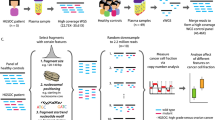
Abstract
Recent progress in the analysis of cell-free DNA fragments (cell-free circulating tumor DNA, ctDNA) now allows monitoring of tumor genomes by non-invasive means. However, previous studies with plasma DNA from patients with cancer demonstrated highly variable allele frequencies of ctDNA. Comprehensive genome-wide analysis of tumor genomes is greatly facilitated when plasma DNA has increased amounts of ctDNA. In order to develop a fast and cost-effective pre-screening method for the identification of plasma samples suitable for further extensive qualitative analysis, we adapted the recently described FAST-SeqS method. We show that our modified FAST-SeqS method (mFAST-SeqS) can be used as a pre-screening tool for an estimation of the ctDNA percentage. Moreover, since the genome-wide mFAST-SeqS z-scores correlate with the actual tumor content in plasma samples, changes in ctDNA levels associated with response to treatment can be easily monitored without prior knowledge of the genetic composition of tumor samples.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
Introduction
Development and access to accurate and sensitive screening methods for the early and fast detection of tumor-specific aberrations and identification of therapy-related changes is of utmost importance for optimal therapy management and patient care. Recent progress in the analysis of cell-free DNA fragments (cell-free circulating tumor DNA , ctDNA) now allows monitoring of tumor genomes by non-invasive means (Bettegowda et al. 2014; Diaz and Bardelli 2014; Heitzer et al. 2013a, b, c, d; Lianidou et al. 2014; Lim et al. 2014). One possibility to make use of a ctDNA is a genome-wide analysis without knowledge of any specific changes that are present in the primary tumor, which is important, in particular, either for tumor entities that lack recurrent genetic changes or patients where no tumor material is available. Most genome-wide methods currently lack sensitivity and the amount of tumor-specific DNA has to be above 5–10 % (Chan et al. 2013a; Forshew et al. 2012; Heitzer et al. 2013d; Murtaza et al. 2013). However, the levels of ctDNA vary dramatically, ranging from less than one to more than 90 % (Bettegowda et al. 2014; Heitzer et al. 2013b; Leary et al. 2012; Thierry et al. 2014). Therefore, the availability of pre-screening methods to assess the amount of tumor DNA and to select samples that are suitable for an extensive qualitative genome-wide analysis would be highly beneficial in order to save costs and time.
To this end, we modified the Fast Aneuploidy Screening Test-Sequencing System described by (Kinde et al. 2012), which is a 2-step PCR amplifying eight subfamilies of long interspersed nucleotide element-1 (L1) retrotransposons that are uniquely mappable across the genome. We used the method to pre-screen plasma DNA samples in order to evaluate whether or not the fraction of tumor-specific DNA is sufficient for our genome-wide plasma-Seq analysis (Heitzer et al. 2013d). While Kinde at al. performed read count and z-score analysis for whole chromosomes (Kinde et al. 2012), we expanded the analysis to the chromosome arms.
Methods
Patient Samples and Cancer Cell Lines
A set of 24 female controls and 18 male controls without malignant disease were collected at the Departments of Urology, Gynecology, Cardiology and Endocrinology at the Medical University of Graz. Blood samples from breast (n = 28) and prostate cancer patients (n = 61) were obtained from either the Departments of Urology and Gynecology or the Division of Clinical Oncology, Department of Internal Medicine at the Medical University of Graz. In order to validate reproducibility of the assay, cancer cell lines HT29 and MCF7 were included (ATCC, Manassas, VA, USA).
Extraction of Genomic DNA and Plasma DNA
High molecular weight genomic DNA from cultured cell lines was extracted using the QIAamp DNA Blood Midi Kit (Qiagen , Hilden, Germany) according to the manufacturer’s protocol. Plasma DNA was prepared using the QIAamp DNA Blood Mini Kit (Qiagen , Hilden, Germany) as previously described (Heitzer et al. 2013b).
mFAST-SeqS Assay
The method was described in detail previously (Belic et al. 2015). Briefly, 20 ng of cell line DNA and 0.1–5 ng of plasma DNA were amplified with Phusion Hot Start II Polymerase in five PCR cycles using target-specific L1 primers. PCR products were purified using AMPure Beads (Beckman Coulter, Brea, CA, USA) and resuspended in 12 μl 1×TE-buffer. 10 μl of purified PCR products were directly used for the second PCR (18 cycles for plasma DNA, 15 cycles for all other samples) in which Illumina specific adaptors and indices were added (for primer sequences please see (Belic et al. 2015)). PCR products were quality checked and quantified on an Agilent Bioanalyzer using a 7500 DNA kit (Agilent, Santa Clara, USA). L1 amplicon libraries were pooled equimolarly and sequenced on an Illumina MiSeq generating 150 bp single reads. Sequence reads were aligned to the hg19 genome using bwa (version 0.7.4). For each chromosome arm, reads having a mapping quality >15 were counted using an in-house script. In order to correct for different sequencing yields, read counts were normalized with the total read count per sample. We applied a z-score statistic to test for over- and under-representation of each chromosome arm. Hence, we calculated mean and standard deviation of normalized read-counts for those controls, of which a minimum of 100.000 reads were obtained after mapping quality filter step (n = 17 for male, n = 19 for female). In order to get a general overview of aneuploidy in our samples, we introduced a genome-wide z-score. Therefore, normalized read counts per chromosome arm were squared and summed. We again applied z-score statistics for the summation of the normalized read-counts by calculating mean and standard deviations for the respective controls.
Plasma-Seq
The method was described in detail previously (Heitzer et al. 2013d). Briefly, shotgun libraries were prepared using the TruSeq DNA LT Sample preparation Kit (Illumina, San Diego, CA, USA) following the manufacturer’s instructions. For plasma samples, we slightly changed the protocol as follows: we used 5–10 ng of input DNA, we omitted the fragmentation step and for selective amplification of the library fragments that have adapter molecules on both ends, we used 20–25 PCR cycles. The libraries were sequenced on an Illumina MiSeq (Illumina, San Diego, CA, USA).
Results
Validation of mFAST-SeqS with DNA from Cancer Cell Lines
First, we validated the reliability and reproducibility of the mFAST-SeqS assay by analyzing different dilutions of HT29 DNA in a background of normal DNA (100 %, 50 %, 10 % 5 %, 0 %). We obtained good reliability and repeatability of the genome-wide z-score with only small inter-assay (r = 0.998) and intra-assay (r = 0.992) variations. Linear regression analysis revealed a good correlation between genome-wide mFAST-SeqS z-scores and the dilution of HT29 cell lines (mean r = 0.974). When we evaluated our mFAST-SeqS assay by analyzing cancer cell lines MCF7 and HT29 with our plasma-Seq approach, copy number alterations (CNAs) were highly concordant. High level amplifications of specific parts of chromosomes resulted in a high overall chromosome-arm specific z-score. For instance, the high level amplifications of chromosome 8q were reflected with 8q-specfic z-scores of 42.2 and 15.0, respectively (Fig. 28.1). Loss of the short arm of chromosome 3 in HT29 with a complete loss of the region close to the centromere resulted in a 3p-specific z-score of −20.1 (Fig. 28.1). For some chromosomes, the chromosome-arm specific z-scores were below the threshold of five although circulating nucleic acids (CNAs) were detected with plasma-Seq . This can be explained by the co-occurrence of gains and losses at the same chromosome arm resulting in a balanced copy number status as exemplified for chromosome 1 of MCF7 (Fig. 28.1).
Comparison of mFAST-SeqS profiles and copy number profiles established with plasma-Seq . Selection of particular chromosomes with high or low chromosome-specific z-score reflecting high level amplifications or loss of chromosomal material in different cell lines. Co-occurrence of gains and losses at the same chromosome arm results in a balanced chromosome specific copy number status established with mFAST-SeqS
mFAST-SeqS for Minimal-Invasive Aneuploidy-Screening in Cancer Patients
In order to evaluate whether this assay can indeed be used as a pre-screening tool for the presence of higher levels of tumor-specific DNA in plasma , we analyzed a total of 89 cancer patients. As a training set, 28 plasma samples from patients with metastatic breast cancer were used. In all samples with a genome-wide mFAST-SeqS above five CNAs could be observed with plasma-Seq, whereas samples with a genome-wide z-score <5 showed balanced plasma-Seq profiles (Fig. 28.2). To validate these results we analyzed a set of 61 prostate cancer patients with various disease stages. Of those, 25 (41.0 %) samples had a genome-wide z-score below five. Not surprisingly, these patients included all patients under surveillance and nine of ten patients with localized cancer after prostectomy, of which a low fraction of tumor-specific DNA was expected. However, 12 metastasized patients also had a z-score below five indicating that the amount of tumor-specific DNA was quite low in these patients. Again, after plasma-Seq, CNAs could be only be observed in samples with a genome-wide z-score higher than five indicating that a high genome-wide z-score reflected a high amount of tumor-specific CNAs and therefore also a high fraction of tumor-specific DNA in plasma . In order to investigate whether the genome-wide z-score is indeed correlated with ctDNA levels, we compared allele frequencies (mAFs) of mutations in driver genes identified by targeted re-sequencing with the mFAST-SeqS z-score and observed a strong correlation. On the basis of linear regression, a z-score of five would predict a mAF of 10.5 %.
mFAST-SeqS assay of plasma samples from patients with metastatic breast cancer. Comparison of mFAST-SeqS profiles (right panels) and copy number profiles established with plasma-Seq (left panels) of selected samples. Samples with genome-wide z-score above five show copy number alterations established with plasma-Seq
mFAST-SeqS as a Monitoring Tool for Changing Levels of Tumor DNA in Plasma
Since the genome-wide z-score correlated with levels of tumor DNA in the circulation, we investigated whether the z-score can also be used for monitoring purposes. Thus, we evaluated the utility of the genome-wide z-score as a monitoring tool for treatment response by analyzing samples from prostate cancer patients before and during ADT. As expected, the genome-wide z-score decreased massively in patients who responded well to the therapy (Fig. 28.3a, b). Consistent with mFAST-SeqS, the mutant allele frequencies of mutations identified with targeted re-sequencing were no longer detectable with deep amplicon sequencing after 2 months of treatment (Fig. 28.3a, b). In addition, plasma-Seq resulted in a balanced copy number profile despite a variety of CNAs in the first samples, also confirming the reduction of tumor DNA (Fig. 28.3a, b). In contrast, in patients who did not respond to ADT treatment, genome-wide z-scores neither changed nor even increased and the same applied to mutant allele frequencies and CNAs (Fig. 28.3c).
Monitoring of patients with metastatic prostate cancer under androgen deprivation therapy (ADT)
(a, b) Genome-wide mFAST-SeqS z-score decreased in patients who responded well to ADT therapy. Consistent with mFAST-SeqS z-scores, the mutant allele frequencies (mAF) of mutations identified with targeted re-sequencing were no longer detectable with deep amplicon sequencing after 2 months of treatment. (c) A patient who did not respond to ADT treatment is shown. Genome-wide Z-scores increased after 6 months of treatment and the same applied to mutant allele frequencies and CNAs
For one patient we received three samples that allowed us to monitor changing clonal patterns. When we obtained the first blood sample the patient was already resistant to androgen deprivation therapy (ADT) because of a high level amplification of the androgen receptor gene (AR) located at the X chromosome (Fig. 28.4). mFAST-SeqS revealed a very high genome-wide z-score of 300. After switching to chemotherapy, the patient responded quite well; this was reflected in decreased tumor DNA levels and a drop in the mFAST-SeqS z-score to six. However, after several months the patient progressed again and the level of ctDNA increased as shown in the mFAST-SeqS z-score. In addition, using plasma-Seq we observed the emergence of novel CNAs, e.g. on chromosomes 2 and 4, while other changes disappeared upon AR amplification. Moreover, we were able to identify two somatic mutations in this patient, one in EP300 and another one in TP53. Both mutations dropped after the switch to chemotherapy, but at the time of progression, only the TP53 increased while the EP300 mutation was no longer detectable further confirming the clonal switch of this tumor. If we had followed the EP300 mutation alone we would have missed the massive increase in tumor DNA associated with progression in this patient (Fig. 28.4).
Monitoring of changing clonal patterns. The patient was already resistant to ADT when the first blood sample was obtained. (a) Changing copy number pattern arose after initial response to chemotherapy. (b, c) mFAST-SeqS r evealed a very high genome-wide z-score of 300. Initial response to chemotherapy was reflected in a decrease of mFAST-SeqS z-score and mutant allele frequencies of TP53 and EP300 mutations. At the time of progression, mFAST-SeqS and TP53 mutation increased while the EP300 mutation was no longer detectable indicating a clonal switch
Discussion
In this study, we describe an extended application of the previously established FAST-SeqS method (Kinde et al. 2012) for fast and cost-effective pre-screening of tumor-specific aneuploidy in plasma samples. We show that mFAST-SeqS serves as a valuable pre-screening tool to identify samples where tumor specific changes are most likely identified with less sensitive methods such as low-coverage whole genome sequencing or exome sequencing. Since genome-wide or exome-wide analysis of ctDNA can lead to high costs, a pre-screening using the mFAST-SeqS assay can help to reduce costs as samples with lower tumor can be identified at an early stage and can be assessed whether the samples is worth analyzing with genome-wide methods or is rather referred to more sensitive approaches (Fig. 28.5).
mFAST-Seq as a decision support tool in order to select the most appropriate analysis strategy
If the mFAST-SeqS z-score is below five, targeted approaches should be used for ctDNA analysis that can detect mutant alleles as low as 0.01–0.5 %. In cases with mFAST-SeqS z-scores above five, untargeted approaches can be used which require a higher fraction of ctDNA
One of the key challenges in the analysis of ctDNA is the fact that the fraction of circulating DNA that is derived from the tumor can range between 0.01 % to more than 90 % (Bettegowda et al. 2014; Heitzer et al. 2013b; Leary et al. 2012; Thierry et al. 2014). Although there exist many promising genome-wide approaches for the analysis of ctDNA (Chan et al. 2013a, b; Heitzer et al. 2013b, d; Leary et al. 2012), most of these methods lack sensitivity and may not be used if the fraction of umor-specific DNA is below a specific thres hold of 5–10 %. With our established plasma-Seq method tumor specific CNAs can be detected in the circulation at levels ≥10 % of circulating tumor DNA with a sensitivity of >80 % and specificity of >80 % (Heitzer et al. 2013d). Using this approach we analyzed a set of colorectal cancer patients under anti-EGFR therapy and cohort of metastasized breast cancer patients. While tumor-specific changes were identified in all CRC patients (Mohan et al. 2014), only 46 % of the breast cancer patients showed CNAs with our plasma-Seq method (Heidary et al. 2014) indicating that for a subset of samples more sensitive methods are required to identify tumor-specific changes in the circulation. Using mFAST-SeqS we are now able to roughly estimate the amount of tumor-specific DNA in plasma without any knowledge of specific aberrations of the primary tumor. In those cases with a genome-wide z-score above five tumor-specific CNAs can be established with plasma-Seq (Fig. 28.5). Furthermore, the obtained results revealed highly concordant copy number profiles of mFAST-SeqS and plasma-Seq indicating that mFAST-Seq can additionally give insights into CNAs at a chromosome arm level.
Since genome-wide z-scores reflect the level of ctDNA in the circulation, mFAST-SeqS might also be a suitable monitoring tool for treatment response. Monitoring of specific mutations might miss clonal changes in the tumor genome occurring during tumor progression or under the selective pressure of therapies as exemplified in Fig. 28.4. mFAST-SeqS is able to reflect changes in ctDNA levels associated with treatment response and progression independently of a prior knowledge of the characteristics of the primary tumor.
Altogether, mFAST-SeqS is a not only a cost-effective screening tool for the evaluation of the fraction of tumor specific DNA, but also an untargeted strategy for the assessment of changes in ctDNA levels as a response to certain therapies .
References
Belic J, Koch M, Ulz P et al (2015) Rapid identification of plasma DNA samples with increased ctDNA levels by a modified FAST-SeqS approach. Clin Chem 61:838–849
Bettegowda C, Sausen M, Leary RJ et al (2014) Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6:224ra24
Chan KC, Jiang P, Chan CW et al (2013a) Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A 110:18761–18768
Chan KC, Jiang P, Zheng YW et al (2013b) Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 59:211–224
Diaz LA Jr, Bardelli A (2014) Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 32:579–586
Forshew T, Murtaza M, Parkinson C et al (2012) Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 4:136ra168.
Heidary M, Ulz P, Heitzer E et al (2014) The dynamic range of circulating tumor DNA in metastatic breast cancer. Breast Cancer Res 16:421–430
Heitzer E, Auer M, Gasch C et al (2013a) Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res 73:2965–2975
Heitzer E, Auer M, Hoffmann EM et al (2013b) Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int J Cancer 133:346–356
Heitzer E, Auer M, Ulz P et al (2013c) Circulating tumor cells and DNA as liquid biopsies. Genome Med 5:73–83
Heitzer E, Ulz P, Belic J et al (2013d) Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med 5:30–45
Kinde I, Papadopoulos N, Kinzler KW et al (2012) FAST-SeqS: a simple and efficient method for the detection of aneuploidy by massively parallel sequencing. PLoS ONE 7:e41162
Leary RJ, Sausen M, Kinde I et al (2012) Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 4:162ra154
Lianidou ES, Strati A, Markou A (2014) Circulating tumor cells as promising novel biomarkers in solid cancers. Crit Rev Clin Lab Sci 51(3):160–171
Lim SH, Becker TM, Chua W et al (2014) Circulating tumour cells and circulating free nucleic acid as prognostic and predictive biomarkers in colorectal cancer. Cancer Lett 346:24–33
Mohan S, Heitzer E, Ulz P et al (2014) Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing. PLoS Genet 10:e1004271
Murtaza M, Dawson S, Tsui DWY (2013) Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497:108–112
Thierry AR, Mouliere F, El Messaoudi S et al (2014) Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 20:430–435
Conflict of Interest
None.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this paper
Cite this paper
Belic, J. et al. (2016). mFast-SeqS as a Monitoring and Pre-screening Tool for Tumor-Specific Aneuploidy in Plasma DNA. In: Gahan, P., Fleischhacker, M., Schmidt, B. (eds) Circulating Nucleic Acids in Serum and Plasma – CNAPS IX. Advances in Experimental Medicine and Biology, vol 924. Springer, Cham. https://doi.org/10.1007/978-3-319-42044-8_28
Download citation
DOI: https://doi.org/10.1007/978-3-319-42044-8_28
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-42042-4
Online ISBN: 978-3-319-42044-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)