or exhaustion of molecules of low concentration. The time taken between sampling and analysis allows for, sometimes abnormal, enzymatic activity and biochemical processing to cope with the changing conditions. Methods such as shock freezing [28], or the immediate addition of organic solvent after sampling, have been used to prevent continual enzymatic activity [22].
To increase high throughput, newer approaches separate and grow single cells in individual wells or innovated trapping arrays. Because they are grown inside these devices, many argue that it is the perfect method for true single-cell metabolomics. On the other hand, others criticize these methods because the cell is not in its natural environment. Think of it this way: would a person in a room full of people act the same if he/she were in a room alone? Cell communication is a natural and essential factor, and completely isolating a cell puts it in an unnatural position, which could be projected into the metabolomic profile.
Moreover, it is worth noting that despite the recent innovations in single-cell isolation techniques, keeping the cell in its natural environment as much as possible until initiating analysis is the most important aspect of analysis. Most current methods require a degree of manipulation to the cell and isolate the cell from cell-cell interactions, thus making it difficult to study certain aspects of cell biology such as communication or signaling. The ideal sampling technique should be noninvasive, include an efficient quenching step, and have a short lysis/analysis time to prevent metabolite decomposition.
3 Methods and Approaches
3.1 Sampling Techniques
The process of single-cell analysis starts with sample preparation. The goal of sample preparation in single-cell analysis is to isolate the target cell in a high-throughput manner without affecting the normal metabolome of the cell. Moreover, to achieve such goal, many sampling techniques have been recently developed, each method having its own unique advantages, disadvantages, and possible applications, which will be discussed in this section.
There are two main approaches for isolation and preparation of the cell for analysis. The first approach is using nanodevices to manipulate the cell and isolate it for further downstream analysis. The second approach utilizes microfluidic devices, which have the advantage of higher throughput, but also comes with its own set of limitations.
3.1.1 Nanodevices
Due to the recent advances in nanoscale fabrication, several techniques for single-cell metabolomics have been developed to isolate, introduce chemicals into the cell, or capture the cell itself by utilizing nanoscale devices.
The use of micropipettes to gently isolate or sample the cells itself is a promising field in it of itself, and this can be done manually using a culture plate and a micromanipulator, which has the obvious challenge of achieving high throughput. In order to increase throughput, several automated systems have been introduced lately.
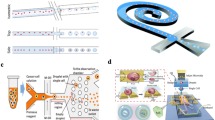
One recent innovation is coupling a micropipette with an automated system that can target certain cell types or regions within the cell according to their visual characteristics by the aid of computer software and image analysis. After selecting the desired cell, a robot picks up the target cell using a glass micropipette with an internal diameter of 30 μm and positioned 5 μm away from the bottom of the Petri dish. The cell is picked up by a vacuum system connected to the micropipette. Finally, the cell is put on 3D printed miniature wells on a Petri dish for downstream analysis. This system succeeded in achieving relatively high throughput compared to other systems and has the inherent advantage of not using markers to isolate the cells (Fig. 13.2) [35]. Moreover, several methods were proposed to introduce chemicals into the cell itself or detect optical signals on a subcellular scale. One such method utilizes a nanowire attached to the end of an optical fiber, which guides visible light into subcellular components. This nanoscale endoscope can also be used to deliver payloads into the cell itself [36].
Panel (a) represents the schematic of the method; the cells are selected by a software and then pipetted to a Petri dish containing miniature multiwall plates that are printed onto 35 mm plastic Petri dishes using a commercial 3D printer to reduce convection. Panel (b, c) show the multiwells (24, 2 × 2 mm2 wells and 4, 5 × 5 mm2 wells). The larger wells are used for stepwise, successive isolation in dense media. Reproduced from Ref 35 with permission from the Nature Publishing Group
Another interesting approach is using electroporation in which an induced electric field is applied to the cells to increase their cell membrane permeability, allowing chemicals or drugs to be introduced into the cell. The device proposed by Boukany et al. consists of two microchannels connected by a nanochannel where electroporation occurs [37]. The target cell is placed on one channel using optical tweezers, and the transfection agents are placed on the other microchannel. Transfection occurs by applying a voltage pulse between the channels resulting in an intense, localized electric field over a small area on the cell membrane, which allows a precise amount of the transfection agent to travel through the nanochannel, cell membrane, and into the cytoplasm by electrophoresis.
Despite the recent innovations, there are still challenges to fully implement nanodevices as the mainstream cell isolation technique. Maintaining a delicate balance between high throughput, low loss percentage, precision, degree of invasiveness, and ease of use is not an easy task. Automated systems have the advantage of being high throughput, but they are also inherently more complex. Manual systems, on the other hand, are simple, but their efficiency depends on human factors and the throughput has a high degree of variability.
3.1.2 Microfluidic Arrays
Microfluidic arrays represent another recent approach for single-cell isolation. The goal of these methods is the same: to transport and isolate single cells for further downstream analysis. The separation or isolation occurs by passing the culture media containing the cells through microfluid channels or arrays that isolate the cells individually in a high-throughput manner, as shown in Fig. 13.3. There are many recent innovations in single-cell isolation by using microfluidic arrays, among these are the integrated microfluidic array plate (iMAP) and dynamic single-cell culture array [38, 39]. Most, if not all, of these methods use polydimethylsiloxane (PDMS) as the construction material, along with utilizing soft lithography in fabrication. The iMAP, proposed by Dimov et al., utilizes gravity to guide the cells into their respective wells and has the advantage of greater capture rates than other methods (close to 100 %) [39]. Its design also allows for reagent addition and single-cell lysis depending on the analytical goals.
Panel (a) shows an overview of the trapping system; cell flow is gently directed into a series of trapping arrays; the scale bar is 500 μm. A single trapping array is shown in panel (b). The traps are molded from PDMS and fixed on a glass surface; trap sizes are optimized to capture one or two cells at most. A bright-field micrograph of the array is shown with cells trapped inside in panel (c) with added magnification of a single cell trapped in the minimum potential of the well. Reproduced from Ref 38 with permission of The Royal Society of Chemistry
One of the challenges of using microfluidic arrays is isolating the cell without causing noticeable damage or changing normal cell behavior. The method established by Carlo et al. shows great promise in this regard [38]. The system works by channeling the cells gently into a branched array system that consists of U-shaped PDMS traps fixed on a glass surface. The geometry of the trap is optimized to isolate one or two cells; once a cell occupies the trap, the altered dynamic flow around it will minimize the chance of other cells joining it, so in a way, the trap is self-regulating. The main advantage of such system is its ability to capture the cells with a high success rate (~90 %) without causing significant disruptions to the normal cell behavior and environment.
3.2 Analytical Methods
Assuming the cell was successfully isolated with minimal pretreatment, the next logical step would be to uncover its metabolome. To this effect, a myriad of analytical methods have been developed to analyze biomolecules. However, there are two main hurdles for single-cell analysis, and both relate to the inherently small sample size. The first challenge is improving the ability of the instrument to distinguish between closely related molecules, i.e., the resolution. Since single-cell analysis deals with small sample size (pico- to nanoliters), conventional separation techniques are ill-equipped to deal with such low sample volume without diluting the sample excessively or causing significant sample loss. The second challenge is increasing the ability to detect lower and lower concentrations reliably, i.e., the sensitivity. As sensitivity increases, the viability of the instrument itself to perform analysis on the single-cell scale increases.
Keeping the previously mentioned challenges in mind, choosing the perfect analysis technique is no easy task, but among the available analytical methods, mass spectrometry has gained prominence lately as one of the best techniques used to analyze the chemical composition at the single-cell level due to its relatively high sensitivity and resolution. There are several approaches used in single-cell mass spectrometric analysis, but this chapter will focus on the two main methods that are matrix-assisted laser desorption ionization (MALDI) and live single-cell mass spectrometry (LSCMS).
3.2.1 Matrix-Assisted Laser Desorption Ionization
MALDI is considered a “soft” ionization method, meaning it does not cause extensive fragmentation to the sample ions. The desired sample is mixed with a solution of low-mass organic compounds called a matrix, which is essential for ionization because it acts as a proton supplier and as a support or scaffold by which ionization can occur. Then the resulting mixture is spotted onto a metal plate called the target, as shown in Fig. 13.4. After spotting, the mixture is left to dry out, and both the sample and the matrix co-crystallize to form a solid deposit on the target. The target plate is then loaded into the mass spectrometer – most commonly using a time-of-flight (TOF) mass analyzer – where it will be subjected to a vacuum while the solid crystals of the mixture are irradiated by a UV laser beam that causes ablation of said crystals into the gas phase, followed by ionization of the sample. TOF mass spectrometry analyzes and detects the ions and gives out signals with ion mass-to-charge ratio (m/z) forming a distinct mass spectrometric profile that can be matched to a database to identify the sample ions [40].
A schematic representation of MS analysis using MALDI-TOF instrument; the sample-matrix mixture is left to dry out and then is irradiated with UV laser beam that causes its sublimation and subsequent sample ionization. The ions are then guided into the TOF by electrostatic attraction where they are analyzed depending on their mass-to-charge ratios (m/z); the MS spectra can then be matched to a database to identify unknown ions
Modern MALDI-TOF instruments have achieved enough sensitivity to be reliably used in single-cell analytical studies. However, there are still challenges associated with using MALDI on the single-cell level. For example, the extensive sample preparation and ionization under vacuum conditions is far away from the natural environment of the cell. Some studies aimed to alleviate this issue by performing the ionization process in normal atmospheric pressure and then transporting the ions by pneumatic assistance of a stream of nitrogen [41]. However, loss of sensitivity is inevitable.
Moreover, the matrix used in the ionization process has extensive molecular signals in the low molecular ranges (<500 Da), which incidentally is the region with most of the small molecule metabolites. This poses a significant challenge to single-cell metabolomics. Several methods have been proposed that forgo the use of a matrix all together and utilize nanophotonic effects for ionization [42]. Lipids, which dominate MALDI ionization, also mask metabolite detection as seen in many MALDI-TOF MS imaging results. Cell membranes contain a high percentage of lipids, also leading to difficulties in single-cell metabolomics. In conclusion, although MALDI-based approaches have a lot of potential in single-cell studies in regard to sensitivity, there are still challenges that need to be addressed to improve this method for use in single-cell analysis.
3.2.2 Live Single-Cell Mass Spectrometry
Understanding cell mechanisms and intracellular dynamics on the single-cell level is a tremendous challenge. The live single-cell mass spectrometry (LSCMS) method played an integral role in overcoming previous limitations, especially in the “omics biology.” Over the past 10 years, the team of T. Masujima has been working on perfecting the live single-cell mass spectrometry protocol to aid in realization of direct and real-time molecular analysis with simultaneous visualization of a reacting single cell, which would elucidate clearly and specifically the molecular mechanisms of living systems [24].
As earlier discussed, there is a limited number of methods that can detect molecular signals, due to the low sensitivity at the cellular and subcellular levels. On the other hand, live single-cell mass spectrometry has succeeded in trapping these miniscule volumes and detecting 100–1000 of molecular peaks from a single living cell while observing the cell under video microscope. Figure 13.5 visually depicts the main steps of the LSCMS methodology.
Scheme of live single-cell mass spectrometry. The analysis is divided into two main parts. First, the cell behavior is monitored via video microscope, and the cell or organelle of interest was directly trapped inside a nanospray tip. Second, the tip’s contents were then directly fed into a nano-ESI mass spectrometer after the addition of ionization solvent. Voltage is applied between the tip and mass spectrometer to obtain the mass spectra
Cells display task-oriented dynamic behavior that can be observed under a microscope [43]. While observing a cell, it exhibits various unanticipated and interesting behaviors, and discovering the mechanism behind those fascinating phenomena of life is very intriguing. Observations of cell behavior revealed that the behavior of each cell is not identical nor synchronized under the same conditions [44].
As discussed earlier, maintaining cells under the most natural conditions possible is paramount. In the LSCMS technique, in which cells are kept in their preferable medium until seconds before trapping and mass spectrometric analysis, this is the case. A metal-coated nanospray tip attached to a micromanipulator is used to suck a whole single cell or cellular contents from a specific micro-region using a tube-connected piston syringe. After capturing the cell contents at the top of the tip, it is very difficult to directly spray it into the mass spectrometer due to the high viscosity of the cells. To solve this, 2 μL of a standard ionization solvent is introduced from the rear end of the tip to aid sample quenching and ionization. Nanospray diameter sizes vary from 1 to 10 μm, which allows for greater flexibility in targeting the whole cell or its subcellular organelles or regions.
The nanospray tip’s contents are fed into a nano-ESI attachment on a mass spectrometer. Nanospray ionization showed to be the most sensitive and exhaustive ionization method. The molecular contents of a cell can be extracted by nL min−1 level stream of an organic solvent through the nanospray tip’s contents and sprayed out to the mass spectrometer [45]. Within minutes, the mass spectrometer detects 100 or 1000 of molecular peaks from the metabolites that were present in the cell under the specific conditions it faced at the time the contents were removed. These can then be identified by matching to databases to detect specific metabolites corresponding to the injected samples, which will be confirmed by their MS/MS fragmentation pattern.
Using this method, we can compare the molecular peaks of cells that are in different stages of growth, different locations, or responding to different circumstances using statistical analyses of the mass spectrometry data. If, for example, we find that certain metabolites are elevated in a specific strain, it implies that the enzyme or protein of this specific metabolic pathway may be the key to the specificity of this strain and could also help us to identify new important pathways. The following sections highlight recent studies from three different fields to demonstrate possible applications of LSCMS.
3.2.2.1 Clinical
Live single-cell mass spectrometry has been mainly applied to adhesive cells, due to difficulty in sampling and isolation of suspended cells. However, live single-cell mass spectrometry succeeded in compiling the comprehensive metabolic profile of a single floating cell. At first, a single floating lymphocyte was directly trapped inside the nanospray tip from a single drop of blood from a healthy human after minimal dilution and sample treatment. Lymphocytes were chosen by visual comparison of their morphological and size differences to red blood cells. Then, efficient homogenization of the trapped cell was established by applying supersonic waves, and the contents were fed into the mass spectrometer, which was conducive to acquiring a wider range of molecular peaks in the single-cell mass spectrum (Fig. 13.6). Molecular detection of higher intensities and larger number of peaks with a wider m/z range was obtained. Speculation would be that super-sonication causes outer lipid cell membrane distortion, which enhances extraction and ionization of the cell contents through mass spectrometry [27].
Discovering and perceiving the significance of cancer cells is one of the numerous potential promising applications of single-cell metabolomics. Detecting cancer cells that exhibit high metabolic rates within populations of normal cells that display normal metabolism, for example, CTCs that lead to metastasis, would be one such application. CTCs are cells shed from the primary tumor that circulate in the blood stream. Their primary function is still not clear, and their concentration in the circulating blood is usually very low (~2–10 cells per 10 mL of blood), which makes them a perfect candidate for single-cell studies. “Direct single-cell metabolomic” method was then applied to a single isolated CTC from a neuroblastoma patient’s blood for a comprehensive detection of the metabolite and lipid profiles. CTCs were separated and sorted using the fluorescence flow cytometry technique. The metabolic profile of a single CTC was acquired along with detection of vital molecules such as amino acids, catecholamine metabolites, which are specific to neuroblastoma cancer and drugs from the patient’s treatment regimen. This indicates that this method could be useful for monitoring drug delivery concentration levels to targeted cells. Site-specific and cell-specific metabolites were identified by matching corresponding peak numbers against the Human Metabolome Database and confirmed by establishing their MS/MS fragmentation patterns. “Direct single-cell metabolomic method” appears to have a role in future molecular diagnosis not only for common cells but also for rare cells like CTCs that present in a very low concentration in the blood [27].
3.2.2.2 Pharmaceutical
There has been increased interest in the analysis of spatial distribution of drugs and their metabolites in various cultured cells, or in a target cell for drug discovery and development. Drug metabolism monitoring and analysis have been mostly carried out by LC-MS, which requires a large number of cells pretreated with sonication and homogenization. This leads to, in most cases, the loss of site-specific molecule identification and drug localization within a cell. However, live single-cell video MS has been developed and applied to the analysis of tamoxifen (anticancer drug) metabolism using a human hepatocellular carcinoma cell line. Cultured HepG2 cells were spiked with 5 μmol L−1 concentration of tamoxifen and then incubated. Using a nanospray tip, multiple organelle suctions took place from several different cells in the same incubator dish (cytoplasm, nucleus, and vacuole). Results showed detection of tamoxifen along with its five metabolites (N-desmethyl tamoxifen, 4-hydroxy tamoxifen, tamoxifen-N-oxide, 3,4-dihydroxy tamoxifen, 4-hydroxy tamoxifen N-oxide). N-Desmethyl tamoxifen, which is mainly metabolized by CYP3A4 enzyme, had the highest intensity and was preferably detected. This corroborates with the discovery that the major metabolite of tamoxifen is N-desmethyl tamoxifen and that CYP3A4 is the most expressed isozyme in P450 subfamilies of HepG2 cells. In vacuoles, tamoxifen was detected but none of its metabolites was detected. However, neither tamoxifen nor its metabolites were detected in the nucleus. Speculation would be that the cytoplasm contains a metabolizing organelle and the transport of metabolites into the nucleus and vacuoles is very limited. This study of monitoring drug metabolism on a single-cell level will pave the way for low-cost, rapid, precise, and site-specific drug monitoring and discovery [26].
Primary cultures of human hepatocytes are mainly used for predicting drug metabolism pathways in humans and detecting the differences between species’ metabolic profiles. Therefore, hepatocytes were chosen as an appropriate cellular system for metabolic studies of tafluprost, an esterified prostaglandin F2α and common drug for glaucoma. 7-Ethoxycoumarin (7-EC) was used to endorse the metabolic activity of hepatocytes in vitro.
LSCMS was applied to the analysis of tafluprost metabolites, including tafluprost acid (TA), dinor-tafluprost (dinor-TA), tetranor-TA and common phase I metabolites, hydroxylated 1,2,3,4-tetranor-TA (tetranor-TA-OH), and hydroxylated 1,2-dinor-TA (dinor-TA-OH) in a single hepatocyte. These data were compared with the averaged results obtained from multiple cells. A picoliter amount of cytoplasm and granules in the cell were captured in a nanospray tip, and the ionization solvent was added. The tip was then introduced to the nano-ESI interface of mass spectrometer to obtain a single-cell spectrum. Tafluprost metabolism results from the multiple hepatocyte analysis using LC-MS showed averaged metabolism to tafluprost acid (TA) and β-oxidized metabolites. On the other hand, LSCMS indicated variation in tafluprost metabolism among individual cells showed significant variation in the quantity of TA and dinor-TA. In contrast, there was no significant variation of 7-ethoxycoumarin metabolism. This method succeeded in detecting the reported metabolic profile in the cytoplasm, and those metabolites matched a metabolic pathway and showed a variety of metabolic functions on the single-cell level. Therefore, LSCMS showed successful detection of drug metabolism heterogeneity in a single living hepatic cell. This approach has the potential for indicating the correlation between drug metabolism and the pharmacological as well as the toxicological effects taking place in cultured cells on single-cellular and subcellular levels [46].
3.2.2.3 Agricultural
Food is life, and maintaining a comprehensive metabolomic image of plant cells will unlock several pathways into improving crop yield, eliminating unwanted pests, and enhancing desired traits in crops. It is important to pursue studies of plant metabolomics and biochemistry because these will provide more insight on the natural molecular mechanisms and dynamic activities taking place inside plant cells. As a result of protein and enzyme activation, plant cell dynamical functioning produces metabolites corresponding to a specific enzyme or protein. Understanding those activities will impart an important outlook on the full image of how plants function, and recognizing plant genotypes will allow the regulation of such processes.
There are many factors to be taken into consideration in the single-cell analysis of plant tissues. Unlike cultured cells, plant tissues have an irregular surface, strong cellulose walls, and higher dilution of biomolecules inside the cell. As a result, LSCMS has been extended to obtain rapid, versatile, and noninvasive direct single-cell plant analysis, which is published in Nature Protocols [22]. This technique provides a molecular profile including metabolites, lipids, hormones, and nutrients of a single plant cell within minutes with minimal pretreatment (Fig. 13.7). If the plant tissues remained intact after analysis, morphological changes could be monitored along with metabolic pathways processes. This method was applied to leaves, stem, and petal from a healthy Pelargonium zonale plant. Collected data showed that there were specific metabolites, which present only in the leaf such as geranic acid, while methyl citronellate was detected in both the leaf and stem, but absent in the petal [21]. With the provided information, site-specific molecules and chemical composition of each site in the cell could be distinguished along with comparing between different plant samples. Furthermore, this method could be useful in several practical and industrial applications such as quality control of crop treatment and medicinal plants, food analysis, and controlling plant diseases. Finally, this will open a new outlook in the research done in agricultural sciences.
Illustrative figure showing the steps of the “live single-cell mass spectrometry” technique. The fresh plant was cut with minimal sample pretreatment and observed under video microscope. The targeted cell was captured by a micropipette. Ionization solvent was added from the rear end, and the tip contents were introduced by electrospray ionization (ESI) to a mass spectrometer, thereby obtaining the metabolic profile of a single plant cell. Reproduced from Ref 22 with permission from the Nature Publishing Group
4 Future Prospects
Single-cell analysis is a rapidly growing field of biology with much room for improvement, but many challenges remain to be addressed; after all, the field is still in its infancy. As previously mentioned, there are several hurdles regarding sample considerations, isolation of single cells, and their subsequent analysis. In order to circumvent said challenges, LSCMS was developed, which combines nanoscale devices, ambient pressure ionization, and sensitive mass spectrometric measurements while maintaining high resolution by the use of Orbitrap technology. Despite all of that, there are still limitations and challenges ahead; since the sampling is performed manually and identification of metabolites by matching with databases is done offline, several concerns have been raised in regard to the throughput of the method itself. It might be argued that obtaining “high-quality” data by insuring that the cell was sampled in its natural environment, i.e., Petri dish, is better than increasing throughput by utilizing more aggressive isolation techniques. However, the fact still remains that improving throughput is a major challenge that needs to be addressed so that the method can be applied to large-scale studies of cell metabolomics.
The sampling phase is the most time-consuming aspect of single-cell analysis, and LSCMS is no exception. In order to increase throughput, the process of selection of suitable cells and sampling was automatized in a collaborative study between the Masujima team and Yokogawa electric company. By coupling a motorized x-y stage to an automated software system and a dispenser robot, the newly developed system is capable of selecting the desired cells according to traits previously inputted and then proceed to automatically pick up the whole cell or an organelle, such as nucleus or cytoplasm. After that, the nanospray tips are stored in a specially made tip rack and can be frozen or analyzed immediately using ESI-MS as shown in Fig. 13.8.
High-content single-cell analysis system is shown; the cell is chosen by an image analysis algorithm according to its morphology or fluorescent markers. Then, the robot proceeds to sample a whole cell or part of a cell by using a specially made hollow nanospray tip. Finally, the trapped cell is stored in a special tip rack for downstream analysis or long-term storage
Another challenge exists in the sample type itself; since single cells are considered as a complex biological matrix, sometimes the need for an efficient separation and enrichment method arises. Due to the low sample volume associated with single cells, it is quite problematic to use a conventional separation method like high-performance liquid chromatography due to sample loss and dilution. It is also worth noting that mass spectrometry by itself cannot differentiate between optical isomers and ions with identical mass-to-charge (m/z) ratios. In the latter case, fragmentation and MS/MS studies can differentiate between identical m/z ions, but a strong signal is required for this to be done successfully – something that depends on the sample and the sensitivity of the instrument.
The ideal method for single-cell analysis would combine the minimal disruptiveness of nanoscale direct sampling of cells in their culture plates along with the high throughput of microfluidic devices coupled with an ionization source that operates in normal atmospheric conditions that also does not cause excessive heating or damage to the biomolecules themselves during the ionization process. The ionization source itself should incorporate a separation step according to ion mobility, for example, or other factors so that it is possible to differentiate between optical isomers. Finally, the mass analyzer used should have the highest sensitivity possible along with a sufficient resolution to differentiate between ions with closely similar mass-to-charge ratio.
Until now, no method proposed for single-cell analysis is perfect in all aspects. Balancing throughput, accuracy, and invasiveness of the isolation method while choosing the perfect analytical technique that combines high sensitivity, resolution, and selectivity is the ultimate goal to be achieved in order to contribute to a wide range of fields such as diagnostics, cancer treatment, agriculture, and many others. In principle, analysis on a single-cell level is an interdisciplinary science, and we hope that this overview will help in encouraging collaborative studies between different fields of life sciences so that we can gain a better understanding of the most important building block in our bodies.
Abbreviations
- 3D:
-
Three dimensional
- 7-EC:
-
7-Ethoxycoumarin
- CTC:
-
Circulating tumor cell
- DNA:
-
Deoxyribonucleic acid
- ESI:
-
Electrospray ionization
- iMAP:
-
Integrated microfluidic array plate
- LC-MS:
-
Liquid chromatography-mass spectrometry
- LCSMS:
-
Live single-cell mass spectrometry
- MALDI:
-
Matrix-assisted laser desorption ionization
- MAMS:
-
Microarrays for mass spectrometry
- mRNA:
-
Messenger ribonucleic acid
- MS:
-
Mass spectrometry
- MS/MS:
-
Tandem mass spectrometry
- PDMA:
-
Polydimethylsiloxane
- TA:
-
Tafluprost acid
- TOF:
-
Time of flight
- UV:
-
Ultraviolet
References
Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–6. doi:10.1093/nar/gkl923.
Dettmer K, Hammock BD. Metabolomics – a new exciting field within the omics sciences. Environ Health Perspect. 2004;112:A396–7.
Patti GJ, Yanes O, Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13:263–9. doi:10.1038/nrm3314.
Wang D, Bodovitz S. Single cell analysis: the new frontier in ‘omics’. Trends Biotechnol. 2010;28:281–90.
Snijder B, Pelkmans L. Origins of regulated cell-to-cell variability. Nat Rev Mol Cell Biol. 2011;12:119–25.
Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–6. doi:10.1126/science.1070919.
Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–26.
Sender R, Fuchs S, Milo R. Preprint on bioRxiv. 2015. doi:10.1101/036103.
Ranjan A, Townsley BT, Ichihashi Y, Sinha NR, Chitwood DH. An intracellular transcriptomic atlas of the giant coenocyte Caulerpa taxifolia. PLoS Genet. 2015;11:e1004900. doi:10.1371/journal.pgen.1004900.
Moss S. Do birds have knees?: All your bird questions answered. London: Bloomsbury Publishing; 2016.
Mattsson A, Kärrman A, Pinto R, Brunström B. Metabolic profiling of chicken embryos exposed to perfluorooctanoic acid (PFOA) and agonists to peroxisome proliferator-activated receptors. PLoS One. 2015;10:e0143780. doi:10.1371/journal.pone.0143780.
Vastag L, Jorgensen P, Peshkin L, Wei R, Rabinowitz JD, Kirschner MW. Remodeling of the metabolome during early frog development. PLoS One. 2011;6:e16881. doi:10.1371/journal.pone.0016881.
Onjiko RM, Moody SA, Nemes P. Single-cell mass spectrometry reveals small molecules that affect cell fates in the 16-cell embryo. Proc Natl Acad Sci U S A. 2015;112:6545–50.
Fuller RR, Moroz LL, Gillette R, Sweedler JV. Single neuron analysis by capillary electrophoresis with fluorescence spectroscopy. Neuron. 1998;20:173–81. doi:10.1016/S0896-6273(00)80446-8.
Page JS, Rubakhin SS, Sweedler JV. Direct cellular assays using off-line capillary electrophoresis with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Analyst. 2000;125:555–61.
Rubakhin SS, Greenough WT, Sweedler JV. Spatial profiling with MALDI MS: distribution of neuropeptides within single neurons. Anal Chem. 2003;75:5374–80.
Rubakhin SS, Sweedler JV. Characterizing peptides in individual mammalian cells using mass spectrometry. Nat Protoc. 2007;2:1987–97.
Romanova EV, Rubakhin SS, Monroe EB, Sweedler JV. Single cell analysis: technologies and applications, vol. 56. Weinheim: Wiley-VCH; 2016.
Zimmerman TA, Rubakhin SS, Romanova EV, Tucker KR, Sweedler JV. MALDI mass spectrometric imaging using the stretched sample method to reveal neuropeptide distributions in aplysia nervous tissue. Anal Chem. 2009;81:9402–9.
Shrestha B, Vertes A. In situ metabolic profiling of single cells by laser ablation electrospray ionization mass spectrometry. Anal Chem. 2009;81:8265–71.
Tejedor ML, Mizuno H, Tsuyama N, Harada T, Masujima T. In situ molecular analysis of plant tissues by live single-cell mass spectrometry. Anal Chem. 2012;84:5221–8. doi:10.1021/ac202447t.
Fujii T, Matsuda S, Tejedor ML, Esaki T, Sakane I, Mizuno H, Tsuyama N, Masujima T. Direct metabolomics for plant cells by live single-cell mass spectrometry. Nat Protoc. 2015;10:1445–56. doi:10.1038/nprot.2015.084.
Tsuyama N, Mizuno H, Tokunaga E, Masujima T. Live single-cell molecular analysis by video-mass spectrometry. Anal Sci. 2008;24:559–61.
Mizuno H, Tsuyama N, Harada T, Masujima T. Live single-cell video-mass spectrometry for cellular and subcellular molecular detection and cell classification. J Mass Spectrom. 2008;43:1692–700. doi:10.1002/jms.1460.
Mizuno H, Tsuyama N, Date S, Harada T, Masujima T. Live single-cell metabolomics of tryptophan and histidine metabolites in a rat basophil leukemia cell. Anal Sci. 2008;24:1525–7.
Date S, Mizuno H, Tsuyama N, Harada T, Masujima T. Direct drug metabolism monitoring in a live single hepatic cell by video mass spectrometry. Anal Sci. 2012;28:201–3.
Hiyama E, Ali A, Amer S, Harada T, Shimamoto K, Furushima R, Abouleila Y, Emara S, Masujima T. Direct lipido-metabolomics of single floating cells for analysis of circulating tumor cells by live single-cell mass spectrometry. Anal Sci. 2015;31:1215–7. doi:10.2116/analsci.31.1215.
Ibáñez AJ, Fagerer SR, Schmidt AM, Urban PL, Jefimovs K, Geiger P, et al. Mass spectrometry-based metabolomics of single yeast cells. Proc Natl Acad Sci U S A. 2013;110:8790–4. doi:10.1073/pnas.1209302110.
Taniguchi Y. Genome-wide analysis of protein and mRNA copy numbers in single Escherichia coli cells with single-molecule sensitivity. Methods Mol Biol. 2015;1346:55–67.
Ohno M, Karagiannis P, Taniguchi Y. Protein expression analyses at the single cell level. Molecules. 2014;19:13932–47.
Taniguchi Y, Choi PJ, Li G, Chen H, Babu M, et al. Quantifying E-coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–8.
Essaka DC, Prendergast J, Keithley RB, Hindsgaul O, Palcic MM, Schnaar RL, Dovichi NJ. Single cell ganglioside catabolism in primary cerebellar neurons and glia. Neurochem Res. 2012;37:1308–14.
Zenobi R. Single-cell metabolomics: analytical and biological perspectives. Science. 2013;342:1243259. doi:10.1126/science.1243259.
O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi:10.1038/nature05372.
Ungai-Salánki R, Gerecsei T, Fürjes P, Orgovan N, Sándor N, Holczer E, et al. Automated single cell isolation from suspension with computer vision. Sci Rep. 2016;9:20375. doi:10.1038/srep20375.
Yan R, Park JH, Choi Y, Heo CJ, Yang SM, Lee LP, et al. Nanowire-based single-cell endoscopy. Nat Nanotechnol. 2011;7:191–6. doi:10.1038/nnano.2011.226.
Boukany PE, Morss A, Liao WC, Henslee B, Jung H, Zhang X, et al. Nanochannel electroporation delivers precise amounts of biomolecules into living cells. Nat Nanotechnol. 2011;6:747–54. doi:10.1038/nnano.2011.164.
Carlo DD, Wu LY, Lee LP. Dynamic single cell culture array. Lab Chip. 2006;6:1445–9. doi:10.1039/B605937F.
Dimov IK, Kijanka G, Park Y, Ducrée J, Kang T, Lee LP. Integrated microfluidic array plate (iMAP) for cellular and molecular analysis. Lab Chip. 2011;11:2701–10. doi:10.1039/c1lc20105k.
Clark AE, Kaleta EJ, Arora A, Wolk DM. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev. 2013;26:547–603. doi:10.1128/CMR.00072-12.
Laiko VV, Baldwin MA, Burlingame AL. Atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem. 2000;72:652–7.
Walker BN, Stolee JA, Vertes A. Nanophotonic ionization for ultratrace and single-cell analysis by mass spectrometry. Anal Chem. 2012;84:7756–62. doi:10.1021/ac301238k.
Masujima T. Visualized single cell dynamics and analysis of molecular tricks. Anal Chim Acta. 1999;400:33–43. doi:10.1016/S0003-2670(99)00704-7.
Masujima T. Live single-cell mass spectrometry. Anal Sci. 2009;25:953–60.
Tejedor LM, Mizuno H, Tsuyama N, Harada T, Masujima T. Direct single-cell molecular analysis of plant tissues by video mass spectrometry. Anal Sci. 2009;25:1053–5.
Fukano Y, Tsuyama N, Mizuno H, Date S, Takano M, Masujima T. Drug metabolite heterogeneity in cultured single cells profiled by pico-trapping direct mass spectrometry. Nanomedicine. 2012;7:1365–74.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Emara, S., Amer, S., Ali, A., Abouleila, Y., Oga, A., Masujima, T. (2017). Single-Cell Metabolomics. In: Sussulini, A. (eds) Metabolomics: From Fundamentals to Clinical Applications. Advances in Experimental Medicine and Biology(), vol 965. Springer, Cham. https://doi.org/10.1007/978-3-319-47656-8_13
Download citation
DOI: https://doi.org/10.1007/978-3-319-47656-8_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-47655-1
Online ISBN: 978-3-319-47656-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)