Abstract
In all living organisms, the flow of genetic information is a two-step process: first DNA is transcribed into RNA, which is subsequently used as template for protein synthesis during translation. In bacteria, archaea and eukaryotes, transcription is carried out by multi-subunit RNA polymerases (RNAPs) sharing a conserved architecture of the RNAP core. RNAPs catalyse the highly accurate polymerisation of RNA from NTP building blocks, utilising DNA as template, being assisted by transcription factors during the initiation, elongation and termination phase of transcription. The complexity of this highly dynamic process is reflected in the intricate network of protein-protein and protein-nucleic acid interactions in transcription complexes and the substantial conformational changes of the RNAP as it progresses through the transcription cycle.
In this chapter, we will first briefly describe the early work that led to the discovery of multisubunit RNAPs. We will then discuss the three-dimensional organisation of RNAPs from the bacterial, archaeal and eukaryotic domains of life, highlighting the conserved nature, but also the domain-specific features of the transcriptional apparatus. Another section will focus on transcription factors and their role in regulating the RNA polymerase throughout the different phases of the transcription cycle. This includes a discussion of the molecular mechanisms and dynamic events that govern transcription initiation, elongation and termination.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
9.1 Multisubunit RNA Polymerases – Discovery of the Enzymes and Their Role Within the Central Dogma of Life
The description of the DNA structure in 1953 (Watson and Crick 1953b), together with early studies in genetics and bacterial transformation, stimulated the question how the repeating mononucleotide units that constitute DNA are assembled in the cell (Olby 2003). This led to the hypothesis that DNA replication would require DNA to function as a template for its own synthesis (Watson and Crick 1953a; Meselson and Stahl 1958). At this point, RNA was only a poorly understood molecule and, consequently, the search for a DNA replicating enzyme rather than an RNA synthesizing enzyme started soon after the double-helical structure of DNA was presented. Arthur Kornberg and colleagues were the first to describe DNA polymerase as the DNA synthesizing enzyme (Lehman et al. 1958). RNA only came into focus when the mechanism of protein synthesis was debated (Campbell and Work 1953; Dounce 1952). In 1955, Grunberg-Manago and Ochoa proposed that polynucleotide phosphorylase (PNP) is the enzyme responsible for RNA synthesis (Grunberg-Manago et al. 1955), an idea that proved wrong. The discovery of tRNAs as acceptor for amino acids (Hoagland et al. 1957) gave the first indication that RNA and protein synthesis are linked and that protein biosynthesis requires a high specificity of the tRNA. However, PNP could not fulfil this requirement for specificity as it only adds nucleotides at the RNA termini in a non-specific fashion. This led to the hypothesis that DNA could serve as a template for RNA synthesis and initiated the search for an enzyme capable of DNA-dependent RNA-polymerisation (Hurwitz 2005). RNA polymerase (RNAP) activity was first described by Weiss and Gladstone in 1959 who demonstrated that all four NTPs were incorporated into RNA when added to rat liver nuclear extracts (Weiss and Gladstone 1959). Shortly after, four laboratories independently reported on DNA-dependent RNA synthesising activity in cellular extracts of bacterial or eukaryotic origin (i.e. from Escherichia coli (Hurwitz and Bresler 1961; Stevens 1960), pea embryos (Huang et al. 1960) and Micrococcus luteus (Weiss and Nakamoto 1961)). Geiduschek et al. (1961) and Chamberlin et al. (1963) used purified RNAPs to demonstrate that in vitro transcribed RNA is complementary to the DNA substrate and that the DNA template remained intact after supporting RNA synthesis. Roeder and Rutter were the first to describe that not one but three different enzymes – RNAP I, II and III – transcribe the eukaryotic genome (Roeder and Rutter 1969, 1970). These early discoveries started a new research field and it became apparent that bacterial and eukaryotic RNAPs differ not only in their subunit composition, but also in their sensitivity towards small molecules, rendering RNAP a prime target for antibiotics. Moreover, the understanding of RNAP subunit composition was of critical importance to establish the archaea as an independent domain of life. Shortly after Woese suggested the existence of the third domain of life in 1977 (Woese and Fox 1977), Stetter and Zillig attempted the isolation and biochemical characterisation of RNAPs from different archaeal organisms (Schnabel et al. 1983; Zillig et al. 1978, 1979). Their work clearly demonstrated that archaeal RNAPs are composed of more than the four subunits prototypical for bacterial RNAPs and cannot be inhibited by the antibiotic Rifampicin, providing evidence that the archaeal RNAP shares more characteristics with eukaryotic than bacterial RNAPs. At this point, the subunit composition and the catalytic activity of RNAPs from all three domains could be explored in detail. But it took another 20 years until the first structure of a multisubunit (bacterial) RNAP was solved (Zhang et al. 1999), closely followed by a crystal structure of the eukaryotic RNAP enzyme from yeast (Gnatt et al. 2001; Cramer et al. 2000) and finally, in 2008, the first crystal structure of an archaeal RNAP was reported (Hirata et al. 2008).
9.2 Multisubunit RNA Polymerases and Transcription Factors in the Three Domains of Life
9.2.1 Overall Subunit Composition and Architecture of RNA Polymerase in Bacteria, Eukaryotes and Archaea
DNA-dependent RNA polymerases either belong to the family of single subunit or to the family of multisubunit RNAPs. Single subunit RNAPs can be found in chloroplasts, mitochondria and in bacteriophages, such as T7 or SP6, and are shaped like a right hand (Cheetham and Steitz 2000). In all living cells, however, DNA-dependent RNA synthesis is carried out by complex multisubunit RNAPs (Werner and Grohmann 2011). While bacteria and archaea employ a single type of RNAP, the eukaryotic genome is transcribed by at least three specialized RNAPs, which are dedicated to different subsets of genes. RNAP I transcribes the ribosomal RNA precursor for the mature 25/28S, 18S and 5.8S rRNAs, RNAP II is responsible for the transcription of messenger RNAs (mRNAs) and many non-coding RNAs, whereas RNAP III synthesises small structured RNAs like transfer RNAs (tRNA), spliceosomal U6 small nuclear RNA (snRNA), ribosomal 5S rRNA and 7 SL RNA (Sentenac 1985; Dieci et al. 2007). In plants, two additional multisubunit RNAPs, RNAP IV and V, are encoded. Both these RNAPs are involved in non-coding RNA-mediated gene silencing processes (Haag and Pikaard 2011).
The minimalistic bacterial RNAP is comprised of four different subunits (β, β′, two copies of α and ω). Homologs of the bacterial subunits can be found in all multisubunit RNAPs (Fig. 9.1, panel a: homologous core subunits are coloured in blue). These subunits form the structural ly conserved RNAP core that harbours the catalytic centre with coordinated magnesium ions at the interface of the two large bacterial subunits β/β′ and archaeal-eukaryotic subunits Rpo1/2, A190/135, RBP1/2 and C160/128, respectively. Even though there is only a low sequence identity among the core subunits across the domains of life, there is a high degree of structural conservation (Fig. 9.1c). The core subunits are arranged like a “crab claw” forming the DNA cleft. The “jaws” are part of the large subunits and interact with the incoming downstream DNA substrate (Fig. 9.1b). The large subunits also comprise a number of flexible elements like the bridge helix, the switch region and trigger loop, which are critically involved in the correct positioning of the next nucleotide and the translocation of RNA and DNA by RNAP (Zhang et al. 2010; Vassylyev et al. 2007; Brueckner et al. 2009a, b; Weinzierl 2010; Tan et al. 2008). Another mobile element of multisubunit RNAPs is the clamp domain, which can close over the DNA binding channel and adopts different conformational states in the different stages of the transcription cycle (Schulz et al. 2016; Chakraborty et al. 2012; Engel et al. 2013). The clamp represents an “interaction hotspot” as regulatory transcription factors like transcription factor E, and the elongation factor Spt4/5 bind the tip of the RNAP clamp region and adjust the position of the clamp (Grohmann et al. 2011; Grunberg et al. 2012; Klein et al. 2011; Schulz et al. 2016).
Subunit composition and overall architecture of multisubunit RNA polymerases. (a) Subunit composition of multisubunit RNA polymerases in the three domains of life. Homologous core subunits are coloured in light blue, archaeal-eukaryotic specific subunits in light orange and subunits specific for the archaeal or specialized eukaryotic RNAPs I, II or II are highlighted in light green. (b) Structure of the bacterial (left, PDB:1I6V) and eukaryotic RNAP II (right, PDB: 2WAQ) with single subunits coloured according to the colour scheme given in the table. (c) The conserved crab-claw like architecture for multisubunit RNA polymerases from bacteria (left, PDB: 1I6V), archaea (PDB: 2WMZ) and eukaroytes (RNAP I: middle, PDB: 4C3I; RNAP II: second right, PDB: 2WAQ; RNAP III: right, PDB: 5FJ9). Universally conserved subunits are shown in blue, archaeal-eukaryotic specific subunits in orange and subunits unique of a specific RNAP type are shown in green
In addition to the core subunits, archaeal-eukaryotic RNAPs have an expanded set of subunits forming macromolecular assemblies up to 0.7 megadalton (MDa) in size (Fig. 9.1). Archaeal RNAPs typically contain 11–13 subunits (Grohmann et al. 2009) and the eukaryotic RNAPs 12–17 subunits (Vannini and Cramer 2012). Some of these subunits can be found in all archaeal-eukaryotic RNAPs (e.g. homologs of RNAP II subunits RPB4/5/7/8/10/12 highlighted in orange in Fig. 9.1a/c). Notably, subunits RPB5/6/8/10/12 are shared between the eukaryotic RNAPs I, II and III. Subunits 3/10/11/12 form the platform crucially important for the correct assembly of the catalytic subunits. A signature module of archaeal-eukaryotic RNAPs is the stalk domain composed of subunits Rpo4/7 (archaea), A14/A43 (RNAP I), RPB4/7 (RNAP II) and C17/C25 (RNAP III), respectively. The stalk protrudes from the core of the enzyme and is involved in a multitude of functions including the stabilisation of the initiation complex and the binding of the nascent RNA thereby increasing the elongation and termination efficiency of RNAPs. It furthermore serves as an interaction site for transcription factor E. In some archaea, subunit Rpo13 is part of the RNAP. No homologue of Rpo13 is encoded in eukaryotes or bacteria. Another example of a domain-specific subunit is RPB9, which is exclusively found in the eukaryotic domain (domain-specific subunits are color-coded in orange in Fig. 9.1c). RBP9 provides an interaction surface for transcription factor TFIIF (Ziegler et al. 2003) and is closely related to RNAP I and III subunits A12 and C11. However, A12 and C11 represent a fusion protein of the N-terminal RPB9 and the C-terminal part of transcript cleavage factor TFIIS (Ruan et al. 2011) and are considered to be “in-built” transcription factors. RNAP I and III contain additional, auxiliary subunits that are located at the surface of the RNAP thereby providing interaction platforms for factors that regulate RNAP activity. Subunits A49/A34.5 and C53/C37 are distantly related heterodimers that exhibit similarities to transcription factor F (TFIIF) (Fig. 9.2a/d) (Vannini and Cramer 2012). These TFIIF-like complexes are implicated to function in initiation complex stabilisation and occupy a location opposite of the stalk domain (Sainsbury et al. 2015). A49/34.5 furthermore enhances transcription elongation and transcript cleavage and C53/37 is important for termination. Just like A49/A34.5 and C53/C37, the protein complex C82/C34/C31 also represents an “in-built” transcription factor. C82/C34 is related to subunits alpha and beta of transcription factor E, which is associated with, but not integrated into, the archaeal RNAP and RNAP II (Fig. 9.2a/c) (Vannini and Cramer 2012; Blombach et al. 2015; Carter and Drouin 2010; Blombach et al. 2016). The archaeal TFE, TFIIE and C82/C34 all map to the RNAP clamp domain and fulfil functions during transcription initiation by stabilisation of the open DNA bubble (Blombach et al. 2016; Grohmann et al. 2011; Grunberg et al. 2012; Engel et al. 2013; Hoffmann et al. 2015). A unique feature of the largest core subunit of RNAP II is the C-terminal domain (CTD) composed of tandem heptad repeats (26 in yeast, 52 in vertebrates). The CTD is subject to extensive posttranslational modifications , most notably phosphorylation , throughout the transcription cycle , which allows a fine-tuned regulation of RNAP II activity and, among others, supports the coupling of transcription and post-transcriptional processing (for an overview see for example (Hsin and Manley 2012; Conaway and Conaway 2015). Taken together, multisubunit RNAPs are multiprotein complexes that are organised in conserved functional domains (e.g. core, assembly platform, stalk, clamp, jaws, DNA cleft) characterised by an increase in complexity and diversity in archaeal-eukaryotic RNAP variants.
Structure function relationship between transcription factors in bacteria, archaea and eukaryotes. (a) Overview of transcription factors implicated in transcription initiation and elongation in bacteria, archaea and eukaryotes. Factors with structural homologies are marked in bold letters. The yeast name and the mammalian name (in squared brackets) of the homologous protein are given. If these proteins are complex components, the name of the respective complex is depicted in round brackets. (b–e) Schematic representation of transcription factors and RNAP subunits sharing structural homologies. Structural domains are depicted as boxes, with similar domains having the same colour. Abbreviations are explained in the legend on the right and aSpt5 and eSpt5 refers to archaeal and eukaryotic Spt5, respectively
9.2.2 Transcription Factors of RNA Polymerases in Bacteria, Eukaryotes and Archaea
Although RNAPs can synthesise RNAs on their own, they require additional factors guiding them to the promoter regions, as well as assisting transcription initiation, elongation and eventually termination. Initially, these factors were identified in different organisms owed to their ability to support different steps of transcription in highly purified in vitro systems. Results from these biochemical studies led to detailed models for the function of the individual factors within the transcription cycle . Many of the hypotheses derived from these biochemical studies were validated and extended by structural characterisation. Genetic analyses and the advent of technologies that allow the monitoring of the association status of the transcription machinery at virtually every gene locus in vivo, further helped to define a set of general factors supporting transcription in living cells.
As described before, RNAPs in the different domains of life share remarkable similarities with regard to their structure and function, without strong conservation at amino acid sequence level. This structure-function relationship is even more pronounced for transcription factors. Thus, proteins unrelated in their amino acid composition may exert similar functions, correlating with similar topology of structural elements. Consistently, in some cases, predicted structural similarities rather than sequence homologies have provided insights in a previously unknown role of a factor. This paragraph aims to put emphasis on the structure-function conservation within transcription factors in the three domains of life and the different transcription systems (Fig. 9.2). The focus will be on the so-called general transcription factors of transcription initiation, as well as on two factors implicated in transcription elongation. Details about the functional interaction of these factors with RNAPs in course of the transcription cycle are provided in paragraph 3.
9.2.2.1 Initiation Factors
RNAP needs transcription factors to: (i) Identify promoter regions, (ii) Mediate its interaction with the DNA, (iii) Determine the direction of transcription, (iv) Melt the double stranded template leading to the open complex, (v) Enter transcription elongation (Sainsbury et al. 2015; Werner and Grohmann 2011)
In bacteria, all of these tasks are accomplished by the protein family of the promoter specific sigma factors (σ factors) (Feklistov et al. 2014). σ factors were discovered as a biochemical fraction required by purified E. coli RNAP to transcribe selective regions of lambda DNA (Burgess et al. 1969). σ factor bound to RNAP recognises up to four out of five DNA motifs marking a prokaryotic promoter (see Sect. 9.3.2) (Bae et al. 2015). This is explained by the structure of σ proteins, organized in a variable number of folded protein domains (e.g. σ1.1, σ2, σ3, σ4 in the housekeeping σ70 in E. coli), which are connected by flexible linkers (Fig. 9.3a). Except σ1.1, all other domains share the ability to bind to specific DNA elements. In solution, σ factors adopt a compact conformation, interfering with DNA-binding of the individual domains (Sorenson and Darst 2006; Sorenson et al. 2004). Additionally, σ70 has been suggested to bear an autoinhibitory N-terminal domain (σ1.1)(Dombroski et al. 1992), which might stabilize the compact conformation. Upon interaction with core RNAP, σ factors undergo structural rearrangements yielding the initiation competent holoenzyme (Murakami et al. 2002a, b). After promoter binding of the holoenzyme, σ factors assist in formation and stabilisation of the nascent transcription bubble, owed to the ssDNA binding ability of the σ2 domain (Bae et al. 2015) (see Sect. 9.3.2, Fig. 9.3a). The nascent RNA produced at the onset of transcription interferes with some of the σ-RNAP contacts resulting in stochastic dissociation of σ factors from the elongating RNAP after promoter escape (Nickels et al. 2005). The arrangement of structured domains and flexible linkers found in σ factors, which undergo functionally relevant structural changes in different phases of the transcription process, are a common feature of many transcription factors (see below).
Architecture of the open initiation complex in the three domains of life. In all cases, the template strand (TS) is inserted into the active site of the RNAP and the non-template strand (NTS) is accommodated outside the DNA binding cleft. (a) The bacterial initiation complex is composed of a sigma factor (here σ70), the RNAP and DNA (PDB: 4YLO). σ70 is a multidomain protein that recognises the −35, −10 and discriminator (disc) motifs of the bacterial promoter and supports DNA melting. The RNAP contacts bases of the core recognition element (CRE) that surrounds the transcription start site (+1). (b) Model of the archaeal open pre-initiation complex (from (Nagy et al. 2015). The transcription initiation factors TBP and TFB recognise the TATA-box and B-recognition element (BRE) upstream of the transcription start site, respectively. The TFB-reader/helix domains are crucially important for promoter melting and transcription bubble stabilisation – a process furthermore supported by TFE. (c) Architecture of the human open pre-initiation complex (images kindly provided by Eva Nogales). EM-reconstruction of the holo-PIC (including TFIIH, left) and model of the core PIC (without TFIIH, middle). The arrangement of the transcription factors and RNAP motifs critically involved in promoter opening is shown (panel on the right). RAP74 is a subunit of TFIIF. Archaeal and eukaryotic promoters (shown here: human promoter) contain the initiator element (Inr) surrounding the transcription start site, and eukaryotic promoters additionally the downstream promoter element (DPE)
The expression of σ variants with different promoter specificities provides prokaryotes with the possibility to express genes under specific environmental conditions (e.g. activation of genes induced under nitrogen stress conditions by σ54) (Gruber and Gross 2003). Archaea and eukaryotes do not have homologs of σ factors for promoter recognition. Instead, they own the TATA-binding protein (TBP), and transcription factor B (TFB/TFIIB), which – similar to σ4 in bacteria – bind to upstream DNA sequences in archaeal and eukaryotic promoters (Littlefield et al. 1999; Nikolov et al. 1995) (Fig. 9.3b/c). In contrast to bacterial σ factors, TBP and TFB/TFIIB may bind to promoter DNA in the absence of RNAPs, which are then recruited to the promoter-bound scaffold (Sainsbury et al. 2015). In some archaea and eukaryotes, the TBP gene underwent gene-duplication resulting in paralogs of TBP with distinct functions in gene regulation (Duttke 2015). Additionally, some archaea express multiple paralogs of TFB (Facciotti et al. 2007). Thus, in all three domains of life, gene duplication events resulting in variants of core promoter binding factors lead to an increase in the regulatory potential of gene expression.
Promoter-specific transcription by eukaryotic RNAPs I and III also strictly depends on TBP-interacting complexes containing TFB-like factors (Fig. 9.2a/b): RNAP I requires yeast core factor (CF), or its mammalian counterpart, selective factor 1 (SL1), whereas RNAP III depends on TFIIIB (Vannini and Cramer 2012). Homology between TFB/TFIIB, and the TFIIIB subunit Brf1 (and its paralog Brf2) have been described earlier (Colbert and Hahn 1992; López-De-León et al. 1992). The homology between TFB/TFIIB and the yeast CF subunit Rrn7, or its human orthologue the SL1 subunit TAF1B, were only recently uncovered (Blattner et al. 2011; Naidu et al. 2011; Knutson and Hahn 2011). The latter was possible using a computational homology and structure prediction tool (Söding et al. 2005), in combination with functional assays. In agreement with the conserved function of σ-factors and TFB-like factors, the topology of secondary structure elements of these proteins within the different pre-initiation complexes (PICs), is remarkably similar (Burton and Burton 2014; Sekine et al. 2012) (Fig. 9.3a), despite the fact that they are unrelated in their amino acid sequence.
In addition to TBP and TFB, TFE is the third general transcription initiation factor common in the archaeal and eukaryotic transcription systems (Fig. 9.2a/c). Archaeal TFE occurs as either a monomeric TFEα (e.g. TFE from Methanocaldococcus jannaschii) or a dimeric TFEα/β variant (e.g. TFE from Sulfolobus solfataricus) (Blombach et al. 2009). TFEα is a homolog to the N-terminal part of the TFEIIEα subunit in the eukaryotic TFIIE heterodimer while the TFEβ subunit found in Sulfolobus is composed of one winged helix domain and one [4Fe-S] cluster containing domain that are homologous to the human RNAP III subunit RPC39 (Blombach et al. 2015, 2016) (Fig. 9.2c). TFE and TFIIE both contain winged helix (WH) domains (Meinhart et al. 2003). Interestingly, a tandem WH domain similar to a pair of WH domains in TFIIEβ has been found in the C-terminal region of the RNAP I subunit A49, and two adjacent WH domains exist in the RNAP III subunit C34 (Geiger et al. 2010). The human homolog of C82 has four WH domains, resembling the extended WH domain in TFE/TFIIEα (Lefèvre et al. 2011). As TFE/TFIIE, the WH domains of A49 and C34 may adopt a topologically similar positon over the cleft (Bischler et al. 2002; Vannini et al. 2010; Jennebach et al. 2012). As mentioned before (see Sect. 9.2.1), RNAP I and RNAP III subunits can structurally and functionally resemble RNAP II transcription factors, and may be considered as “in-built” transcription factors (Vannini and Cramer 2012).
Several transcription initiation factors are unique to the eukaryotic transcription machinery, some of which have paralogs in the different RNAP machineries. The transcription factor TFIIA is not required for RNAP II transcription in vitro, but it can stimulate RNA synthesis (Kang et al. 1995). This essential factor, composed of two subunits, has functions in stabilising the TBP-DNA interaction (Imbalzano et al. 1994). No TFIIA-like factor has to date been identified in the RNAP I and RNAP III transcription machineries.
Instead, RNAPs I and III contain subunits, structurally resembling TFIIF in the RNAP II system (Vannini and Cramer 2012) (see Sect. 9.2.1, Fig. 9.2a/d). Human TFIIF is a dimer of TFIIFα and TFIIFβ (Burton et al. 1988), with homologs in yeast (Chafin et al. 1991), acting together with an additional, non-essential TFIIF subunit (Henry et al. 1992). TFIIFα and TFIIFβ both contain a C-terminal WH domain (Groft et al. 1998; Kamada et al. 2001) (Chen et al. 2007; Eichner et al. 2010) (Fig. 9.2d). TFIIF binds to the lobe of RNAP II via a dimerisation module formed by the two subunits (Gaiser et al. 2000). A similar dimerization module is found in the A49/34.5 subunits of RNAP I and the C37/53 subunits of RNAP III (Geiger et al. 2010; Kuhn et al. 2007). In both polymerases, the respective dimerization modules bind close to the lobe (Engel et al. 2013; Fernandez-Tornero et al. 2013) (Fig. 9.3c) and – as TFIIF – might be implicated in diverse steps of transcription initiation. Interestingly, two subunits of the RNAP III-specific transcription factor TFIIIC establish a structure with a TFIIF-like dimerisation module, and a WH domain (Taylor et al. 2013). It has been hypothesised that these subunits assist PIC formation, while stabilising the single stranded non-template strand (NTS) DNA. There is evidence that the DNA binding mode of the TFIIIC subunit might resemble the one observed for the bacterial σ2 domain to the single stranded −10 region (see also Sect. 9.3.2). A similar mechanism, supporting promoter opening, was suggested for TFIIF, which shows weak homology to σ2 (Tan et al. 1994).
The core RNAP I and III machineries lack TFIIH-related factors. This 10-subunit transcription factor is recruited by TFIIE to the RNAP II PIC, where a translocase activity is required for OC formation in an ATP-consuming step (see Sect. 9.3.2) (Grunberg et al. 2012; Kim et al. 2000). Other enzymatic activities of holo-TFIIH include the phosphorylation of the CTD of the largest subunit of RNAP II (Feaver et al. 1991; Serizawa et al. 1995). A TFIIH subcomplex has an important role in DNA repair processes, mediated by two subunits with helicase activities (Coin et al. 2007). TFIIH has been visualized by cryo EM in both, the human and the yeast PIC, and the overall topology of the complex was consistent with its suggested functions during transcription initiation (He et al. 2016; Murakami et al. 2015; Murakami et al. 2013b).
Two other megadalton-sized regulatory complexes, TFIID and Mediator, are important for the regulation and modulation of RNAP II transcription initiation (Cler et al. 2009; Poss et al. 2013; Sikorski and Buratowski 2009; Thomas and Chiang 2006). TBP was originally identified as a component of TFIID, a large complex with 13 canonical TBP associated factors (TAFs), supporting activated transcription in vitro. In vivo, TBP in the context of TFIID can be recruited to genes lacking a TATA box, due to the recognition of additional promoter elements by TAFs (Baumann et al. 2010; Kadonaga 2012). Interestingly, interaction of the transcription factor TFIIA with TBP displaces the TAF1 N-terminal domain 1 from the TBP DNA-binding surface, enabling its interaction with promoter DNA (Kokubo et al. 1994). TFIID supports activated transcription from chromatin templates in vitro, suggesting that this might be important for its in vivo function in the chromosomal context (Wu and Hampsey 1999). Several high-resolution structures of individual TAF subcomplexes are available (Sainsbury et al. 2015) helping to interpret recent cryo EM structures of the full complex in solution or bound to a promoter DNA (Bieniossek et al. 2013; Cianfrocco et al. 2013; Elmlund et al. 2009; Louder et al. 2016; Papai et al. 2010).
Mediator was identified in search of a complex that transmits regulatory signals of DNA-binding factors to the RNAP II basal transcription machinery (Casamassimi and Napoli 2007; Thomas and Chiang 2006). Mediator also supports basal transcription by assisting PIC assembly in vitro, and regulates CTD phosphorylation by TFIIH (Sikorski and Buratowski 2009). As for TFIID, X-ray structures are available for individual Mediator subunits and sub-complexes and EM analyses have been performed with both the free complex and RNAP II-bound Mediator (Larivière et al. 2012). Recently, EM structures of the human Mediator-RNAP II-TFIIF complex (Bernecky et al. 2011), and a 15 subunit core Mediator interacting with an RNAP II core initiation complex, containing TFIIB, TBP, and the Pol II–TFIIF (Plaschka et al. 2015) have been obtained. The latter analysis suggested that Mediator might enhance CTD-phosphorylation by orienting the TFIIH kinase module and part of the CTD in the PIC. It also provided insights in how Mediator might interact with the upstream DNA and RNAP II to communicate signals from regulatory DNA binding proteins (Plaschka et al. 2016b).
Interestingly, the binding of the Mediator head module to RNAP II might be topologically related to the binding of the essential transcription factor Rrn3 (TIF-IA in mammals) to RNAP I (Blattner et al. 2011). Yeast Rrn3 and CF are sufficient to support RNAP I transcription in vitro even in absence of TBP (Bedwell et al. 2012; Keener et al. 1998; Merkl et al. 2014). The RNAP I-Rrn3 complex is the initiation-competent form of the polymerase, which might be recruited by CF bound to the RNAP I promoter (Peyroche et al. 2000). RNAP I-Rrn3 complex formation might be important to stabilise the interaction of the polymerase with Rrn7 driving transcription initiation (Blattner et al. 2011; Knutson et al. 2014).
TFIIIA represents a transcription factor, which is conserved in all eukaryotes investigated so far and is exclusively involved in RNAP III-dependent transcription initiation (Layat et al. 2013). It was the first eukaryotic transcription factor purified to homogeneity (Engelke et al. 1980; Segall et al. 1980). TFIIIA is needed for 5S rDNA transcription by RNAP III in vitro and in vivo (Andrews and Brown 1987; Rollins et al. 1993). The protein contains multiple C2H2 zinc finger repeats and binds to a specific DNA-sequence upstream of the 5S rDNA. Using its zinc fingers, TFIIIA might also bind to rRNA (Brow and Geiduschek 1987). Conservation between TFIIIAs from different species is largely restricted to the zinc finger domain (Huang and Maraia 2001), and the structural basis for its interaction with nucleic acids has been provided by X-ray crystallography (Lu et al. 2003).
9.2.2.2 Elongation Factors
After initiation, RNAPs start the elongation of the nascent transcript (see Sect. 9.3.3). Specific factors have evolved that directly interact with the RNAP and assist RNAPs to efficiently promote elongation. Interestingly, two general mechanisms of elongation factor operation appear to be conserved in the three domains of life (see Sect. 9.3.3):
-
I.
Factors binding to the RNAP clamp domain, closing the RNAP active centre cleft and thereby stabilising the elongation complex (Fig. 9.5a)
-
II.
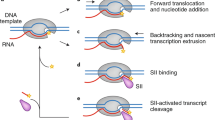
Factors entering the RNAP active site via the secondary channel catalysing cleavage of backtracked transcripts and thus release arrested RNAPs (Fig. 9.5b)
Secondary channel binding proteins in bacteria belong to the GreA/B family and are unrelated in sequence and structure to the archaeal and eukaryotic secondary channel binding TFIIS-like proteins (Nickels and Hochschild 2004; Werner and Grohmann 2011). GreA/B and TFIIS stimulate elongation in vitro by releasing arrested, backtracked RNAPs (see Sect. 9.3.3) (Borukhov et al. 1992; Rutherford et al. 2007; Thomas et al. 1998). As observed for σ factors and TFB-like factors, those factors share striking similarity in their overall topology in complex with RNAP in good correlation with their related function (Cheung and Cramer 2011; Tagami et al. 2010; Wang et al. 2009). TFIIS-like proteins are conserved in archaea and in RNAP I and III subunits A12.2, and C11 (see Sect. 9.2.1) (Vannini and Cramer 2012; Werner and Grohmann 2011). Apart from their function in transcription elongation, secondary channel binding proteins in bacteria and eukaryotes may play a role in transcription initiation (Sikorski and Buratowski 2009).
The NusG/Spt5 RNAP clamp binding elongation factor is the only transcription factor conserved in sequence and structure in all three domains of life (Werner and Grohmann 2011; Yakhnin and Babitzke 2014). Bacterial NusG and archaeal Spt5 bear a NusG N-terminal domain (NGN) and a C-terminal Kyprides-Onzonis-Woese (KOW) domain (Knowlton et al. 2003; Mooney et al. 2009) (Figs. 9.2e and 9.5b). In eukaryotes, Spt5 may carry multiple KOW domains and extensions at the N- and C-termini. Archaeal and eukaryotic Spt5 further form a heterodimer with Spt4 (Hirtreiter et al. 2010a; Klein et al. 2011).
Taken together, transcription factors in the three domains of life seem to fall in at least three different categories:
-
I.
Factors highly similar in sequence and structure (e.g. NusG/Spt5; TBP; TFB/TFIIB, TFE/TFIIE)
-
II.
Factors with high structural homology (e.g. TFIIF/A34.5–49/C37–53/TFIIIC)
-
III.
Factors, which do not share obvious sequence or structural similarities but are related in the topology of secondary structure elements in complex with RNAPs (e.g. GreA-B and TFS/TFIIS, σ-factor and TFB/TFIIB)
Thus, within the highly conserved RNAP active centre (see Sect. 9.2.1), transcription factors are required to adopt similar topologies to efficiently stimulate and regulate the catalytic process. The increasing complexity of the transcription machinery in eukaryotes, when compared to bacteria and archaea, culminates eventually in the evolution of complexes like TFIID and Mediator, comparable in size to RNAP II with associated general transcription factors. This complex network of protein-protein interactions may allow RNAP to integrate a large variety of different signals from DNA-bound regulators and from components of chromatin, the DNA-template of eukaryotic transcription.
9.3 Structural Dynamics of RNA Polymerase Throughout the Transcription Cycle
9.3.1 Overview
In the course of transcription, RNAPs repeatedly cycle through three distinct functional phases termed initiation, elongation and termination. Throughout the transcription cycle, RNAPs are associated with and regulated by transcription initiation, elongation and termination factors. During initiation, RNAP is specifically recruited to the promoter DNA located upstream of the transcription start site (TSS). Transcription initiation factors like σ factors (bacteria) or TBP and TFB-like factors (archaea/eukaryotes) specifically recognise and bind sequences in the promoter DNA, recruit the RNAP to the promoter and establish the directionality for transcription. Following RNAP recruitment, the DNA is locally melted at the TSS to allow loading of the template (TS) DNA strand into the active site of the RNAP. This process – called open complex formation – eventually yields the stable transcription bubble. RNAPs are able to start RNA synthesis de novo requiring no primer for RNA polymerisation. Once the RNAP escapes from the promoter, the phase of productive RNA synthesis (elongation) starts and the RNAP translocates along the TS with each new nucleotide added to the growing transcript chain. Elongation is a discontinuous process frequently interrupted by pausing events that can lead to backtracking of the RNAP. The elongation factors reduce pausing or recover backtracked arrested transcription complexes, thereby increasing the processivity of the RNAP. Eventually, the RNAP will encounter a termination signal leading to the dissociation of the elongation complex and release of the newly synthesized RNA.
9.3.2 Initiation
9.3.2.1 Bacteria
In bacteria, the RNAP core is transiently bound to a member of the σ factor family forming the bacterial RNAP holoenzyme. The number of encoded σ factors differs between bacterial species but one of the σ factor often serves a “housekeeping” factor that supports transcription initiation from the majority of promoters (Feklistov et al. 2014). E.coli encodes seven σ factors among which σ 70 (σ70, also known as RpoD in E.coli and as σA in many other bacteria) is the “housekeeping” factor (Paget 2015). A well-studied alternative σ factors is σ54 (also known as RpoN or σN), which directs transcription under stress conditions and in response to environmental signals. σ70 and σ54 are structurally unrelated and belong to two distinct classes of σ factor that differ in their mode of action during transcription initiation (Paget 2015; Yang et al. 2015).
σ70 is organized in four functional domains termed σ1.1, σ2, σ3 and σ4 that are connected by flexible linkers (see Sect. 9.2.2). Sigma as part of the holoenzyme (but not by itself) undergoes specific interactions with conserved sequence elements of the bacterial core promoter on the one hand side and with the RNAP to perform sigma’s three main functions:
-
I.
Sequence-specific recognition of the promoter
-
II.
Recruitment of the RNAP to the promoter
-
III.
Promoting strand separation for initial transcription bubble formation (Paget 2015)
A helix-turn-helix motif in the highly conserved σ region 4 undergoes specific interactions with five nucleotides in the −35 region of the promoter and bends the DNA in a 30° angle (Basu et al. 2014; Murakami et al. 2002a; Zhang et al. 2012; Zuo and Steitz 2015). σ region 3 and 2 interact with the extended −10 motif and the −10 and discriminator region of the NTS, respectively (Fig. 9.3a). These interactions are critical for the site-specific recognition of the promoter DNA and only occur if the binding interfaces of sigma are exposed as a result of its interaction with the core RNAP. σ recognises the −35 and −10 motif in the double-stranded form and the nucleotides in the extended −10 and discriminator motif are “read” upon strand separation. In contrast, the bases of the NTS of the core recognition element (CRE) interact directly with the RNAP β subunit between positions −4 to +2. Here, a G at position +2 (G+2) is unstacked and inserted into a deep pocket of the β subunit in a manner in which σ interacts with the bases A−11, T−7 and G−6 (Zhang et al. 2012). It is thought that DNA opening starts at position A−11 followed by an extension of the initial melted region to yield an initial transcription bubble of 12–14 bases resulting in the open initiation complex (RPo). The interaction of σ2 with the −10 motif results in a 90° bent in the DNA that directs the downstream DNA toward the RNAP active site cleft allowing the loading of the TS into the active site (Feklistov and Darst 2011). Upon sequence-specific recognition of the promoter DNA and loading of the DNA, σ1.1 is displaced from the cleft. Relocation of σ1.1 – a sigma domain that is only present in “housekeeping” sigma factors – does not occur if DNA is non-specifically bound to the RNAP rendering σ1.1 a “gatekeeper” for the RNAP active site. Part of the linker region that connects σ3 and σ4 specifically interacts with the RNAP inserting a loop region (termed the sigma finger, σ3.2) deep into the active site of the RNAP occupying the pathway of the growing RNA chain. Insertion of σ3.2 stimulates binding of NTPs in the active site but when the RNA chain extends to a length of only four to five nucleotides, σ3.2 contacts the nascent RNA. While this potentially stabilises short RNA-DNA hybrids (Zuo and Steitz 2015), it eventually results in a stressed intermediate state when further NTPs are added to the RNA. Single-molecule studies showed that during initial transcription, the RNAP does not translocate but remains connected to the promoter and downstream DNA is “scrunched” into the RNAP further adding to the accumulated stress (Kapanidis et al. 2006; Revyakin et al. 2006). The occurrence of the stressed intermediate state provides the mechanistic rational for an observation called “abortive initiation”. Initially, RNAP repeatedly synthesises and releases short RNA transcript 2–15 nucleotides in length (Goldman et al. 2009) without entering the productive elongation phase, as σ blocks the RNA exit pathway. RNA release instead of productive initiation beyond nucleotide 15 is one possibility to escape from the stressed intermediate state while staying connected to the promoter DNA. Alternatively, the accumulated stress provides the driving force for promoter escape leading to productive initiation. The sigma finger and σ4 are only displaced if the RNA reaches a length of 16 nucleotides and the RNA successfully competes for the space occupied by σ3.2 (Nickels et al. 2005). Open complex formation is a spontaneous energy-independent process in σ70 containing holoenzymes, but σ54 holoenzymes require an additional transcriptional activator protein that belongs to the AAA+ ATPase family before DNA melting can commence (Wigneshweraraj et al. 2008; Saecker et al. 2011; Yang et al. 2015). The coupling of ATP hydrolysis to transcriptional activation allows a tighter control of the promoter and overall transcriptional output thereby allowing a swift and precise response to environmental change. Loading and unwinding of the DNA also leads to structural changes in the RNAP. The mobile RNAP clamp is predominantly in an open conformation in the DNA-free state of the bacterial RNAP. Upon open complex formation, the clamp closes securing the DNA in the nucleic acid cleft (Chakraborty et al. 2012).
9.3.2.2 Archaea
In archaea, transcription initiation is mediated by two transcription initiation factors, TBP and TFB. While the function of TBP and TFB – e.g. the sequence-specific positioning of the RNAP at the TSS – is comparable to that of the bacterial σ factors, TBP and TFB are not homologous to sigma. TBP recognises an AT-rich region termed the TATA-box centred at position −26/−27 with respect to the TSS (Soppa 1999). Upon binding, TBP induces a severe bend of approximately 90°in the DNA (Littlefield et al. 1999; Gietl et al. 2014). The B recognition element (BRE) is another archaeal promoter element located at the upstream end of the TATA-box (Fig. 9.3b). BRE is specifically recognised by the C-terminal cyclin domain of TFB (Werner and Weinzierl 2005). The TBP-DNA interaction is of transient nature and in some archaeal species (e.g. the crenarchaeon Sulfolobus acidocaldarius) TFB is required to stabilise the TBP-DNA interaction (Gietl et al. 2014) and recruits the RNAP via its N-terminal domains to the promoter DNA to form the functional pre-initiation complex (PIC) (Bell and Jackson 2000; Bell et al. 1999). As TBP is composed of two symmetric repeats (Brindefalk et al. 2013), TFBs orientation at the promoter determines the directionality of the transcription initiation complex on the DNA. TFB furthermore fulfils functions at the post-recruitment stage aiding in start site selection and promoter opening (Kostrewa et al. 2009; Wiesler et al. 2013; Wiesler and Weinzierl 2011). TBP and TFB are necessary and sufficient to drive promoter-directed transcription in the archaeal transcription system and transcription bubble formation does not necessitate ATP hydrolysis (Hausner et al. 1996; Werner et al. 2006; Werner and Weinzierl 2002; Bell et al. 1998; Qureshi et al. 1997). However, TFE stabilises the PIC and aids open complex formation (Bell et al. 2001; Grohmann et al. 2011; Grünberg et al. 2007; Werner and Weinzierl 2005). Both, TFB and TFE are comprised of multiple domains connected by flexible linkers (Fig. 9.2b/c). A high-resolution structure of the archaeal PIC is not available but structures of archaeal and homologous eukaryotic subcomplexes exist, providing insights into the structural organisation of TFE and TFB and their respective RNAP binding sites. The N-terminal part of TFB is composed of the B-ribbon, −reader and –linker domains. The B-ribbon domain interacts with the RNAP dock domain while the reader and linker protrude deep into the cleft contacting the RNAP clamp and partly occupying the RNA exit channel (Fig. 9.3b) (Kostrewa et al. 2009; Liu et al. 2010; Sainsbury et al. 2013). The B-reader follows a similar path as σ3.2 that likewise results in a clash with the emerging RNA chain. TFB contacts the TS in a way that allows the organisation of the transcription bubble and prevents a tilting of short DNA-RNA hybrids thereby stimulating transcription (Werner and Weinzierl 2005; Sainsbury et al. 2013).
TFEα/β enhance open complex formation and stimulate productive initiation (Blombach et al. 2015; Grohmann et al. 2011). Archaeal TFEα is composed of an N-terminal WH domain that interacts with the RNAP clamp and a C-terminal zinc ribbon domain that contacts the RNAP at the base of the stalk – an interaction network conserved in the eukaryotic domain (Grohmann et al. 2011; Grunberg et al. 2012; Nagy et al. 2015; Engel et al. 2013). The stalk domain is fundamentally important for the recruitment and function of TFEα. The contribution of TFEβ to TFE function is less well understood. The C-terminal domain of TFEβ from S. solfataricus contains a [4Fe-S] cluster that is important for dimerization with TFEα and the interaction with the RNAP clamp. TFEα executes its function by contacting the NTS at position −12 thereby stabilising the upstream edge of the transcription bubble and it is likely that TFEβ (like its eukaryotic counterpart) reaches towards the cleft to assist with the handling of DNA strands.
Due to its high flexibility, the structure of the archaeal initiation complex could not be captured at high resolution. Nevertheless, using the distance constraints derived from single-molecule FRET measurements in combination with the known partial structures allowed the modelling of the archaeal open PIC (Nagy et al. 2015). The DNA is melted between position −12 and +2 and the TS is fully loaded into the active site while the NTS is deposited outside the cleft (Fig. 9.3b). The location of the downstream DNA is comparable to the bacterial and eukaryotic initiation complex. The upstream edge of the transcription bubble is located near the RNAP clamp and WH domain of TFEα suggesting that these structural elements collaborate to achieve the stabilisation of the transcription bubble. As observed for the bacterial RNAP clamp, a conformational change of the RNAP clamp accompanies the transition from the closed (dsDNA association without loading into the RNAP) to open complex. This transition is stimulated by TFE resulting in an equilibrium shift towards the open clamp if the PIC contains TFE (Schulz et al. 2016).
9.3.2.3 Eukaryotes
The basal transcription machineries of the archaeal and eukaryotic RNAP II transcription system are highly conserved. In both systems, TBP and TFIIB (homologous to the archaeal TBP and TFB) are sufficient to drive transcription from strong promoters and negatively supercoiled templates (Parvin and Sharp 1993). Archaeal TFE has the eukaryotic TFIIE as a functional counterpart. However, RNAP II initiation complexes contain additional general transcription factors TFIIA, TFIIF and TFIIH. Moreover, TBP is mainly part of the multiprotein complex TFIID (see Sect. 9.2.2). Like the archaeal PIC, the eukaryotic initiation complex most likely forms from individual factors in a stepwise manner in vivo (Fig. 9.3c). Initially, TBP (as part of TFIID) recognises the promoter DNA approximately 30 base pairs upstream of the TSS (in humans). While TBP is also required for transcription from all eukaryotic promoters, the presence of a TATA box is not mandatory for TBP binding. In fact, only 10–15% of the mammalian promoters contain a TATA-box (Huisinga and Pugh 2004; Lee et al. 2000). Nevertheless, TBP is found to be associated with the majority of TATA-containing and TATA-less promoters in yeast (Rhee and Pugh 2012; Juo et al. 2003) demonstrating the general role of TBP in transcription initiation. The mechanism of DNA-bending differs from the archaeal system, as eukaryotic TBP bends the DNA in a two-step process (Blair et al. 2012; Gietl et al. 2014; Tolic-Norrelykke et al. 2006; Wu and Hampsey 1999). Eukaryotic TBP forms long-lived complexes with TATA-containing promoter DNAs that are stable for minutes to hours. TFIIB stabilises the TBP-DNA complex in its fully bend form (Gietl et al. 2014). The auxiliary transcription factor TFIIA binds to the upstream side of the TATA-box and stabilises the TBP-DNA complex without changing the overall architecture of the complex (Blair et al. 2012). Like archaeal TFB, TFIIB associates with the TBP-DNA complex via sequence-specific interactions with the BRE element. In eukaryotes, BRE sequences line both sides of the TATA-box, termed upstream and downstream BRE (Deng and Roberts 2006). TFIIB is also responsible for the recruitment of RNAP in the eukaryotic system executing its role in a highly analogous manner to the archaeal system. When RNAP II engages with the TFIID-TFIIB-TFIIA-DNA complex it is already associated with TFIIF, a heterodimeric complex formed by an alpha and beta subunit (also known as Tfg1 and Tfg2 in yeast where a third protein, Tfg3, is part of the complex) (Chen et al. 2007). Around 50% of RNAP II are in complex with TFIIF (Rani et al. 2004), which may help to recruit the polymerase to the promoter by interfering with its non-specific DNA binding (Conaway et al. 1991). TFIIF fulfils various functions including the reinforcement of the PIC via a stabilisation of TFIIB within the PIC and stabilisation of the transcription bubble. The WH domain of TFIIFβ appears to be mobile and may adopt different positions to interact with the DNA during PIC formation (Chen et al. 2007; Eichner et al. 2010). TFIIF is also involved in TSS selection and stimulates early RNA synthesis (Khaperskyy et al. 2008; Ren et al. 1999; Tan et al. 1995; Yan et al. 1999; Rani et al. 2004; Fishburn and Hahn 2012). Association of TFIIE and TFIIH completes the PIC to form the closed initiation complex (Murakami et al. 2013a; Sainsbury et al. 2015). TFIIH and TFIIE seem to bind the initiation complex in a cooperative mode (He et al. 2016). No X-ray structure of the complete eukaryotic PIC is available but electron microscopy and crosslinking studies revealed the overall organisation of the human and yeast PIC (Fishburn and Hahn 2012; He et al. 2016; Murakami et al. 2013b, 2015; Plaschka et al. 2015) (Fig. 9.3c). TFIIE occupies a comparable position as archaeal TFE at the clamp coiled coil and another binding site close to the stalk domain. TFIIEβ stretches over the cleft. In contrast to σ70-dependent bacterial initiation and initiation in archaea, open complex formation is an energy-dependent process in the RNAP II system. The ATPase activity resides in TFIIH, a 10-subunit factor that harbours an ATPase (XPB in humans, Ssl2 in yeast). TFIIH contacts the PIC in proximity to TFIIE and interacts with the downstream DNA but not with the transcription bubble. Mechanistically, TFIIH appears to act as an ATP-dependent dsDNA translocase that rotates the downstream DNA leading to torsional stress and the unwinding of dsDNA as the upstream DNA is fixed due to the tight interaction of the TATA/BRE-TFIIB-TBP complex with the RNAP (Fishburn et al. 2015). The RNAP II initiation complex unwinds approximately 11–15 bp of DNA. Recent cryo-EM reconstruction showed that the TFIIB linker is disordered in the closed human PIC but becomes ordered in the open PIC and directly contacts the NTS (He et al. 2016). A comparable scenario is found in the structures of PIC from S. cerevisiae. Here, the B-linker shows weak density and the B-reader is mobile (Plaschka et al. 2016a). An intricate network of TFIIB, the clamp coiled coil, the TFIIE “E-ribbon” domain and the RNAP rudder is likely to stabilise the transcription bubble. TFIIF and TFIIE bind the promoter from the opposite sites of the cleft working in concert to encircle, retain and open the DNA (He et al. 2016; Plaschka et al. 2016a). The RAP30 subunit of TFIIF, which contacts Rpb2 (external 2 and protrusion domain), TFIIB, TBP and the downstream BRE, is another factor critically involved in bubble stabilization. Just as observed for the bacterial and eukaryotic system, the clamp domain of RNAP II adopts different conformations at the different stages of PIC assembly (He et al. 2013, 2016; Plaschka et al. 2016a). In the closed complex, the clamp is found in an open state while the clamp is closed over the DNA cleft in the open PIC. Notably, the tip of the clamp coiled coil changes its interaction partner when progressing from the open to closed transition. In the closed PIC the tip contacts WH2 of TFIIEβ but relocates to interact with the WH domain of TFIIEα during open complex formation (personal communication Eva Nogales).
The process of initiation complex assembly and open complex formation is well described for the eukaryotic RNAP II system but less well understood for RNAP I and RNAP III transcription. TBP seems to be associated with all eukaryotic initiation complexes and TFIIB-like factors are integral parts of the RNAP I and III initiation complexes (Vannini and Cramer 2012; Knutson and Hahn 2013) (see Sect. 9.2.2). In the RNAP I system, Rrn7 (TAF1 B in humans) represents the TFIIB-like factor. Rrn7, together with Rrn11 and Rrn6, is part of the CF which recruits RNAP I to the core element (CE) of the promoter. In yeast, recruitment of RNAP I additionally requires the recognition of a sequence stretch upstream of the CE, the upstream element (UE), by the upstream activation factor (UAF). UAF is comprised of histones H3 and H4, UAF30 and the factors Rrn5, Rrn9 and Rrn10. In higher eukaryotes, the unrelated HMG-box protein upstream binding factor (UBF) might at least in part have a similar function (Sanij and Hannan 2009). RNAP I itself is associated with Rrn3 (TIF-IA in humans), which contacts subunit A43 and the CF fulfilling a bridging function.
In the RNAP III system, Brf1 represents the TFIIB-like factor when the RNAP III machinery is assembled at type I (e.g. 5S rRNA) and type II promoters (tRNAs). Here, Brf1 is found in complex with TBP and Bdp1, which together form the TFIIIB complex. Initiation at these promoter types also requires TFIIIC, a 6-subunit complex (Male et al. 2015). TFIIIC contains subunits Sfc1/Sfc7, which contains similar domains and an overall domain architecture comparable to the RNAP II-specific general transcription factor TFIIF Rap30/Rap74 (Taylor et al. 2013). At the 5S rRNA promoter, TFIIIA recruits the TFIIIC complex, whereas TFIIIC directly recognises the A and B box in tRNA promoters. RNAP III utilises a third promoter type represented for example by the human promoter for the U6 snRNA gene. Instead of Brf1, Brf2 is integral part of TFIIIB and the multisubunit SNAPc complex is crucially involved in promoter recognition upstream of the TATA-box. A structural characterisation of the Brf2/TBP/promoter DNA complex revealed the high degree of structural conservation as compared to the equivalent archaeal and RNAP II complex (Gouge et al. 2015).
Homologs of TFIIF (A49/A34.5 in RNAP I, C37/C53 in RNAP III) and TFIIE (A49 is homologous to TFIIEβ in RNAP I and C82/C34 in RNAP III) are stably integrated into RNAP I and RNAP III (see Sect. 9.2.2 and Fig. 9.2) and play a comparable role in open complex stabilisation (Hoffmann et al. 2015; Engel et al. 2013; Fernandez-Tornero et al. 2013). In the RNAP I and RNAP III system, open complex formation occurs without TFIIH-like factors rendering the melting of DNA independent of ATP hydrolysis.
9.3.3 Elongation
9.3.3.1 Promoter Clearance
Before a fully committed transcription elongation complex is established, an intermediate phase of transcription occurs in which RNAPs loose contact with initiation factors. Additionally, as noted above, the ternary RNAP-DNA-RNA complex is gradually stabilised when the first phosphodiester-bonds are synthesized. On the other hand, a few nucleotides (nt) long transcripts of bacterial RNAP are blocked by the σ3.2 loop positioned at the RNA exit channel, which might lead to abortive transcription. To enter elongation, the σ3.2 loop is displaced by the nascent RNA leading to its release when the DNA-RNA hybrid reaches a length of about 12 nt. The displacement of the σ3.2 loop marks the initial state of promoter escape, triggering the release of RNAP from the promoter, which is required for RNAP translocation during transcription elongation (Murakami et al. 2002a).
The 5′end of eukaryotic RNAP II generated transcripts starts to enter the RNA exit channel when the DNA-RNA hybrid reaches a length of 8 nt (Westover et al. 2004b). While RNAP II scans for a transcription start site, the upstream edge of the separated strands remains fixed and the resulting transcription bubble extends downstream until it spans a length of 17–18 nt (Giardina and Lis 1993; Kuehner and Brow 2006). Immediately afterwards, the upstream region closes (Holstege et al. 1997; Pal et al. 2005) resulting in an approximately 10 nt long transcription bubble, which migrates with the translocating RNAP. Displacement of transcription factor TFIIB and initial RNA synthesis coincides with bubble collapse. It is a matter of discussion, how TFIIB destabilisation or the action of TFIIF and/or the elongation factor Spt4/Spt5 (see below) are implicated in this process (Andrecka et al. 2008; Blombach et al. 2013; Bushnell et al. 2004; Cabart et al. 2011; Fishburn and Hahn 2012; Kostrewa et al. 2009; Tran and Gralla 2008; Zawel et al. 1995). Furthermore, closing of the RNAP clamp accompanies the transition from the initiation to elongation phase (Chakraborty et al. 2012; Engel et al. 2013; Schulz et al. 2016; Gnatt et al. 2001). The stalk domain, which is present in archaea and eukaryotes but not in bacteria, is involved in this movement. It is formed by subunits homologous to the RNAP II subunits Rpb4/Rpb7 and promotes open complex formation and the processivity of the polymerase (Grohmann and Werner 2011; Hirtreiter et al. 2010b).
Transcription bubble collapse correlates with the dissociation of TFIIH and can occur after different RNA lengths have been reached, depending on the sequence of the promoter (Pal et al. 2005). Recent single-molecule analyses suggest that TFIIH-driven scanning for a TSS could result in largely extended transcription bubbles, which might either collapse back to a 10 nt containing DNA-RNA hybrid and allow promoter escape, or lead to repeated scanning, or cause dissociation of the RNAP machinery (Fazal et al. 2015).
Apparently, promoter escape can be modulated by different factors in the different RNAP systems, for example initiation factor UBF for RNAP I (Panov et al. 2006), or transcription factors TFIIF, FBP, FIR (Liu et al. 2001) and PC4 (Fukuda et al. 2004) for RNAP II. Promoter-proximal pausing represents an important regulatory step of transcription (Adelman and Lis 2012a; Jonkers and Lis 2015; Kugel and Goodrich 1998; Kwak and Lis 2013), which is relevant for a vast majority of genes in many organisms. Promoter-proximal nucleosomes or local DNA and/or RNA sequences might be implicated in this phenomenon, which might be overcome with the help of specific transcription (elongation) factors. Several detailed reviews give an overview about these important, but more specific gene-regulatory mechanisms (Liu et al. 2010, 2015, 2016; Adelman and Lis 2012b; Yamaguchi et al. 2013; Jonkers and Lis 2015).
9.3.3.2 Overview of the RNA Polymerisation Reaction
Once the promoter is cleared, RNA synthesis proceeds through repeating steps of nucleotide addition. Cooperation between RNAP subunits and precise structural refinements of the DNA-RNA hybrid in the active centre enables the RNAP to travel along the DNA, to incorporate the correct nucleotide, to remove wrongly added nucleotides and to re-enter productive transcription elongation if the enzyme is backtracked or stalled at a transcriptional barrier. Transcriptional barriers might be DNA-bound proteins like histones, or RNA-DNA hybrid structures, called R-loops. In this book chapter, we will mainly focus on the basic mechanism of RNAPs in RNA elongation and structural dynamics of RNAP domains during RNAP-movement along the naked DNA template. In addition, the function of three evolutionary conserved RNAP-associated elongation factors, (TFIIF, Spt4/5 like factors and TFIIS) will be described. Transcription elongation on a native chromatin template and the multiple interactions of RNAPs with many other factors involved in this process has been reviewed earlier in great detail (Dangkulwanich et al. 2014; Formosa 2013; Kwak and Lis 2013; Van Lijsebettens and Grasser 2014; Zhou et al. 2012).
9.3.3.3 The Nucleotide Addition Cycle
The active centre of all RNAPs is highly conserved and catalyses RNA chain elongation following a common basic mechanism, here described for RNAP II. The nucleotide addition cycle is defined as the condensation of a single nucleotide, which is incorporated as NMP in the growing RNA chain under release of pyrophosphate. Chain elongation is accompanied by the translocation of the enzyme along the DNA and by the movement of the transcription bubble. This involves both opening and closure at the 3′ and 5′ends of the 11–12 nucleotides long melted DNA strands, respectively, and annealing of 8–9 nucleotides of the nascent RNA.
In stable elongation complexes (EC), the incoming dsDNA unwinds in the downstream region, which allows the TS to contact the active centre (Gnatt et al. 2001). Positively charged residues in the RNAP II switch 2 region probably together with repellent negative charged amino acids of switch 1 and the fork loop 1 cooperate to separate the TS from the NTS (Kettenberger et al. 2004; Kireeva et al. 2011; Naji et al. 2008). Within the transcription bubble, eight to nine consecutive nucleotides hybridize with the nascent RNA, which is extended at its 3′end. Three RNAP loops- rudder, lid and fork loop – are considered to be involved in RNA-DNA strand separation (Gnatt et al. 2001; Kettenberger et al. 2004; Westover et al. 2004a). While the DNA-RNA hybrid is formed, the NTS makes a turn of about 90° near the catalytic centre and reanneals upstream with the TS to form the exiting DNA (Kornberg 2007).
Based on X-ray crystallography structures (Cheung and Cramer 2011, 2012; Kettenberger et al. 2004; Wang et al. 2006, 2009; Westover et al. 2004a; Zhang et al. 1999) six different stages can be distinguished in the nucleotide addition cycle (NAC), which are accompanied or driven by structural rearrangements of the RNAP (Fig. 9.4) (see also Nudler 2009; Zhang et al. 2010; Cheung and Cramer 2012; Martinez-Rucobo and Cramer 2013; Dangkulwanich et al. 2014). The crucial conformational changes which are conserved through evolution (see also Bar-Nahum et al. 2005; Epshtein et al. 2002; Weinzierl 2011) concern especially two RNAP domains of the active centre, the trigger loop and the bridge helix (Fig. 9.4a). Before the nucleotide substrate (NTP) diffuses into the enzyme, RNAP II is in the post-translocation state (state I), which is characterised by an empty active site, an open trigger loop and a completely folded bridge helix (Fig. 9.4b). The NTP enters the active site between the RNA 3′end, the bridge helix and the mobile trigger loop and binds in a non-catalytic pre-insertion state (state II). A correct pairing NTP induces closure of the active centre because the trigger loop adopts an α-helical hairpin structure (state III). This conformational change brings the NTP in contact with all residues required for catalysis. Non-correct base pairing results in an equilibrium, which favours the open trigger loop formation and leads finally to dissociation of the non-pairing NTP. A two metal ion mechanism is used for catalysis (state IV). One metal ion A (Mg2+) is permanently kept in the proximity of the RNA 3′end by three conserved aspartate side chains, whereas metal B (Mg2+) contacts the NTP near conserved aspartate and glutamate residues of both the largest and second largest RNAP subunit. The RNA 3′hydroxyl group is deprotonated and attacks as a nucleophile the NTP-α-phosphate, which results in the formation of a new phosphodiester bond and the release of pyrophosphate (PPi ) (Carvalho et al. 2011; Svetlov and Nudler 2013). Subsequently, the trigger loop opens and the elongation complex enters the pre-translocation state (state V). In this state, the nucleotide insertion site is occupied by the extended 3′end of the RNA. To incorporate the next nucleotide, the active site has to be emptied by translocation of the elongation complex by one nucleotide from the pre- to the post-translocation state (state I). Partial unfolding of the bridge helix in cooperation with trigger loop movement was suggested to push the nascent hybrid base pair out of the active centre first. In a second step, refolding of the bridge helix could guide the next DNA template base into the active centre by twisting it by 90°. However, the significance of the possible different bridge helix conformations for translocation is still discussed, especially since structural information confirming the predicted bridge helix movements is still missing (Svetlov and Nudler 2008).
Nucleotide addition cycle. (a) Overall organisation of the RNAP II transcription elongation complex (PDB: 1Y1W) including the TS (template strand), NTS (non-template strand), RNAP (RNA polymerase). Flexible elements important for the nucleotide addition cycle are highlighted. (b) Six distinguishable stages of the nucleotide addition cycle are shown (PDBs: 1Y1W (stage I), 1Y77 (II), 2E2H (III), 16IH (IV), 1VUM (V) and 3GTG (VI)). Dynamics of the trigger loop (TL) and the bridge helix (BH) according to published X-ray structures are indicated. Stage V represents a backtracked elongation complex. See text for details
To ensure transcriptional fidelity, RNAPs can move backwards (state VI). Backtracking of RNAP is a prerequisite to remove misincorporated nucleotides either by intrinsic RNAP-mediated cleavage or by cleavage supported by TFIIS-like factors.
9.3.3.4 Dynamics of Elongation
To unravel the exact molecular mechanism of transcription, crystallographic studies are not sufficient since X-ray structures are static and represent only single snapshots of the transcription cycle . Single-molecule analysis and molecular dynamics simulation help to understand the temporal and spatial dynamics of biomolecules by considering both specific atomic interactions and the biological relevant kinetics (Zhang et al. 2016). Among other conclusions, these studies support a Brownian ratchet model to explain the dynamics of RNAP translocation. In principal, the mechano-chemical coupling that drives RNAP movement is still under debate and includes three models (Kireeva et al. 2010): a power-stroke mechanism, allosteric models and the Brownian ratchet model. Based on crystallographic analyses the power-stroke mechanism suggests that PPi release after nucleotide addition induces a structural change in the enzyme resulting in translocation and strand separation for 1 nt (Yin and Steitz 2004). Accordingly, the energy derived from the chemical reaction of NTP hydrolysis drives the motor forward. The allosteric model suggests that RNAP can exist in an activated and in a less activated state, which catalyse phosphodiester bond formation at different rates. Transition from the slow to the fast state is induced by binding of the templated NTP to the allosteric site (Foster et al. 2001; Holmes and Erie 2003; Nedialkov et al. 2003).
The Brownian ratchet model proposes that the enzyme oscillates between pre- and post-translocation states in a Brownian motion consuming thermal energy. NTP binding and formation of the phosphodiester bond favours the movement from the pre-to the post-translocation position (forward movement), but this step does not require a chemical reaction. In contrast, hydrolysis of both the pyrophosphate and the RNA transcript influences RNAP movement in the opposite direction (Abbondanzieri et al. 2005; Guajardo and Sousa 1997; Komissarova and Kashlev 1997a; Bar-Nahum et al. 2005). This model can explain the ability of RNAP to slide backwards on the DNA template without NTP utilisation when elongation is paused by specific DNA sequences or when NTPs become limiting. But it is still discussed whether such a back and forward movement occurs at all template positions during elongation. Investigating RNAP elongation kinetics via molecular dynamics simulation and single-molecule analyses using different experimental conditions clearly support the Brownian ratchet model (Kireeva et al. 2010; Zhang et al. 2016).
Another important aspect is that elongation speed is influenced by the underlying DNA sequence. RNAPs do not move at a constant speed over DNA templates with varying ATGC contents. Some sequences can cause a longer dwell time of the enzyme and two mechanisms – long and short pausing – can be distinguished. RNAP tends to backtrack during long pauses, which means that transcription is blocked until the 3′end of the nascent RNA is moved back in the active centre either by RNAP forward movement or by internal cleavage of the RNA. DNA sites provoking pauses have been identified for bacterial and human RNAPs (Imashimizu et al. 2013; Hawryluk et al. 2004; Hein et al. 2011; Herbert et al. 2006). Genome-wide analyses revealed almost 20,000 pausing sites on the E. coli chromosome, which are characterised by a distinct consensus sequence (Larson et al. 2014; Vvedenskaya et al. 2014).This sequence could cause pausing by interaction between the RNAP core enzyme and the 3′end of the RNA-DNA-hybrid. Accordingly, RNAPs might directly sense the shape or identity of basepairs, which could then delay enzyme translocation (Bochkareva et al. 2012).
9.3.3.5 Function of TFIIF-, Spt4/5- and TFIIS-Related Elongation Factors
One of the first factors found to be involved in RNAP II elongation was transcription initiation factor TFIIF, which associates with paused elongation complexes. During elongation, TFIIF stimulates the rate of elongation and suppresses pausing by stabilising the post-translocated elongation complex (Zhang and Burton 2004; Zhang et al. 2005). In RNAP I the heterodimer A49/34.5 and in RNAP III the subcomplex C37/53 share structural similarities, which correlates with their functional relatedness to TFIIF (Engel et al. 2013; Fernandez-Tornero et al. 2013; Geiger et al. 2010; Hoffmann et al. 2015).
Spt5/NusG-related factors are involved in elongation in all three domains of life indicating that they play an important role in supporting a basic mechanism of transcription. A common model emerged how the Spt4-Spt5-RNAP II architecture could influence transcription elongation (Fig. 9.5a). The NGN/Spt4 complex binds to the clamp, spans over the cleft and encircles the DNA in the enzyme. Binding of Spt4/5 induces closure of the clamp domain (Schulz et al. 2016). The downstream edge of the transcription bubble could be stabilised by interactions between the non-transcribed DNA strand and the NGN domain of Spt4/5. Biochemical and mutational analyses suggest that NusG speeds up the oscillation of RNAP between the pre- and post-translocation state (Fig. 9.4b) (Bar-Nahum et al. 2005; Borukhov and Nudler 2008). Taken together, these studies underline that the dissociation of the ternary transcription complex and release of the DNA template is prevented by Spt5/NusG elongation factors, which leads to an increase in polymerase processivity. Interestingly, cryo-EM studies indicated a shift of the Spt4/Spt5 location towards the opposite site of the cleft, interacting with the protrusion and lobe domains of RNAP II (Klein et al. 2011). This could mean that RNAP II domains (clamp and/or lobe) and the NGN domain of Spt5 interact in a dynamic fashion.
RNAP II elongation complexes. (a) Structural model of a Spt5NGN/Spt4 containing RNAPII elongation complex (Model from Martinez-Rucobo et al. 2011). TS (template strand), NTS (non-template strand), RNAP (RNA polymerase), clamp coiled coil (Rpb1 domain interacting with Spt5NGN/Spt4). Spt5NGN/Spt4 bridges the cleft securing the DNA in the DNA binding channel, which contributes to RNAP processivity. (b) Structure of a TFIIS-containing elongation complex (PDB: 3PO3). The enlarged section shows components involved in the cleavage reaction of a backtracked RNA. See text for details
Recent structural investigations suggest that Spt4/Spt5/NusG proteins are associated with RNAP regions to which also transcription initiation factors bind. For instance, bacterial sigma factor or eukaryotic TFIIB binds to the coiled coil domain of the clamp (Arthur et al. 2000; Vassylyev et al. 2002; Kostrewa et al. 2009; Sevostyanova et al. 2008). Archaeal TFE and eukaryotic TFIIE interact with the clamp at a site that putatively overlaps with NGN domain-binding. Finally, the two large subunits of yeast TFIIF interact with RNAP domains that form one side of the cleft, but only at the periphery (Chen et al. 2007; Eichner et al. 2010). This raises the question whether competition of elongation and initiation factors for access at the clamp is a general mechanism to switch from initiation to elongation (Blombach et al. 2013). In fact, functional evidence was provided that archaeal TFE competes with Spt4/Spt5 for binding at the clamp coiled coil domain, and that this competition determines the transcription mode of the polymerase: TFE binding abolishes the inhibitory role of Spt4/Spt5 for initiation and Spt4/Spt5 removes TFE from the elongation complex (Grohmann et al. 2011). The latter could then facilitate the exchange between initiation factors and later acting RNAP-associated factors.
Pausing can lead to backtracking of the polymerase (Kireeva et al. 2005; Nudler et al. 1997; Komissarova and Kashlev 1997b). When the polymerase moves backwards along the DNA and RNA, the 3′RNA end dissociates from the DNA-RNA hybrid and is expelled from the active site through the pore underneath. Thus, RNA chain elongation is impaired. Transcription can only resume if the 3′end of the RNA is relocated into the active centre, which is mainly achieved through factor-stimulated RNA cleavage reactions. Arrested elongation complexes could be visualised by X-ray crystallography and revealed the exact position of the backtracked RNA (Cheung and Cramer 2011; Wang et al. 2009). In principal, a gating tyrosine residue Y769 can restrict the extent of backtracking, allowing the preferential cleavage at the level of dinucleotides. However, if RNAP is arrested, the DNA-RNA hybrid can become so instable that the RNA can easily backtrack beyond the gating tyrosine into the pore keeping the trigger loop in an inactive conformation (Fig. 9.4b, stage VI). Furthermore, the bending of the bridge helix, which apparently depends on mismatches in the RNA-DNA hybrid, was reported to promote RNAP II backtracking (Da et al. 2016). Backtracked RNAPs can be rescued by intrinsic RNA cleavage activity and with the help of factors that stimulate transcript cleavage at the active RNAP site. Upon RNA cleavage, 3–18 nucleotides long RNA stretches are released and the newly generated 3′OH of the RNA is aligned in the active site to become competent for RNA chain elongation. Bacteria, archaea and eukaryotic RNAP II have accessory factors Gre(B), TFS and TFIIS, respectively, to stimulate RNA cleavage of paused complexes (Fish and Kane 2002). Although GreB and TFIIS-related factors share no known structural homology (see Sect. 9.2.2), they function according to a similar principle (Martinez-Rucobo and Cramer 2013). In contrast to RNAP II, eukaryotic RNAP I and RNAP III employ the TFIIS-related polymerase subunits A12.2 and C11, respectively, to execute RNA cleavage (Chedin et al. 1998; Landrieux et al. 2006; Ruan et al. 2011; Jennebach et al. 2012; Kuhn et al. 2007; Lisica et al. 2016). Bacterial GreB binds to the jaw domain of the polymerase through its C-terminal globular domain while its N-terminal antiparallel α-helical coiled-coil domain approaches the active centre through the secondary channel and contacts the RNA (Opalka et al. 2003). TFIIS contains three domains. Domain II interacts with the polymerase at the RNAP jaw, and is connected via a linker domain with domain III, which is inserted into the active centre through the secondary channel (Fig. 9.5b) (Kettenberger et al. 2003, 2004). Apparently, the active site of RNAP II can be switched from polymerisation to cleavage mode and the transiently interacting TFIIS induces structural rearrangements of RNAP, which finally allow RNA processing and realign RNA in the active centre. The C-terminal parts of the RNAP I and III subunits A12.2 and C11 and the archaeal cleavage factor TFS, are homologous to the C-terminal domain of TFIIS, which is required for RNA processing. This molecular arrangement allows probably more efficient backtrack recovery and probably better proofreading (Lisica et al. 2016) for RNAP I and III. RNAP II and archaeal transcription might benefit from a dissociable cleavage factor since reversible association of TFIIS-related factors adds another level of transcriptional regulation (Ruan et al. 2011).
The mechanism of endonucleolytic cleavage is likely similar in all known TFIIS and GreB-related factors (Cheung and Cramer 2011; Kettenberger et al. 2003; Sosunov et al. 2003). Two conserved acidic residues (D290 and E291) are located at the foremost tip of the TFIIS-hairpin, which is inserted into the active centre of RNAP II. These residues position one of two Mg2+ ions involved in the cleavage reaction. The other Mg2+ ion is persistently bound to the RPB1 aspartate loop of the active site and binds the +1 RNA phosphate to align the hydrolysable phosphodiester bond (scissile bond, Fig. 9.5b). With the aid of the two acidic residues at the hairpin, the Mg2+ ion positions a water molecule, which then serves as a nucleophile. After RNA cleavage, the downstream RNA oligonucleotide is liberated, a new RNA 3′ end is associated to the active centre, the polymerase switches into the polymerisation mode and elongation can resume.
9.3.4 Termination
Correct transcription termination results in a length restricted transcript and in the release of RNAP and the transcript from the template. Termination of transcription requires common features for all transcription machineries, first, recognizing the end of the transcribed region, and second, overcoming related structural and energetic constraints to release the nascent transcript. Typically, terminator-complexes destabilise and release both, elongating RNA polymerases and associated transcripts. In addition, co-transcriptional RNA processing can also have a major impact on termination. No detailed structures of RNAP-containing termination complexes are yet available. Therefore, the possible mechanisms of termination are only briefly summarised.
In bacteria, two termination mechanisms are known: intrinsic termination induced by a palindromic sequence in the RNA that forms an RNA hairpin. A stem loop in the nascent RNA in which a series of U-residues follow a G-C rich element leads to pausing and destabilisation of transcription complexes (Larson et al. 2008; Peters et al. 2011; Washburn and Gottesman 2015). Mutations in the bridge helix can prevent termination, pointing at a crucial role of this RNAP domain also in the termination process (Nedialkov et al. 2013). The RNA hairpin has to be correctly folded when RNAP reaches the termination point. Furthermore, the sequence, size and length of the RNA loop influence termination efficiency.
Alternatively, termination in bacteria is mediated by the termination factor Rho (ρ). This termination factor is an RNA-dependent ATPase with helicase activity and forms a homohexameric ring. Two domains bind the nascent RNA and triggers dissociation of the RNAP from the DNA in an ATP-dependent manner. One RNA binding site binds RNA in the absence of ATP, the other site is stimulated to bind RNA transiently after the primary site is occupied. To reach the second binding site, the RNA is threaded to the central pore of Rho and the hexamer is closed when the RNA is attached. Subsequently, the transient interactions of RNA with the secondary binding site propel Rho along the RNA until it finally reaches the elongating RNAP at the release site. Dissociation of RNAP from the template can probably be achieved by unwinding of the RNA-DNA hybrid (Skordalakes and Berger 2003, 2006; Richardson 1982; Rabhi et al. 2011). The speed of elongation influences Rho dependent termination efficiency (Jin et al. 1992). In general, slowing down the elongation speed seems to be an important feature for most of the postulated termination mechanisms.
Termination in archaea and eukaryotes, however, is less well understood. Archaea and RNAP III terminate transcription after polymerizing a series of U residues (Campbell and Setzer 1992; Arimbasseri et al. 2013; Grohmann et al. 2010; Grohmann and Werner 2011; Hirtreiter et al. 2010b). This resembles the intrinsic bacteria-like termination mechanism. Whether the formation of an RNA hairpin upstream of the termination site of RNAP III is important for the mechanism of RNAP III termination is currently debated (Arimbasseri et al. 2014).
RNAP I termination involves a polymerase-specific termination factor that binds to a specific DNA sequence to pause elongation shortly upstream of its binding site, leading to transcript release (Németh et al. 2013; Jaiswal et al. 2016). In the yeast S. cerevisiae, this binding is sufficient to induce termination in vitro and in vivo (Merkl et al. 2014; Reiter et al. 2012).
Additionally, it was reported that factors involved in rRNA 3′end processing also participate in termination. Accordingly, co-transcriptional cleavage by rRNA processing factors generates a 5′RNA entry point for an exonuclease, which progressively degrades the nascent transcript associated with the elongating RNAP. After the exonuclease reaches the polymerase, which is slowed down at the termination site, it disrupts the ternary complex probably with the help of a helicase in a process termed the “torpedo model” (El Hage et al. 2008; Kawauchi et al. 2008; Rondon et al. 2009).
For RNAP II dependent termination either RNA 3′end processing factors or RNA-DNA helicases were suggested to disrupt the ternary transcription complexes (Kuehner et al. 2011; Mischo and Proudfoot 2013; Arndt and Reines 2015; Porrua and Libri 2015). RNAP II transcripts are often cleaved at a specific site (AAUAA) after the coding sequence and subsequently polyadenylated, which plays an important role in termination of protein-coding genes (Logan et al. 1987; Connelly and Manley 1988; Whitelaw and Proudfoot 1986). According to the “torpedo model” described above, the endoribonucleolytic cleavage allows entry of an exonuclease and rapid 5′-3′degradation, which leads to a clash between the exonuclease and the elongating RNAP II. This releases the polymerase from the template (Kuehner et al. 2011; Tollervey 2004). Alternatively, RNAP II transcription through the polyA-signal could induce structural changes of the elongation complex leading to its release from the template (Nag et al. 2006; Orozco et al. 2002). The involvement of additional factors is possible. Termination of RNAP II genes, which contain no polyA-tail, require factors containing RNA/DNA helicase activity. These factors destabilise the ternary elongation complex, thus, resembling bacterial Rho-dependent termination (Kuehner et al. 2011; Mischo and Proudfoot 2013; Porrua and Libri 2015).
References
Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM (2005) Direct observation of base-pair stepping by RNA polymerase. Nature 438(7067):460–465. doi:10.1038/nature04268
Adelman K, Lis JT (2012a) Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13(10):720–731. doi:10.1038/nrg3293
Adelman K, Lis JT (2012b) Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13(10):720–731. doi:10.1038/nrg3293
Andrecka J, Lewis R, Bruckner F, Lehmann E, Cramer P, Michaelis J (2008) Single-molecule tracking of mRNA exiting from RNA polymerase II. Proc Natl Acad Sci U S A 105(1):135–140. doi:10.1073/pnas.0703815105
Andrews MT, Brown DD (1987) Transient activation of oocyte 5S RNA genes in Xenopus embryos by raising the level of the trans-acting factor TFIIIA. Cell 51(3):445–453
Arimbasseri AG, Rijal K, Maraia RJ (2013) Transcription termination by the eukaryotic RNA polymerase III. Biochim Biophys Acta 1829(3–4):318–330. doi:10.1016/j.bbagrm.2012.10.006
Arimbasseri AG, Rijal K, Maraia RJ (2014) Comparative overview of RNA polymerase II and III transcription cycles, with focus on RNA polymerase III termination and reinitiation. Transcription 5(1):e27639. doi:10.4161/trns.27369
Arndt KM, Reines D (2015) Termination of transcription of short noncoding RNAs by RNA Polymerase II. Annu Rev Biochem 84:381–404. doi:10.1146/annurev-biochem-060614-034457
Arthur TM, Anthony LC, Burgess RR (2000) Mutational analysis of beta ′260-309, a sigma 70 binding site located on Escherichia coli core RNA polymerase. J Biol Chem 275(30):23113–23119. doi:10.1074/jbc.M002040200
Bae B, Feklistov A, Lass-Napiorkowska A, Landick R, Darst SA (2015) Structure of a bacterial RNA polymerase holoenzyme open promoter complex. Elife 4. doi:10.7554/eLife.08504
Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E (2005) A ratchet mechanism of transcription elongation and its control. Cell 120(2):183–193. doi:10.1016/j.cell.2004.11.045
Basu RS, Warner BA, Molodtsov V, Pupov D, Esyunina D, Fernandez-Tornero C, Kulbachinskiy A, Murakami KS (2014) Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J Biol Chem 289(35):24549–24559. doi:10.1074/jbc.M114.584037
Baumann M, Pontiller J, Ernst W (2010) Structure and basal transcription complex of RNA polymerase II core promoters in the mammalian genome: an overview. Mol Biotechnol 45(3):241–247. doi:10.1007/s12033-010-9265-6
Bedwell GJ, Appling FD, Anderson SJ, Schneider DA (2012) Efficient transcription by RNA polymerase I using recombinant core factor. Gene 492(1):94–99. doi:10.1016/j.gene.2011.10.049
Bell SD, Jackson SP (2000) The role of transcription factor B in transcription initiation and promoter clearance in the archaeon Sulfolobus acidocaldarius. J Biol Chem 275(17):12934–12940
Bell SD, Jaxel C, Nadal M, Kosa PF, Jackson SP (1998) Temperature, template topology, and factor requirements of archaeal transcription. Proc Natl Acad Sci U S A 95(26):15218–15222
Bell SD, Kosa PL, Sigler PB, Jackson SP (1999) Orientation of the transcription preinitiation complex in archaea. Proc Natl Acad Sci U S A 96(24):13662–13667
Bell SD, Brinkman AB, van der Oost J, Jackson SP (2001) The archaeal TFIIEalpha homologue facilitates transcription initiation by enhancing TATA-box recognition. EMBO Rep 2(2):133–138. doi:10.1093/embo-reports/kve021
Bernecky C, Grob P, Ebmeier CC, Nogales E, Taatjes DJ (2011) Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly. PLoS Biol 9(3):e1000603. doi:10.1371/journal.pbio.1000603
Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I (2013) The architecture of human general transcription factor TFIID core complex. Nature 493(7434):699–702. doi:10.1038/nature11791
Bischler N, Brino L, Carles C, Riva M, Tschochner H, Mallouh V, Schultz P (2002) Localization of the yeast RNA polymerase I-specific subunits. EMBO J 21(15):4136–4144
Blair RH, Goodrich JA, Kugel JF (2012) Single-molecule fluorescence resonance energy transfer shows uniformity in TATA binding protein-induced DNA bending and heterogeneity in bending kinetics. Biochemistry 51(38):7444–7455. doi:10.1021/bi300491j
Blattner C, Jennebach S, Herzog F, Mayer A, Cheung ACM, Witte G, Lorenzen K, Hopfner K-P, Heck AJR, Aebersold R, Cramer P (2011) Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth. Genes Dev 25(19):2093–2105. doi:10.1101/gad.17363311
Blombach F, Makarova KS, Marrero J, Siebers B, Koonin EV, van der Oost J (2009) Identification of an ortholog of the eukaryotic RNA polymerase III subunit RPC34 in Crenarchaeota and Thaumarchaeota suggests specialization of RNA polymerases for coding and non-coding RNAs in Archaea. Biol Direct 4:39. doi:10.1186/1745-6150-4-39
Blombach F, Daviter T, Fielden D, Grohmann D, Smollett K, Werner F (2013) Archaeology of RNA polymerase: factor swapping during the transcription cycle. Biochem Soc Trans 41(1):362–367. doi:10.1042/BST20120274
Blombach F, Salvadori E, Fouqueau T, Yan J, Reimann J, Sheppard C, Smollett KL, Albers SV, Kay CW, Thalassinos K, Werner F (2015) Archaeal TFEalpha/beta is a hybrid of TFIIE and the RNA polymerase III subcomplex hRPC62/39. Elife 4:e08378. doi:10.7554/eLife.08378
Blombach F, Smollett KL, Grohmann D, Werner F (2016) Molecular mechanisms of transcription initiation – structure, function and evolution of TFE/TFIIE-like factors and open complex formation. J Mol Biol. doi:10.1016/j.jmb.2016.04.016
Bochkareva A, Yuzenkova Y, Tadigotla VR, Zenkin N (2012) Factor-independent transcription pausing caused by recognition of the RNA-DNA hybrid sequence. EMBO J 31(3):630–639. doi:10.1038/emboj.2011.432
Borukhov S, Nudler E (2008) RNA polymerase: the vehicle of transcription. Trends Microbiol 16(3):126–134. doi:10.1016/j.tim.2007.12.006
Borukhov S, Polyakov A, Nikiforov V, Goldfarb A (1992) GreA protein: a transcription elongation factor from Escherichia coli. Proc Natl Acad Sci U S A 89(19):8899–8902
Brindefalk B, Dessailly BH, Yeats C, Orengo C, Werner F, Poole AM (2013) Evolutionary history of the TBP-domain superfamily. Nucleic Acids Res 41(5):2832–2845. doi:10.1093/nar/gkt045
Brow DA, Geiduschek EP (1987) Modulation of yeast 5 S rRNA synthesis in vitro by ribosomal protein YL3. A possible regulatory loop. J Biol Chem 262(29):13953–13958
Brueckner F, Armache KJ, Cheung A, Damsma GE, Kettenberger H, Lehmann E, Sydow J, Cramer P (2009a) Structure-function studies of the RNA polymerase II elongation complex. Acta Crystallogr D Biol Crystallogr 65(Pt 2):112–120. doi:10.1107/S0907444908039875
Brueckner F, Ortiz J, Cramer P (2009b) A movie of the RNA polymerase nucleotide addition cycle. Curr Opin Struct Biol 19(3):294–299. doi:10.1016/j.sbi.2009.04.005
Burgess RR, Travers AA, Dunn JJ, Bautz EKF (1969) Factor stimulating transcription by RNA polymerase. Nature 221(5175):43–46. doi:10.1038/221043a0
Burton SP, Burton ZF (2014) The σ enigma: bacterial σ factors, archaeal TFB and eukaryotic TFIIB are homologs. Transcription 5(4):e967599. doi:10.4161/21541264.2014.967599
Burton ZF, Killeen M, Sopta M, Ortolan LG, Greenblatt J (1988) RAP30/74: a general initiation factor that binds to RNA polymerase II. Mol Cell Biol 8(4):1602–1613
Bushnell DA, Westover KD, Davis RE, Kornberg RD (2004) Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science 303(5660):983–988. doi:10.1126/science.1090838
Cabart P, Ujvari A, Pal M, Luse DS (2011) Transcription factor TFIIF is not required for initiation by RNA polymerase II, but it is essential to stabilize transcription factor TFIIB in early elongation complexes. Proc Natl Acad Sci U S A 108(38):15786–15791. doi:10.1073/pnas.1104591108
Campbell FE Jr, Setzer DR (1992) Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol Cell Biol 12(5):2260–2272
Campbell PN, Work TS (1953) Biosynthesis of proteins. Nature 171(4362):997–1001
Carter R, Drouin G (2010) The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol Biol Evol 27(5):1035–1043. doi:10.1093/molbev/msp316
Carvalho AT, Fernandes PA, Ramos MJ (2011) The catalytic mechanism of RNA polymerase II. J Chem Theory Comput 7(4):1177–1188. doi:10.1021/ct100579w
Casamassimi A, Napoli C (2007) Mediator complexes and eukaryotic transcription regulation: an overview. Biochimie 89(12):1439–1446. doi:10.1016/j.biochi.2007.08.002
Chafin DR, Claussen TJ, Price DH (1991) Identification and purification of a yeast protein that affects elongation by RNA polymerase II. J Biol Chem 266(14):9256–9262
Chakraborty A, Wang D, Ebright YW, Korlann Y, Kortkhonjia E, Kim T, Chowdhury S, Wigneshweraraj S, Irschik H, Jansen R, Nixon BT, Knight J, Weiss S, Ebright RH (2012) Opening and closing of the bacterial RNA polymerase clamp. Science 337(6094):591–595. doi:10.1126/science.1218716
Chamberlin M, Baldwin RL, Berg P (1963) An enzymically synthesized RNA of alternating base sequence: physical and chemical characterization. J Mol Biol 7:334–349
Chedin S, Riva M, Schultz P, Sentenac A, Carles C (1998) The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev 12(24):3857–3871
Cheetham GM, Steitz TA (2000) Insights into transcription: structure and function of single-subunit DNA-dependent RNA polymerases. Curr Opin Struct Biol 10(1):117–123
Chen HT, Warfield L, Hahn S (2007) The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat Struct Mol Biol 14(8):696–703. doi:10.1038/nsmb1272
Cheung AC, Cramer P (2011) Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471(7337):249–253. doi:10.1038/nature09785
Cheung AC, Cramer P (2012) A movie of RNA polymerase II transcription. Cell 149(7):1431–1437. doi:10.1016/j.cell.2012.06.006
Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, Nogales E (2013) Human TFIID binds to core promoter DNA in a reorganized structural state. Cell 152(1–2):120–131. doi:10.1016/j.cell.2012.12.005
Cler E, Papai G, Schultz P, Davidson I (2009) Recent advances in understanding the structure and function of general transcription factor TFIID. Cell Mol Life Sci 66(13):2123–2134. doi:10.1007/s00018-009-0009-3
Coin F, Oksenych V, Egly J-M (2007) Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell 26(2):245–256. doi:10.1016/j.molcel.2007.03.009
Colbert T, Hahn S (1992) A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev 6(10):1940–1949
Conaway RC, Conaway JW (2015) Orchestrating transcription with the Pol II CTD. Nat Rev Mol Cell Biol 16(3):128. doi:10.1038/nrm3956
Conaway RC, Garrett KP, Hanley JP, Conaway JW (1991) Mechanism of promoter selection by RNA polymerase II: mammalian transcription factors alpha and beta gamma promote entry of polymerase into the preinitiation complex. Proc Natl Acad Sci U S A 88(14):6205–6209
Connelly S, Manley JL (1988) A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev 2(4):440–452
Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD (2000) Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288(5466):640–649
Da LT, Pardo-Avila F, Xu L, Silva DA, Zhang L, Gao X, Wang D, Huang X (2016) Bridge helix bending promotes RNA polymerase II backtracking through a critical and conserved threonine residue. Nat Commun 7:11244. doi:10.1038/ncomms11244
Dangkulwanich M, Ishibashi T, Bintu L, Bustamante C (2014) Molecular mechanisms of transcription through single-molecule experiments. Chem Rev 114(6):3203–3223. doi:10.1021/cr400730x
Deng W, Roberts SG (2006) Core promoter elements recognized by transcription factor IIB. Biochem Soc Trans 34(Pt 6):1051–1053. doi:10.1042/BST0341051
Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A (2007) The expanding RNA polymerase III transcriptome. Trends Genet 23(12):614–622. doi:10.1016/j.tig.2007.09.001
Dombroski AJ, Walter WA, Record MT, Siegele DA, Gross CA (1992) Polypeptides containing highly conserved regions of transcription initiation factor sigma 70 exhibit specificity of binding to promoter DNA. Cell 70(3):501–512
Dounce AL (1952) Duplicating mechanism for peptide chain and nucleic acid synthesis. Enzymologia 15(5):251–258
Duttke SHC (2015) Evolution and diversification of the basal transcription machinery. Trends Biochem Sci 40(3):127–129. doi:10.1016/j.tibs.2015.01.005
Eichner J, Chen H-T, Warfield L, Hahn S (2010) Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J 29(4):706–716. doi:10.1038/emboj.2009.386
El Hage A, Koper M, Kufel J, Tollervey D (2008) Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev 22(8):1069–1081. doi:10.1101/gad.463708
Elmlund H, Baraznenok V, Linder T, Szilagyi Z, Rofougaran R, Hofer A, Hebert H, Lindahl M, Gustafsson CM (2009) Cryo-EM reveals promoter DNA binding and conformational flexibility of the general transcription factor TFIID. Structure (London, England: 1993) 17(11):1442–1452. doi:10.1016/j.str.2009.09.007
Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P (2013) RNA polymerase I structure and transcription regulation. Nature 502(7473):650–655. doi:10.1038/nature12712
Engelke DR, Ng SY, Shastry BS, Roeder RG (1980) Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell 19(3):717–728
Epshtein V, Mustaev A, Markovtsov V, Bereshchenko O, Nikiforov V, Goldfarb A (2002) Swing-gate model of nucleotide entry into the RNA polymerase active center. Mol Cell 10(3):623–634
Facciotti MT, Reiss DJ, Pan M, Kaur A, Vuthoori M, Bonneau R, Shannon P, Srivastava A, Donohoe SM, Hood LE, Baliga NS (2007) General transcription factor specified global gene regulation in archaea. Proc Natl Acad Sci U S A 104(11):4630–4635. doi:10.1073/pnas.0611663104
Fazal FM, Meng CA, Murakami K, Kornberg RD, Block SM (2015) Real-time observation of the initiation of RNA polymerase II transcription. Nature 525(7568):274–277. doi:10.1038/nature14882
Feaver WJ, Gileadi O, Li Y, Kornberg RD (1991) CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell 67(6):1223–1230
Feklistov A, Darst SA (2011) Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell 147(6):1257–1269. doi:10.1016/j.cell.2011.10.041
Feklistov A, Sharon BD, Darst SA, Gross CA (2014) Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 68:357–376. doi:10.1146/annurev-micro-092412-155737
Fernandez-Tornero C, Moreno-Morcillo M, Rashid UJ, Taylor NM, Ruiz FM, Gruene T, Legrand P, Steuerwald U, Muller CW (2013) Crystal structure of the 14-subunit RNA polymerase I. Nature 502(7473):644–649. doi:10.1038/nature12636
Fish RN, Kane CM (2002) Promoting elongation with transcript cleavage stimulatory factors. Biochim Biophys Acta 1577(2):287–307
Fishburn J, Hahn S (2012) Architecture of the yeast RNA polymerase II open complex and regulation of activity by TFIIF. Mol Cell Biol 32(1):12–25. doi:10.1128/Mcb.06242-11
Fishburn J, Tomko E, Galburt E, Hahn S (2015) Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc Natl Acad Sci U S A 112(13):3961–3966. doi:10.1073/pnas.1417709112
Formosa T (2013) The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta 1819(3–4):247–255
Foster JE, Holmes SF, Erie DA (2001) Allosteric binding of nucleoside triphosphates to RNA polymerase regulates transcription elongation. Cell 106(2):243–252
Fukuda A, Nakadai T, Shimada M, Tsukui T, Matsumoto M, Nogi Y, Meisterernst M, Hisatake K (2004) Transcriptional coactivator PC4 stimulates promoter escape and facilitates transcriptional synergy by GAL4-VP16. Mol Cell Biol 24(14):6525–6535. doi:10.1128/MCB.24.14.6525-6535.2004
Gaiser F, Tan S, Richmond TJ (2000) Novel dimerization fold of RAP30/RAP74 in human TFIIF at 1.7 A resolution. J Mol Biol 302(5):1119–1127. doi:10.1006/jmbi.2000.4110
Geiduschek EP, Nakamoto T, Weiss SB (1961) The enzymatic synthesis of RNA: complementary interaction with DNA. Proc Natl Acad Sci U S A 47(9):1405–1415
Geiger SR, Lorenzen K, Schreieck A, Hanecker P, Kostrewa D, Heck AJ, Cramer P (2010) RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol Cell 39(4):583–594. doi:10.1016/j.molcel.2010.07.028
Giardina C, Lis JT (1993) DNA melting on yeast RNA polymerase II promoters. Science 261(5122):759–762
Gietl A, Holzmeister P, Blombach F, Schulz S, von Voithenberg LV, Lamb DC, Werner F, Tinnefeld P, Grohmann D (2014) Eukaryotic and archaeal TBP and TFB/TF(II)B follow different promoter DNA bending pathways. Nucleic Acids Res 42(10):6219–6231. doi:10.1093/nar/gku273
Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD (2001) Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292(5523):1876–1882. doi:10.1126/science.1059495
Goldman SR, Ebright RH, Nickels BE (2009) Direct detection of abortive RNA transcripts in vivo. Science 324(5929):927–928. doi:10.1126/science.1169237
Gouge J, Satia K, Guthertz N, Widya M, Thompson AJ, Cousin P, Dergai O, Hernandez N, Vannini A (2015) Redox signaling by the RNA polymerase III TFIIB-related factor Brf2. Cell 163(6):1375–1387. doi:10.1016/j.cell.2015.11.005
Groft CM, Uljon SN, Wang R, Werner MH (1998) Structural homology between the Rap30 DNA-binding domain and linker histone H5: implications for preinitiation complex assembly. Proc Natl Acad Sci U S A 95(16):9117–9122
Grohmann D, Werner F (2011) Cycling through transcription with the RNA polymerase F/E (RPB4/7) complex: structure, function and evolution of archaeal RNA polymerase. Res Microbiol 162(1):10–18. doi:10.1016/j.resmic.2010.09.002
Grohmann D, Hirtreiter A, Werner F (2009) Molecular mechanisms of archaeal RNA polymerase. Biochem Soc Trans 37(Pt 1):12–17. doi:10.1042/BST0370012
Grohmann D, Klose D, Klare JP, Kay CW, Steinhoff HJ, Werner F (2010) RNA-binding to archaeal RNA polymerase subunits F/E: a DEER and FRET study. J Am Chem Soc 132(17):5954–5955. doi:10.1021/ja101663d
Grohmann D, Nagy J, Chakraborty A, Klose D, Fielden D, Ebright RH, Michaelis J, Werner F (2011) The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol Cell 43(2):263–274. doi:10.1016/j.molcel.2011.05.030
Gruber TM, Gross CA (2003) Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol 57:441–466. doi:10.1146/annurev.micro.57.030502.090913
Grünberg S, Bartlett MS, Naji S, Thomm M (2007) Transcription factor E is a part of transcription elongation complexes. J Biol Chem 282(49):35482–35490. doi:10.1074/jbc.M707371200
Grunberg S, Warfield L, Hahn S (2012) Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat Struct Mol Biol 19(8):788–796. doi:10.1038/nsmb.2334
Grunberg-Manago M, Oritz PJ, Ochoa S (1955) Enzymatic synthesis of nucleic acidlike polynucleotides. Science 122(3176):907–910
Guajardo R, Sousa R (1997) A model for the mechanism of polymerase translocation. J Mol Biol 265(1):8–19. doi:10.1006/jmbi.1996.0707
Haag JR, Pikaard CS (2011) Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 12(8):483–492. doi:10.1038/nrm3152
Hausner W, Wettach J, Hethke C, Thomm M (1996) Two transcription factors related with the eucaryal transcription factors TATA-binding protein and transcription factor IIB direct promoter recognition by an archaeal RNA polymerase. J Biol Chem 271(47):30144–30148
Hawryluk PJ, Ujvari A, Luse DS (2004) Characterization of a novel RNA polymerase II arrest site which lacks a weak 3′ RNA-DNA hybrid. Nucleic Acids Res 32(6):1904–1916. doi:10.1093/nar/gkh505
He Y, Fang J, Taatjes DJ, Nogales E (2013) Structural visualization of key steps in human transcription initiation. Nature 495(7442):481–486. doi:10.1038/nature11991
He Y, Yan C, Fang J, Inouye C, Tjian R, Ivanov I, Nogales E (2016) Near-atomic resolution visualization of human transcription promoter opening. Nature 533(7603):359–365. doi:10.1038/nature17970
Hein PP, Palangat M, Landick R (2011) RNA transcript 3′-proximal sequence affects translocation bias of RNA polymerase. Biochemistry 50(32):7002–7014. doi:10.1021/bi200437q
Henry NL, Sayre MH, Kornberg RD (1992) Purification and characterization of yeast RNA polymerase II general initiation factor g. J Biol Chem 267(32):23388–23392
Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM (2006) Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell 125(6):1083–1094. doi:10.1016/j.cell.2006.04.032
Hirata A, Klein BJ, Murakami KS (2008) The X-ray crystal structure of RNA polymerase from Archaea. Nature 451(7180):851–854. doi:10.1038/nature06530
Hirtreiter A, Damsma GE, Cheung AC, Klose D, Grohmann D, Vojnic E, Martin AC, Cramer P, Werner F (2010a) Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res 38(12):4040–4051. doi:10.1093/nar/gkq135
Hirtreiter A, Grohmann D, Werner F (2010b) Molecular mechanisms of RNA polymerase--the F/E (RPB4/7) complex is required for high processivity in vitro. Nucleic Acids Res 38(2):585–596. doi:10.1093/nar/gkp928
Hoagland MB, Zamecnik PC, Stephenson ML (1957) Intermediate reactions in protein biosynthesis. Biochim Biophys Acta 24(1):215–216
Hoffmann NA, Jakobi AJ, Moreno-Morcillo M, Glatt S, Kosinski J, Hagen WJ, Sachse C, Muller CW (2015) Molecular structures of unbound and transcribing RNA polymerase III. Nature 528(7581):231–236. doi:10.1038/nature16143
Holmes SF, Erie DA (2003) Downstream DNA sequence effects on transcription elongation. Allosteric binding of nucleoside triphosphates facilitates translocation via a ratchet motion. J Biol Chem 278(37):35597–35608. doi:10.1074/jbc.M304496200
Holstege FC, Fiedler U, Timmers HT (1997) Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J 16(24):7468–7480. doi:10.1093/emboj/16.24.7468
Hsin JP, Manley JL (2012) The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 26(19):2119–2137. doi:10.1101/gad.200303.112
Huang Y, Maraia RJ (2001) Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res 29(13):2675–2690
Huang RC, Maheshwari N, Bonner J (1960) Enzymatic synthesis of RNA. Biochem Biophys Res Commun 3:689–694
Huisinga KL, Pugh BF (2004) A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell 13(4):573–585
Hurwitz J (2005) The discovery of RNA polymerase. J Biol Chem 280(52):42477–42485. doi:10.1074/jbc.X500006200
Hurwitz J, Bresler AE (1961) The incorporation of ribonucleotides into ribonucleic acid. J Biol Chem 236:542–548
Imashimizu M, Kireeva ML, Lubkowska L, Gotte D, Parks AR, Strathern JN, Kashlev M (2013) Intrinsic translocation barrier as an initial step in pausing by RNA polymerase II. J Mol Biol 425(4):697–712. doi:10.1016/j.jmb.2012.12.002
Imbalzano AN, Zaret KS, Kingston RE (1994) Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J Biol Chem 269(11):8280–8286
Jaiswal R, Choudhury M, Zaman S, Singh S, Santosh V, Bastia D, Escalante CR (2016) Functional architecture of the Reb1-Ter complex of Schizosaccharomyces pombe. Proc Natl Acad Sci U S A 113(16):E2267–E2276. doi:10.1073/pnas.1525465113
Jennebach S, Herzog F, Aebersold R, Cramer P (2012) Crosslinking-MS analysis reveals RNA polymerase I domain architecture and basis of rRNA cleavage. Nucleic Acids Res 40(12):5591–5601. doi:10.1093/nar/gks220
Jin DJ, Burgess RR, Richardson JP, Gross CA (1992) Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci U S A 89(4):1453–1457
Jonkers I, Lis JT (2015) Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16(3):167–177. doi:10.1038/nrm3953
Juo ZS, Kassavetis GA, Wang J, Geiduschek EP, Sigler PB (2003) Crystal structure of a transcription factor IIIB core interface ternary complex. Nature 422(6931):534–539. doi:10.1038/nature01534
Kadonaga JT (2012) Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip Rev Dev Biol 1(1):40–51. doi:10.1002/wdev.21
Kamada K, De Angelis J, Roeder RG, Burley SK (2001) Crystal structure of the C-terminal domain of the RAP74 subunit of human transcription factor IIF. Proc Natl Acad Sci U S A 98(6):3115–3120. doi:10.1073/pnas.051631098
Kang JJ, Auble DT, Ranish JA, Hahn S (1995) Analysis of the yeast transcription factor TFIIA: distinct functional regions and a polymerase II-specific role in basal and activated transcription. Mol Cell Biol 15(3):1234–1243
Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH (2006) Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314(5802):1144–1147. doi:10.1126/science.1131399
Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ (2008) Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev 22(8):1082–1092. doi:10.1101/gad.463408
Keener J, Josaitis CA, Dodd JA, Nomura M (1998) Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J Biol Chem 273(50):33795–33802
Kettenberger H, Armache KJ, Cramer P (2003) Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114(3):347–357
Kettenberger H, Armache KJ, Cramer P (2004) Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell 16(6):955–965. doi:10.1016/j.molcel.2004.11.040
Khaperskyy DA, Ammerman ML, Majovski RC, Ponticelli AS (2008) Functions of Saccharomyces cerevisiae TFIIF during transcription start site utilization. Mol Cell Biol 28(11):3757–3766. doi:10.1128/mcb.02272-07
Kim TK, Ebright RH, Reinberg D (2000) Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science (New York, NY) 288(5470):1418–1422
Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M (2005) Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell 18(1):97–108. doi:10.1016/j.molcel.2005.02.027
Kireeva M, Kashlev M, Burton ZF (2010) Translocation by multi-subunit RNA polymerases. Biochim Biophys Acta 1799(5–6):389–401. doi:10.1016/j.bbagrm.2010.01.007
Kireeva ML, Domecq C, Coulombe B, Burton ZF, Kashlev M (2011) Interaction of RNA polymerase II fork loop 2 with downstream non-template DNA regulates transcription elongation. J Biol Chem 286(35):30898–30910. doi:10.1074/jbc.M111.260844
Klein BJ, Bose D, Baker KJ, Yusoff ZM, Zhang X, Murakami KS (2011) RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci U S A 108(2):546–550. doi:10.1073/pnas.1013828108
Knowlton JR, Bubunenko M, Andrykovitch M, Guo W, Routzahn KM, Waugh DS, Court DL, Ji X (2003) A spring-loaded state of NusG in its functional cycle is suggested by X-ray crystallography and supported by site-directed mutants. Biochemistry 42(8):2275–2281. doi:10.1021/bi0272508
Knutson BA, Hahn S (2011) Yeast Rrn7 and human TAF1B are TFIIB-related RNA polymerase I general transcription factors. Science (New York, NY) 333(6049):1637–1640. doi:10.1126/science.1207699
Knutson BA, Hahn S (2013) TFIIB-related factors in RNA polymerase I transcription. Biochim Biophys Acta 1829(3–4):265–273. doi:10.1016/j.bbagrm.2012.08.003
Knutson BA, Luo J, Ranish J, Hahn S (2014) Architecture of the Saccharomyces cerevisiae RNA polymerase I Core Factor complex. Nat Struct Mol Biol 21(9):810–816. doi:10.1038/nsmb.2873
Kokubo T, Yamashita S, Horikoshi M, Roeder RG, Nakatani Y (1994) Interaction between the N-terminal domain of the 230-kDa subunit and the TATA box-binding subunit of TFIID negatively regulates TATA-box binding. Proc Natl Acad Sci U S A 91(9):3520–3524
Komissarova N, Kashlev M (1997a) RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J Biol Chem 272(24):15329–15338
Komissarova N, Kashlev M (1997b) Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci U S A 94(5):1755–1760
Kornberg RD (2007) The molecular basis of eukaryotic transcription. Proc Natl Acad Sci U S A 104(32):12955–12961. doi:10.1073/pnas.0704138104
Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P (2009) RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462(7271):323–330. doi:10.1038/nature08548
Kuehner JN, Brow DA (2006) Quantitative analysis of in vivo initiator selection by yeast RNA polymerase II supports a scanning model. J Biol Chem 281(20):14119–14128. doi:10.1074/jbc.M601937200
Kuehner JN, Pearson EL, Moore C (2011) Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol 12(5):283–294. doi:10.1038/nrm3098
Kugel JF, Goodrich JA (1998) Promoter escape limits the rate of RNA polymerase II transcription and is enhanced by TFIIE, TFIIH, and ATP on negatively supercoiled DNA. Proc Natl Acad Sci U S A 95(16):9232–9237
Kuhn CD, Geiger SR, Baumli S, Gartmann M, Gerber J, Jennebach S, Mielke T, Tschochner H, Beckmann R, Cramer P (2007) Functional architecture of RNA polymerase I. Cell 131(7):1260–1272. doi:10.1016/j.cell.2007.10.051
Kwak H, Lis JT (2013) Control of transcriptional elongation. Annu Rev Genet 47:483–508. doi:10.1146/annurev-genet-110711-155440
Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C (2006) A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J 25(1):118–128. doi:10.1038/sj.emboj.7600915
Larivière L, Seizl M, Cramer P (2012) A structural perspective on Mediator function. Curr Opin Cell Biol 24(3):305–313. doi:10.1016/j.ceb.2012.01.007
Larson MH, Greenleaf WJ, Landick R, Block SM (2008) Applied force reveals mechanistic and energetic details of transcription termination. Cell 132(6):971–982. doi:10.1016/j.cell.2008.01.027
Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS (2014) A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344(6187):1042–1047. doi:10.1126/science.1251871
Layat E, Probst AV, Tourmente S (2013) Structure, function and regulation of Transcription Factor IIIA: From Xenopus to Arabidopsis. Biochim Biophys Acta 1829(3–4):274–282. doi:10.1016/j.bbagrm.2012.10.013
Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA (2000) Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405(6787):701–704. doi:10.1038/35015104
Lefèvre S, Dumay-Odelot H, El-Ayoubi L, Budd A, Legrand P, Pinaud N, Teichmann M, Fribourg S (2011) Structure-function analysis of hRPC62 provides insights into RNA polymerase III transcription initiation. Nat Struct Mol Biol 18(3):352–358. doi:10.1038/nsmb.1996
Lehman IR, Bessman MJ, Simms ES, Kornberg A (1958) Enzymatic synthesis of deoxyribonucleic acid. I. Preparation of substrates and partial purification of an enzyme from Escherichia coli. J Biol Chem 233(1):163–170
Lisica A, Engel C, Jahnel M, Roldan E, Galburt EA, Cramer P, Grill SW (2016) Mechanisms of backtrack recovery by RNA polymerases I and II. Proc Natl Acad Sci U S A. doi:10.1073/pnas.1517011113
Littlefield O, Korkhin Y, Sigler PB (1999) The structural basis for the oriented assembly of a TBP/TFB/promoter complex. Proc Natl Acad Sci U S A 96(24):13668–13673
Liu J, Akoulitchev S, Weber A, Ge H, Chuikov S, Libutti D, Wang XW, Conaway JW, Harris CC, Conaway RC, Reinberg D, Levens D (2001) Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell 104(3):353–363
Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD (2010) Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science 327(5962):206–209. doi:10.1126/science.1182015
Liu X, Kraus WL, Bai X (2015) Ready, pause, go: regulation of RNA polymerase II pausing and release by cellular signaling pathways. Trends Biochem Sci 40(9):516–525. doi:10.1016/j.tibs.2015.07.003
Liu B, Zuo Y, Steitz TA (2016) Structures of E. coli sigmaS-transcription initiation complexes provide new insights into polymerase mechanism. Proc Natl Acad Sci U S A. doi:10.1073/pnas.1520555113
Logan J, Falck-Pedersen E, Darnell JE Jr, Shenk T (1987) A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc Natl Acad Sci U S A 84(23):8306–8310
López-De-León A, Librizzi M, Puglia K, Willis IM (1992) PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell 71(2):211–220
Louder RK, He Y, Lopez-Blanco JR, Fang J, Chacon P, Nogales E (2016) Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature. doi:10.1038/nature17394
Lu D, Searles MA, Klug A (2003) Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature 426(6962):96–100. doi:10.1038/nature02088
Male G, von Appen A, Glatt S, Taylor NM, Cristovao M, Groetsch H, Beck M, Muller CW (2015) Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly. Nat Commun 6:7387. doi:10.1038/ncomms8387
Martinez-Rucobo FW, Cramer P (2013) Structural basis of transcription elongation. Biochim Biophys Acta 1829(1):9–19. doi:10.1016/j.bbagrm.2012.09.002
Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P (2011) Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J 30(7):1302–1310. doi:10.1038/emboj.2011.64
Meinhart A, Blobel J, Cramer P (2003) An extended winged helix domain in general transcription factor E/IIE alpha. J Biol Chem 278(48):48267–48274. doi:10.1074/jbc.M307874200
Merkl P, Perez-Fernandez J, Pilsl M, Reiter A, Williams L, Gerber J, Bohm M, Deutzmann R, Griesenbeck J, Milkereit P, Tschochner H (2014) Binding of the termination factor Nsi1 to its cognate DNA site is sufficient to terminate RNA polymerase I transcription in vitro and to induce termination in vivo. Mol Cell Biol 34(20):3817–3827. doi:10.1128/MCB.00395-14
Meselson M, Stahl FW (1958) The replication of DNA in Escherichia Coli. Proc Natl Acad Sci U S A 44(7):671–682
Mischo HE, Proudfoot NJ (2013) Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochim Biophys Acta 1829(1):174–185. doi:10.1016/j.bbagrm.2012.10.003
Mooney RA, Schweimer K, Rösch P, Gottesman M, Landick R (2009) Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol 391(2):341–358. doi:10.1016/j.jmb.2009.05.078
Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA (2002a) Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science (New York, NY) 296(5571):1285–1290. doi:10.1126/science.1069595
Murakami KS, Masuda S, Darst SA (2002b) Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science (New York, NY) 296(5571):1280–1284. doi:10.1126/science.1069594
Murakami K, Calero G, Brown CR, Liu X, Davis RE, Boeger H, Kornberg RD (2013a) Formation and fate of a complete 31-protein RNA polymerase II transcription preinitiation complex. J Biol Chem 288(9):6325–6332. doi:10.1074/jbc.M112.433623
Murakami K, Elmlund H, Kalisman N, Bushnell DA, Adams CM, Azubel M, Elmlund D, Levi-Kalisman Y, Liu X, Gibbons BJ, Levitt M, Kornberg RD (2013b) Architecture of an RNA polymerase II transcription pre-initiation complex. Science 342(6159):1238724. doi:10.1126/science.1238724
Murakami K, Tsai KL, Kalisman N, Bushnell DA, Asturias FJ, Kornberg RD (2015) Structure of an RNA polymerase II preinitiation complex. Proc Natl Acad Sci U S A 112(44):13543–13548. doi:10.1073/pnas.1518255112
Nag A, Narsinh K, Kazerouninia A, Martinson HG (2006) The conserved AAUAAA hexamer of the poly(A) signal can act alone to trigger a stable decrease in RNA polymerase II transcription velocity. RNA 12(8):1534–1544. doi:10.1261/rna.103206
Nagy J, Grohmann D, Cheung AC, Schulz S, Smollett K, Werner F, Michaelis J (2015) Complete architecture of the archaeal RNA polymerase open complex from single-molecule FRET and NPS. Nat Commun 6:6161. doi:10.1038/ncomms7161
Naidu S, Friedrich JK, Russell J, Zomerdijk JCBM (2011) TAF1B is a TFIIB-like component of the basal transcription machinery for RNA polymerase I. Science (New York, NY) 333(6049):1640–1642. doi:10.1126/science.1207656
Naji S, Bertero MG, Spitalny P, Cramer P, Thomm M (2008) Structure-function analysis of the RNA polymerase cleft loops elucidates initial transcription, DNA unwinding and RNA displacement. Nucleic Acids Res 36(2):676–687. doi:10.1093/nar/gkm1086
Nedialkov YA, Gong XQ, Hovde SL, Yamaguchi Y, Handa H, Geiger JH, Yan H, Burton ZF (2003) NTP-driven translocation by human RNA polymerase II. J Biol Chem 278(20):18303–18312. doi:10.1074/jbc.M301103200
Nedialkov YA, Opron K, Assaf F, Artsimovitch I, Kireeva ML, Kashlev M, Cukier RI, Nudler E, Burton ZF (2013) The RNA polymerase bridge helix YFI motif in catalysis, fidelity and translocation. Biochim Biophys Acta 1829(2):187–198. doi:10.1016/j.bbagrm.2012.11.005
Németh A, Perez-Fernandez J, Merkl P, Hamperl S, Gerber J, Griesenbeck J, Tschochner H (2013) RNA polymerase I termination: where is the end? Biochim Biophys Acta 1829(3–4):306–317. doi:10.1016/j.bbagrm.2012.10.007
Nickels BE, Hochschild A (2004) Regulation of RNA polymerase through the secondary channel. Cell 118(3):281–284. doi:10.1016/j.cell.2004.07.021
Nickels BE, Garrity SJ, Mekler V, Minakhin L, Severinov K, Ebright RH, Hochschild A (2005) The interaction between sigma70 and the beta-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc Natl Acad Sci U S A 102(12):4488–4493. doi:10.1073/pnas.0409850102
Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Roeder RG, Burley SK (1995) Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 377(6545):119–128. doi:10.1038/377119a0
Nudler E (2009) RNA polymerase active center: the molecular engine of transcription. Annu Rev Biochem 78:335–361. doi:10.1146/annurev.biochem.76.052705.164655
Nudler E, Mustaev A, Lukhtanov E, Goldfarb A (1997) The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell 89(1):33–41
Olby R (2003) Quiet debut for the double helix. Nature 421(6921):402–405. doi:10.1038/nature01397
Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA (2003) Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell 114(3):335–345
Orozco IJ, Kim SJ, Martinson HG (2002) The poly(A) signal, without the assistance of any downstream element, directs RNA polymerase II to pause in vivo and then to release stochastically from the template. J Biol Chem 277(45):42899–42911. doi:10.1074/jbc.M207415200
Paget MS (2015) Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules 5(3):1245–1265. doi:10.3390/biom5031245
Pal M, Ponticelli AS, Luse DS (2005) The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol Cell 19(1):101–110. doi:10.1016/j.molcel.2005.05.024
Panov KI, Friedrich JK, Russell J, Zomerdijk JC (2006) UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J 25(14):3310–3322. doi:10.1038/sj.emboj.7601221
Papai G, Tripathi MK, Ruhlmann C, Layer JH, Weil PA, Schultz P (2010) TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature 465(7300):956–960. doi:10.1038/nature09080
Parvin JD, Sharp PA (1993) DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell 73(3):533–540
Peters JM, Vangeloff AD, Landick R (2011) Bacterial transcription terminators: the RNA 3′-end chronicles. J Mol Biol 412(5):793–813. doi:10.1016/j.jmb.2011.03.036
Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M (2000) The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J 19(20):5473–5482. doi:10.1093/emboj/19.20.5473
Plaschka C, Lariviere L, Wenzeck L, Seizl M, Hemann M, Tegunov D, Petrotchenko EV, Borchers CH, Baumeister W, Herzog F, Villa E, Cramer P (2015) Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 518(7539):376–380. doi:10.1038/nature14229
Plaschka C, Hantsche M, Dienemann C, Burzinski C, Plitzko J, Cramer P (2016a) Transcription initiation complex structures elucidate DNA opening. Nature 533(7603):353–358. doi:10.1038/nature17990
Plaschka C, Nozawa K, Cramer P (2016b) Mediator architecture and RNA polymerase II interaction. J Mol Biol. doi:10.1016/j.jmb.2016.01.028
Porrua O, Libri D (2015) Transcription termination and the control of the transcriptome: why, where and how to stop. Nat Rev Mol Cell Biol 16(3):190–202. doi:10.1038/nrm3943
Poss ZC, Ebmeier CC, Taatjes DJ (2013) The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol 48(6):575–608. doi:10.3109/10409238.2013.840259
Qureshi SA, Bell SD, Jackson SP (1997) Factor requirements for transcription in the Archaeon Sulfolobus shibatae. EMBO J 16(10):2927–2936. doi:10.1093/emboj/16.10.2927
Rabhi M, Gocheva V, Jacquinot F, Lee A, Margeat E, Boudvillain M (2011) Mutagenesis-based evidence for an asymmetric configuration of the ring-shaped transcription termination factor Rho. J Mol Biol 405(2):497–518. doi:10.1016/j.jmb.2010.11.006
Rani PG, Ranish JA, Hahn S (2004) RNA polymerase II (Pol II)-TFIIF and Pol II-mediator complexes: the major stable Pol II complexes and their activity in transcription initiation and reinitiation. Mol Cell Biol 24(4):1709–1720
Reiter A, Hamperl S, Seitz H, Merkl P, Perez-Fernandez J, Williams L, Gerber J, Nemeth A, Leger I, Gadal O, Milkereit P, Griesenbeck J, Tschochner H (2012) The Reb1-homologue Ydr026c/Nsi1 is required for efficient RNA polymerase I termination in yeast. EMBO J 31(16):3480–3493. doi:10.1038/emboj.2012.185
Ren D, Lei L, Burton ZF (1999) A region within the RAP74 subunit of human transcription factor IIF is critical for initiation but dispensable for complex assembly. Mol Cell Biol 19(11):7377–7387
Revyakin A, Liu C, Ebright RH, Strick TR (2006) Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314(5802):1139–1143. doi:10.1126/science.1131398
Rhee HS, Pugh BF (2012) Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483(7389):295–301. doi:10.1038/nature10799
Richardson JP (1982) Activation of rho protein ATPase requires simultaneous interaction at two kinds of nucleic acid-binding sites. J Biol Chem 257(10):5760–5766
Roeder RG, Rutter WJ (1969) Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature 224(5216):234–237
Roeder RG, Rutter WJ (1970) Specific nucleolar and nucleoplasmic RNA polymerases. Proc Natl Acad Sci U S A 65(3):675–682
Rollins MB, Del Rio S, Galey AL, Setzer DR, Andrews MT (1993) Role of TFIIIA zinc fingers in vivo: analysis of single-finger function in developing Xenopus embryos. Mol Cell Biol 13(8):4776–4783
Rondon AG, Mischo HE, Kawauchi J, Proudfoot NJ (2009) Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol Cell 36(1):88–98. doi:10.1016/j.molcel.2009.07.028
Ruan W, Lehmann E, Thomm M, Kostrewa D, Cramer P (2011) Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J Biol Chem 286(21):18701–18707. doi:10.1074/jbc.M111.222273
Rutherford ST, Lemke JJ, Vrentas CE, Gaal T, Ross W, Gourse RL (2007) Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J Mol Biol 366(4):1243–1257. doi:10.1016/j.jmb.2006.12.013
Saecker RM, Record MT Jr, Dehaseth PL (2011) Mechanism of bacterial transcription initiation: RNA polymerase – promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol 412(5):754–771. doi:10.1016/j.jmb.2011.01.018
Sainsbury S, Niesser J, Cramer P (2013) Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature 493(7432):437–440. doi:10.1038/nature11715
Sainsbury S, Bernecky C, Cramer P (2015) Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol 16(3):129–143. doi:10.1038/nrm3952
Sanij E, Hannan RD (2009) The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics 4(6):374–382
Schnabel R, Thomm M, Gerardy-Schahn R, Zillig W, Stetter KO, Huet J (1983) Structural homology between different archaebacterial DNA-dependent RNA polymerases analyzed by immunological comparison of their components. EMBO J 2(5):751–755
Schulz S, Kramm K, Werner F, Grohmann D (2015) Fluorescently labeled recombinant RNAP system to probe archaeal transcription initiation. Methods 86:10–18. doi:10.1016/j.ymeth.2015.04.017
Schulz S, Gietl A, Smollett K, Tinnefeld P, Werner F, Grohmann D (2016) TFE and Spt4/5 open and close the RNA polymerase clamp during the transcription cycle. Proc Natl Acad Sci U S A. doi:10.1073/pnas.1515817113
Segall J, Matsui T, Roeder RG (1980) Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem 255(24):11986–11991
Sekine S-i, Tagami S, Yokoyama S (2012) Structural basis of transcription by bacterial and eukaryotic RNA polymerases. Curr Opin Struct Biol 22(1):110–118. doi:10.1016/j.sbi.2011.11.006
Sentenac A (1985) Eukaryotic RNA polymerases. CRC Crit Rev Biochem 18(1):31–90
Serizawa H, Mäkelä TP, Conaway JW, Conaway RC, Weinberg RA, Young RA (1995) Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature 374(6519):280–282. doi:10.1038/374280a0
Sevostyanova A, Svetlov V, Vassylyev DG, Artsimovitch I (2008) The elongation factor RfaH and the initiation factor sigma bind to the same site on the transcription elongation complex. Proc Natl Acad Sci U S A 105(3):865–870. doi:10.1073/pnas.0708432105
Sikorski TW, Buratowski S (2009) The basal initiation machinery: beyond the general transcription factors. Curr Opin Cell Biol 21(3):344–351. doi:10.1016/j.ceb.2009.03.006
Skordalakes E, Berger JM (2003) Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell 114(1):135–146
Skordalakes E, Berger JM (2006) Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell 127(3):553–564. doi:10.1016/j.cell.2006.08.051
Söding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33(Web Server issue):W244–W248. doi:10.1093/nar/gki408
Soppa J (1999) Normalized nucleotide frequencies allow the definition of archaeal promoter elements for different archaeal groups and reveal base-specific TFB contacts upstream of the TATA box. Mol Microbiol 31(5):1589–1592
Sorenson MK, Darst SA (2006) Disulfide cross-linking indicates that FlgM-bound and free sigma28 adopt similar conformations. Proc Natl Acad Sci U S A 103(45):16722–16727. doi:10.1073/pnas.0606482103
Sorenson MK, Ray SS, Darst SA (2004) Crystal structure of the flagellar sigma/anti-sigma complex sigma(28)/FlgM reveals an intact sigma factor in an inactive conformation. Mol Cell 14(1):127–138
Sosunov V, Sosunova E, Mustaev A, Bass I, Nikiforov V, Goldfarb A (2003) Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J 22(9):2234–2244. doi:10.1093/emboj/cdg193
Stevens A (1960) Incorporation of the adenine ribonucleotide into RNA by cell fractions from E. coli. Biochem Biophys Res Commun 3:92–96
Svetlov V, Nudler E (2008) Jamming the ratchet of transcription. Nat Struct Mol Biol 15(8):777–779. doi:10.1038/nsmb0808-777
Svetlov V, Nudler E (2013) Basic mechanism of transcription by RNA polymerase II. Biochim Biophys Acta 1829(1):20–28. doi:10.1016/j.bbagrm.2012.08.009
Tagami S, Sekine S-I, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S (2010) Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 468(7326):978–982. doi:10.1038/nature09573
Tan S, Garrett KP, Conaway RC, Conaway JW (1994) Cryptic DNA-binding domain in the C terminus of RNA polymerase II general transcription factor RAP30. Proc Natl Acad Sci U S A 91(21):9808–9812
Tan S, Conaway RC, Conaway JW (1995) Dissection of transcription factor TFIIF functional domains required for initiation and elongation. Proc Natl Acad Sci U S A 92(13):6042–6046
Tan L, Wiesler S, Trzaska D, Carney HC, Weinzierl RO (2008) Bridge helix and trigger loop perturbations generate superactive RNA polymerases. J Biol 7(10):40. doi:10.1186/jbiol98
Taylor NMI, Baudin F, von Scheven G, Müller CW (2013) RNA polymerase III-specific general transcription factor IIIC contains a heterodimer resembling TFIIF Rap30/Rap74. Nucleic Acids Res 41(19):9183–9196. doi:10.1093/nar/gkt664
Thomas MC, Chiang C-M (2006) The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41(3):105–178. doi:10.1080/10409230600648736
Thomas MJ, Platas AA, Hawley DK (1998) Transcriptional fidelity and proofreading by RNA polymerase II. Cell 93(4):627–637
Tolic-Norrelykke SF, Rasmussen MB, Pavone FS, Berg-Sorensen K, Oddershede LB (2006) Stepwise bending of DNA by a single TATA-box binding protein. Biophys J 90(10):3694–3703. doi:10.1529/biophysj.105.074856
Tollervey D (2004) Molecular biology: termination by torpedo. Nature 432(7016):456–457. doi:10.1038/432456a
Tran K, Gralla JD (2008) Control of the timing of promoter escape and RNA catalysis by the transcription factor IIb fingertip. J Biol Chem 283(23):15665–15671. doi:10.1074/jbc.M801439200
Van Lijsebettens M, Grasser KD (2014) Transcript elongation factors: shaping transcriptomes after transcript initiation. Trends Plant Sci 19(11):717–726. doi:10.1016/j.tplants.2014.07.002
Vannini A, Cramer P (2012) Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell 45(4):439–446. doi:10.1016/j.molcel.2012.01.023
Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P (2010) Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143(1):59–70. doi:10.1016/j.cell.2010.09.002
Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S (2002) Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 417(6890):712–719. doi:10.1038/nature752
Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I (2007) Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448(7150):157–162. doi:10.1038/nature05932
Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, Ebright RH, Nickels BE (2014) Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science 344(6189):1285–1289. doi:10.1126/science.1253458
Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD (2006) Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127(5):941–954. doi:10.1016/j.cell.2006.11.023
Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD (2009) Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science 324(5931):1203–1206. doi:10.1126/science.1168729
Washburn RS, Gottesman ME (2015) Regulation of transcription elongation and termination. Biomolecules 5(2):1063–1078. doi:10.3390/biom5021063
Watson JD, Crick FH (1953a) Genetical implications of the structure of deoxyribonucleic acid. Nature 171(4361):964–967
Watson JD, Crick FH (1953b) Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171(4356):737–738
Weinzierl RO (2010) The nucleotide addition cycle of RNA polymerase is controlled by two molecular hinges in the Bridge Helix domain. BMC Biol 8:134. doi:10.1186/1741-7007-8-134
Weinzierl RO (2011) The Bridge Helix of RNA polymerase acts as a central nanomechanical switchboard for coordinating catalysis and substrate movement. Archaea 2011:608385. doi:10.1155/2011/608385
Weiss SB, Gladstone L (1959) A mammalian system for the incorporation of cytidine triphosphate into ribonucleic ACID1. J Am Chem Soc 81(15):4118–4119. doi:10.1021/ja01524a087
Weiss SB, Nakamoto T (1961) Net synthesis of ribonucleic acid with a microbial enzyme requiring deoxyribonucleic acid and four ribonucleoside triphosphates. J Biol Chem 236:PC18–PC20
Werner F, Grohmann D (2011) Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol 9(2):85–98. doi:10.1038/nrmicro2507
Werner F, Weinzierl RO (2002) A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol Cell 10(3):635–646
Werner F, Weinzierl RO (2005) Direct modulation of RNA polymerase core functions by basal transcription factors. Mol Cell Biol 25(18):8344–8355. doi:10.1128/MCB.25.18.8344-8355.2005
Werner F, Wiesler S, Nottebaum S, Weinzierl RO (2006) Modulation of RNA polymerase core functions by basal transcription factor TFB/TFIIB. Biochem Soc Symp 73:49–58
Westover KD, Bushnell DA, Kornberg RD (2004a) Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell 119(4):481–489. doi:10.1016/j.cell.2004.10.016
Westover KD, Bushnell DA, Kornberg RD (2004b) Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science 303(5660):1014–1016. doi:10.1126/science.1090839
Whitelaw E, Proudfoot N (1986) Alpha-thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human alpha 2 globin gene. EMBO J 5(11):2915–2922
Wiesler SC, Weinzierl RO (2011) The linker domain of basal transcription factor TFIIB controls distinct recruitment and transcription stimulation functions. Nucleic Acids Res 39(2):464–474. doi:10.1093/nar/gkq809
Wiesler SC, Werner F, Weinzierl RO (2013) Promoter independent abortive transcription assays unravel functional interactions between TFIIB and RNA polymerase. Methods Mol Biol 977:217–227. doi:10.1007/978-1-62703-284-1_17
Wigneshweraraj S, Bose D, Burrows PC, Joly N, Schumacher J, Rappas M, Pape T, Zhang X, Stockley P, Severinov K, Buck M (2008) Modus operandi of the bacterial RNA polymerase containing the sigma54 promoter-specificity factor. Mol Microbiol 68(3):538–546. doi:10.1111/j.1365-2958.2008.06181.x
Woese CR, Fox GE (1977) Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A 74(11):5088–5090
Wu W-H, Hampsey M (1999) An activation-specific role for transcription factor TFIIB in vivo. PNAS 96(6):2764–2769. doi:10.1073/pnas.96.6.2764
Yakhnin AV, Babitzke P (2014) NusG/Spt5: are there common functions of this ubiquitous transcription elongation factor? Curr Opin Microbiol 18:68–71. doi:10.1016/j.mib.2014.02.005
Yamaguchi Y, Shibata H, Handa H (2013) Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim Biophys Acta 1829(1):98–104. doi:10.1016/j.bbagrm.2012.11.007
Yan Q, Moreland RJ, Conaway JW, Conaway RC (1999) Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J Biol Chem 274(50):35668–35675
Yang Y, Darbari VC, Zhang N, Lu D, Glyde R, Wang YP, Winkelman JT, Gourse RL, Murakami KS, Buck M, Zhang X (2015) TRANSCRIPTION. Structures of the RNA polymerase-sigma54 reveal new and conserved regulatory strategies. Science 349(6250):882–885. doi:10.1126/science.aab1478
Yin YW, Steitz TA (2004) The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 116(3):393–404
Zawel L, Kumar KP, Reinberg D (1995) Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev 9(12):1479–1490
Zhang C, Burton ZF (2004) Transcription factors IIF and IIS and nucleoside triphosphate substrates as dynamic probes of the human RNA polymerase II mechanism. J Mol Biol 342(4):1085–1099. doi:10.1016/j.jmb.2004.07.070
Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA (1999) Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell 98(6):811–824
Zhang C, Zobeck KL, Burton ZF (2005) Human RNA polymerase II elongation in slow motion: role of the TFIIF RAP74 alpha1 helix in nucleoside triphosphate-driven translocation. Mol Cell Biol 25(9):3583–3595. doi:10.1128/MCB.25.9.3583-3595.2005
Zhang J, Palangat M, Landick R (2010) Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol 17(1):99–104. doi:10.1038/nsmb.1732
Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH (2012) Structural basis of transcription initiation. Science 338(6110):1076–1080. doi:10.1126/science.1227786
Zhang L, Pardo-Avila F, Unarta IC, Cheung PP, Wang G, Wang D, Huang X (2016) Elucidation of the dynamics of transcription elongation by RNA polymerase II using kinetic network models. Acc Chem Res. doi:10.1021/acs.accounts.5b00536
Zhou Q, Li T, Price DH (2012) RNA polymerase II elongation control. Annu Rev Biochem 81:119–143. doi:10.1146/annurev-biochem-052610-095910
Ziegler LM, Khaperskyy DA, Ammerman ML, Ponticelli AS (2003) Yeast RNA polymerase II lacking the Rpb9 subunit is impaired for interaction with transcription factor IIF. J Biol Chem 278(49):48950–48956. doi:10.1074/jbc.M309656200
Zillig W, Stetter KO, Tobien M (1978) DNA-dependent RNA polymerase from Halobacterium halobium. Eur J Biochem 91(1):193–199
Zillig W, Stetter KO, Janekovic D (1979) DNA-dependent RNA polymerase from the archaebacterium Sulfolobus acidocaldarius. Eur J Biochem 96(3):597–604
Zuo Y, Steitz TA (2015) Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol Cell 58(3):534–540. doi:10.1016/j.molcel.2015.03.010
Acknowledgements
The authors apologise to those whose work could not be discussed owing to space limitation. The authors would like to thank Klaus Grasser for critical reading of the manuscript. We thank Eva Nogales for sharing and discussing unpublished information and for providing images used for the preparation of Fig. 9.3. We would like to thank the members of the Tschochner/Griesenbeck and Grohmann laboratory for their courage and hard work. D.G. would like to thank Finn Werner for passing on his enthusiasm for transcription and the many fruitful discussions.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Griesenbeck, J., Tschochner, H., Grohmann, D. (2017). Structure and Function of RNA Polymerases and the Transcription Machineries. In: Harris, J., Marles-Wright, J. (eds) Macromolecular Protein Complexes. Subcellular Biochemistry, vol 83. Springer, Cham. https://doi.org/10.1007/978-3-319-46503-6_9
Download citation
DOI: https://doi.org/10.1007/978-3-319-46503-6_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46501-2
Online ISBN: 978-3-319-46503-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)