Abstract
Trichoderma reesei (Teleomorph Hypocrea jecorina) RUT-C30 and QM9414 strains are the principal industrial producers of enzymes that hydrolyze lignocellulosic biomass into simple sugars, such as glucose and xylose. These fungi were generated from the wild isolate QM6a via multiple rounds of random mutagenesis for enhanced cellulase production and/or catabolite derepression. Accumulating evidence indicates that their genomes have acquired multiple chromosomal alternations (including nucleotide mutations, segmental deletions and rearrangements) compared with that of QM6a. We found that RUT-C30 and QM9414, such as QM6a, efficiently mated with the H. jecorina CBS999.97 mating partner, including completed sexual development and meiosis. However, they generated more non-viable segmental aneuploidy (SAN) ascospores than the sexual crossing of CBS999.97 with QM6a. Our results indicate that extensive mutagenesis during strain improvements resulted in speciation, i.e., RUT-C30 and QM9414 are no longer the same species as QM6a and CBS999.97. Our finding is consistent with the classic chromosomal speciation model stating that chromosome rearrangements contribute to heterozygous hybrid infertility and serve as a genetic barrier between recently diverged species. We suggest that RUT-C30 and QM9414 are ideal models for deciphering molecular mechanisms by which new genetic barriers impeding reproduction arise. Hybridization between the industrial strains and the CBS999.97 wild isolates may generate SAN progeny that harbor existing or new beneficial phenotypes for economic applications.
Authors’ Contributions:
W.C.L. performed the RUT-C30 and QM6a sexual crossing experiments and generated the figures. Y.C.C., W.C.L. and C.L.C. performed experiments and analyzed the data. W.C.L. and T.F.W. wrote the manuscript. All authors read and approved the manuscript.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Trichoderma reesei
- Hypocrea jecorina
- Genome evolution
- Sexual development
- Meiosis
- Hybrid infertility
- Aneuploidy
Abbreviations
- aCGH:
-
Array-based comparative genomic hybridization
- RTU:
-
Return to euploid
- SAN:
-
Segmental aneuploidy
- WCA:
-
Whole chromosome aneuploidy
1 Strain Improvement Programs Have Resulted in Multiple Chromosomal Alternations in the Genomes of T. reesei Hypersecreting Mutants
QM6a was first collected on the Solomon Islands during the Second World War (Mandels and Reese 1957). It was later renamed as Trichoderma reesei in honor of Elwyn T Reese, the lead investigator responsible for the identification of QM6a as a good producer of cellulases (Peterson and Nevalainen 2012; Mukherjee et al. 2013). QM6a had been considered to reproduce asexually for many years. To enhance cellulase production, several hypersecreting mutants were generated via random mutagenesis. For example, QM9414 was selected after two rounds of irradiation in a linear particle accelerator (Vitikainen et al. 2010). RUT-C30 was generated via three mutagenesis steps (UV, N-nitrosoguanidine and UV). These mutants produced several times more cellulase than QM6a. RUT-C30 also exhibits a carbon catabolite derepression phenotype, as the glucose repressor cre1 gene was found encode a truncated protein with only one of the two zinc finger motifs in the wild-type CRE1 protein (Ilmen et al. 1996).
QM9414 and RUT-C30 not only displayed an increase in cellulase production but also acquired many genetic alterations via the random mutagenesis programs. Electrophoretic karyotyping and Southern hybridization revealed extensive alterations (i.e., deletions and rearrangements) on the chromosomes of QM6a and the hypersecreting mutants (Carter et al. 1992; Mantyla et al. 1992). Multiple genomic alternations have been identified in the genomes of RUT-C30, QM9414 and several other hypersecreting mutants via high-resolution array-based comparative genomic hybridization (aCGH) (Vitikainen et al. 2010) and comparative genome sequence sequencing (Peterson and Nevalainen 2012; Martinez et al. 2008; Seidl et al. 2008; Le Crom et al. 2009; Koike et al. 2013). For example, the results of an aCGH experiment revealed at least four and five translocation breakpoints in the genomes of QM9414 and RUT-C30, respectively (Vitikainen et al. 2010). An in-depth alignment of RUT-C30 and QM6a scaffold sequences suggested that the RUT-C30 genome might contain 11 translocations, 65 deletions, 455 nonhomologous regions and >2000 nucleotide mutations (Koike et al. 2013).
2 Breaking the Barriers of Sexual Crossing
QM6a is a haploid strain with a MAT1-2 mating type locus . The opposite mating type locus was recently found in the H. jecorina CBS999.97 diploid, which is the only T. reesei wild isolate strain that is sexually competent under laboratory conditions (Seidl et al. 2009). CBS999.97 sexual development generates two sexually compatible haploids, CBS999.97(1-1) and CBS999.97(1-2), which harbor the MAT1-1 and MAT1-2 mating-type locus, respectively. Although QM6a is a sterile female, it can act as a male gamete to mate with CBS999.97(1-1). Sexual crossing of CBS999.97(1-1) with either CBS999.97(1-2) or QM6a produces fruiting bodies containing asci with 16 linearly arranged ascospores (the sexual spores specific to ascomycetes) (Seidl et al. 2009). Using classical genetic and molecular biology approaches, we have recently shown that the 16 ascospores in an ascus are generated via meiosis followed by two rounds of postmeiotic mitosis (Chuang et al. 2015). Meiosis is a special type of cell division in sexually reproductive organisms that gives rise to genetic diversity via interhomolog recombination between non-sister homologous chromosomes. The identification of a QM6a mating partner has created new opportunities to further understand and improve this biotechnologically important fungus (Seidl et al. 2009).
3 Sexual Crossing of CBS999.97(1-1) with Either CBS999.97(1-2) or QM6a Frequently Produces Segmental Aneuploidy (SAN) Ascospores
Our results (Chuang et al. 2015) have also revealed that sexual crossing of wild type CBS999.97(1-1) with wild type CBS999.97(1-2) or QM6a produced three different types of ascospores: (1) Euploidy ascospores that can germinate to form mycelia and produce green conidia (i.e., asexual spores). CBS999.97(1-1), CBS999.97(1-2) and QM6a are also euploidy and produce green conidia. (2) Viable SAN ascospores that germinate to form mycelia but produce white conidia; their genomes contain a ~523 kb segmental duplication and a ~33 kb segmental deletion. The corresponding segments are referred to as the “D (duplicated)” segment and the “L (loss)” segment, respectively (Fig. 1). Notably, the T. reesei polyketide synthase 4 gene (tpks4) is located within the L segment. The PKS4 protein products are responsible for the green conidial pigmentation in the wild type strains (Atanasova et al. 2013). We also found that duplication of the D segment results in an increase in xylanase (but not cellulase) production in these viable SAN progeny. Intriguingly, there is a carbohydrate-active enzyme (CAZyme) gene cluster in the D segment. This CAZyme gene cluster harbors a set of hemicellulases or xylanases (abf1, bga1, cip2, cel74a and xyn3) (Hakkinen et al. 2014). (3) Non-viable SAN ascospores, which are unable to germinate. Their genomes likely contain two L segments but lack the D segment, which causes meiotic-drive ascospore lethality. The D segments in the genomes of QM6a and CBS999.97(1-2) consist of the 5′ terminus of scaffold 36 (15 genes, ~52 kb), scaffold 27 (130 genes and ~431 kb) and the 3′ terminus of scaffold 28 (12 genes, ~40 kb). The D segment contains at least 113 annotated genes, including several putative essential genes such as an actin-like protein (protein ID 111468; http://genome.jgi-psf.org/Trire2).
These three types of ascospores are differentially distributed in three types of asci generated by sexual crossing of CBS999.97(1-1) with CBS999.97(1-1) or QM6a. The type I asci (~10 %) have 16 viable euploidy ascospores. The type II asci (~80 %) have eight viable euploidy ascospores, four viable SAN ascospores and four non-viable SAN ascospores. Finally, the type III asci (~10 %) have eight viable SAN ascospores, eight non-viable SAN ascospores, but they lack euploidy ascospore.
4 Identification of a Novel Chromosome Arrangement in CBS999.97(1-1)
Investigation of the genomes of QM6a and CBS999.97(1-2) revealed that the three scaffolds (36, 27 and 28) are contiguous segments and together form a larger scaffold “M”. Scaffold M is divided into the D segment and the “S” segment. Unlike the D segment, the S segment (~452 kb) is not duplicated in the viable SAN ascospores. The L segment is located at the 5′ portion of scaffold 33, whereas the 3′ portion (~173 kb) of scaffold 33 is referred to as the “N” segment (Fig. 1).
Notably, compared with the genomes of CBS999.97(1-2) and QM6a(1-2), a large chromosomal translocation or reciprocal exchange (re) was found at this region of the CBS999.97(1-1) genome. (1) The L segment connects with the S segment to form a new scaffold “X” in CBS999.97(1-1), and (2) the N segment physically links to the D segment, thereby forming another new scaffold “F” in CBS999.97(1-1) (Fig. 1). Accordingly, the two CBS999.97 haploid strains and QM6a are hereafter referred to as CBS999.97(1-1, re), CBS999.97(1-2, wt [wild type]) and QM6a(1-2, wt).
5 The Ancestral T. reesei Genome Contains Scaffold M and Scaffold 33
The sexually competent CBS999.97 diploid strain was isolated from a storage lake in French Guiana (Lieckfeldt et al. 2000). It was used to produce the CBS999.97(1-1, re) haploid strain and the CBS999.97(1-2, wt) haploid strain via sexual development (Seidl et al. 2009). QM6a(1-2, wt) was isolated on the Solomon Islands during the Second World War (Mandels and Reese 1957). Several other non-CBS999.97 haploid strains were isolated from different geographical locations (Seidl et al. 2009). Using PCR and classic genetic approaches, we have shown that scaffold M and scaffold 33, but not scaffold X and scaffold F, were present in all nine non-CBS999.97 isolates examined (Chuang et al. 2015). These results indicate that the ancestral T. reesei genome likely contained scaffold M and scaffold 33 and that scaffold F and scaffold X evolved later in French Guiana via a chromosomal translocation between scaffold M and scaffold 33.
6 Chromosome Arrangement Is Responsible for the Production of SAN Ascospores
Our findings can be explained by classical genetics. The type I asci are “parental ditype” (PD). Eight ascospores harbor scaffold M and scaffold 33, whereas the other eight ascospores harbor scaffold F and scaffold X. The type II asci and the type III asci are “tetratype” (TT) and “non-parental ditype” (NPD), respectively. When meiotic interhomolog recombination (or crossover) fails to occur at the translocational breakpoints between these four scaffolds (M, 33, X and F), PD and NPD can be produced simply by random chromosome segregation during MI. In contrast, TT is likely generated via a single crossover between scaffold M and scaffold X or between scaffold 33 and scaffold F. NPD can also arise from two crossovers between two of these four scaffolds. As our single ascospore isolation experiments revealed more type II “TT” asci (>80 %) than type I “PD” asci (~10 %) and type III “NPD” asci (~10 %), we suggest that meiotic interhomolog recombination occurs at a high frequency between scaffold M and scaffold X or between scaffold 33 and scaffold F.
This hypothetic model had also been confirmed by sexual crossing experiments using two homozygous haploids. We first isolated two new haploid strains, CBS999.97(1-1, wt) and CBS999.97(1-2, re), from the type I asci generated from sexually crossing the two parental haploids, CBS999.97(1-1, re) and CBS999.97(1-2, wt). Sexually crossing CBS999.97(1-1, re) with CBS999.97(1-2, re), CBS999.97(1-1, wt) with CBS999.97(1-2, wt) or CBS999.97(1-1, wt) with QM6a (1-2, wt) generated asci with 16 viable euploidy. We conclude that genome heterozygosity caused by chromosome rearrangement is responsible for the production of viable and non-viable SAN ascospores (Chuang et al. 2015).
Our results are consistent with the classic chromosome speciation model proposed by Michael J. D. White in 1978, which states that chromosome rearrangement between populations can efficiently cause or contribute to heterozygote infertility (White 1978). Similarly, sexually crossing of two Saccharomyces strains (i.e., S. cerevisiae vs. S. mikatae or S. paradoxus) (Delneri et al. 2003) or two Schizosaccharomyces strains (i.e., S. pombe vs. S. kombucha) (Zanders et al. 2014) gave rise to viable hybrid diploids that efficiently completed meiosis, but frequently generated non-viable spores. Chromosomal rearrangements (inversions) also contribute to reproduction isolation in two occasionally hybridizing North American Drosophila species, D. pseudoobscura and D. persimilis (Noor et al. 2001). Cryptococcus neoformans (a human fungal pathogen) meiosis generates SAN progeny with a large segmental duplication via telomere-telomere fusion and chromosomal translocation between two different chromosomes (Fraser et al. 2005; Ni et al. 2013). In these scenarios, chromosomal rearrangement and related recombination defects are the major cause of non-viable gametes. Intriguingly, it has been reported that two spore killer elements (Sk-2 and Sk-3) are located near a chromosome rearrangement site in Neurospora crassa (Harvey et al. 2014). It remains unclear whether chromosome rearrangement can be a cause of meiotic drive. The molecular mechanism of spore killer in Neurospora crassa remains a mystery. Based on our T. reesei sexual crossing experiments described above, we hypothesized that most (if not all) of the non-viable gametes are SAN and that their infertility defects are likely caused by a loss of genes required for either fungal ascospore germination or for the formation of normal zygotes (e.g., Drosophila).
7 Mutagenesis Programs for Strain Improvement Resulted in Speciation
Genome sequencing and aCGH analyses have indicated that the genomes of RUT-C30 and QM9414 acquired multiple chromosomal changes (including translocations, deletions and nucleotide mutations) after multiple rounds of random mutagenesis. We found that RUT-C30 and QM9419 could mate with either CBS999.97(1-1, re) or CBS999.97(1-1, wt) and subsequently efficiently formed fruiting bodies and completed meiosis. However, the majority of asci have either no (70–80 %) or only four (20–30 %) viable ascospores (Fig. 2). These results indicate that multiple rounds of random mutagenesis apparently generated genetic barriers against the CBS999.97 wild isolate. Therefore, we hypothesize that high levels of ascospore lethality is due to hybrid infertility.
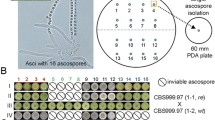
Hexadecad dissection. CBS999.97(1-1, re) (a, b) and CBS999.97(1-1, wt) (c, d) were sexually crossed with RUT-C30 (a, c) or QM9414 (b, d). Sixteen ascospores from a hexadecad ascus were sequentially separated applying the yeast tetrad dissection method and then grown on individual 100-mm malt-extract agar plates. A single colony from one ascospore was isolated and transferred individually to a 60 mm potato dextrose agar plate to determine the spore viability, spore color and colony morphology. Sixteen single-ascospore colonies were aligned sequentially according to the ascospore order. The non-viable ascospores are indicated by a black circle
We noted a higher number of asci with 8 or 12 viable ascospores when CBS999.97(1-1, wt), but not CBS999.97(1-1, re), was used to mate with RUT-C30 or QM9414. In all cases, we found no asci containing 16 viable ascospores (Fig. 2). Compared with CBS999.97(1-1, wt), CBS999.97(1-1, re) has an additional chromosomal rearrangement between scaffold M and scaffold 33. It seems that an extra chromosome rearrangement in CBS999.97(1-1, re), compared with CBS999.97(1-1, wt), resulted in more severe hybrid infertility phenotypes. However, chromosome arrangement may not be the only cause of hybrid infertility. Other chromosomal alternations may also result in meiotic-drive lethality, e.g., deletions or nucleotide mutations in genes that specifically affect meiotic recombination (Hunter et al. 1996) or ascospore germination (Lambou et al. 2008; Strich et al. 2011).
8 Conclusion
Our results suggest that speciation was artificially promoted via extensive random mutagenesis and selection regimes during strain improvements. RUT-C30 and QM9414, the two widely used industrial stains, are no longer the same species as QM6a and the CBS999.97 wild isolate. This supposition is consistent with the findings of comparative genome sequencing and aCGH experiments, which highlight many chromosomal alternations (i.e., translocations, deletions and nucleotide mutations) in these fungal genomes. According to the classic chromosome speciation model (White 1978), chromosome rearrangement and related meiotic-drive recombination can provide a simple and rapid method to create genetic barriers. Because of their economic importance in the production of cellulolytic enzymes and recombinant proteins, these two industrial strains and related hypersecreting mutants have been extensively studied using genetic, molecular, biochemical, physiological, transcriptomic and proteomic approaches. Therefore, we suggest that they are ideal models to investigate the evolutionary process by which new biological species arise.
References
Atanasova L, Knox BP, Kubicek CP, Druzhinina IS, Baker SE. The polyketide synthase gene pks4 of Trichoderma reesei provides pigmentation and stress resistance. Eukaryot Cell. 2013;12:1499–508.
Carter GL, Allison D, Rey MW, Dunn-Coleman NS. Chromosomal and genetic analysis of the electrophoretic karyotype of Trichoderma reesei: mapping of the cellulase and xylanase genes. Mol Microbiol. 1992;6:2167–74.
Chuang YC, Li WC, Chen CL, Hsu PWC, Tung SY, Kuo HC, et al. Trichoderma reesei meiosis generates segmentally aneuploid progeny with higher xylanase-producing capability. Biotechnol Biofuels. 2015;8:30.
Delneri D, Colson I, Grammenoudi S, Roberts IN, Louis EJ, Oliver SG. Engineering evolution to study speciation in yeasts. Nature. 2003;422:68–72.
Fraser JA, Huang JC, Pukkila-Worley R, Alspaugh JA, Mitchell TG, Heitman J. Chromosomal translocation and segmental duplication in Cryptococcus neoformans. Eukaryot Cell. 2005;4:401–6.
Hakkinen M, Valkonen MJ, Westerholm-Parvinen A, Aro N, Arvas M, Vitikainen M, et al. Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol Biofuels. 2014;7:14.
Harvey AM, Rehard DG, Groskreutz KM, Kuntz DR, Sharp KJ, Shiu PK, et al. A critical component of meiotic drive in Neurospora is located near a chromosome rearrangement. Genetics. 2014;197:1165–74.
Hunter N, Chambers SR, Louis EJ, Borts RH. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 1996;15:1726–33.
Ilmen M, Onnela ML, Klemsdal S, Keranen S, Penttila M. Functional analysis of the cellobiohydrolase I promoter of the filamentous fungus Trichoderma reesei. Mol Gen Genet. 1996;253:303–14.
Koike H, Aerts A, LaButti K, Grigoriev IV, Baker SE. Comparative genomics analysis of Trichoderma reesei strains. Ind Biotechnol. 2013;9:352–67.
Lambou K, Malagnac F, Barbisan C, Tharreau D, Lebrun MH, Silar P. The crucial role of the Pls1 tetraspanin during ascospore germination in Podospora anserina provides an example of the convergent evolution of morphogenetic processes in fungal plant pathogens and saprobes. Eukaryot Cell. 2008;7:1809–18.
Le Crom S, Schackwitz W, Pennacchio L, Magnuson JK, Culley DE, Collett JR, et al. Tracking the roots of cellulase hyperproduction by the fungus Trichoderma reesei using massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:16151–6.
Lieckfeldt E, Kullnig C, Samuels GJ, Kubicek CP. Sexually competent, sucrose- and nitrate-assimilating strains of Hypocrea jecorina (Trichoderma reesei) from South American soils. Mycologia. 2000;92:374–80.
Mandels M, Reese ET. Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. J Bacteriol. 1957;73:269–78.
Mantyla AL, Rossi KH, Vanhanen SA, Penttila ME, Suominen PL, Nevalainen KM. Electrophoretic karyotyping of wild-type and mutant Trichoderma longibrachiatum (reesei) strains. Curr Genet. 1992;21:471–7.
Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat Biotechnol. 2008;26:553–60.
Mukherjee PK, Horwitz BA, Herrera-Estrella A, Schmoll M, Kenerley CM. Trichoderma research in the genome era. Annu Rev Phytopathol. 2013;51:105–29.
Ni M, Feretzaki M, Li W, Floyd-Averette A, Mieczkowski P, Dietrich FS, et al. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol. 2013;11:e1001653.
Noor MA, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci U S A. 2001;98:12084–8.
Peterson R, Nevalainen H. Trichoderma reesei RUT-C30—thirty years of strain improvement. Microbiology. 2012;158:58–68.
Seidl V, Gamauf C, Druzhinina IS, Seiboth B, Hartl L, Kubicek CP. The Hypocrea jecorina (Trichoderma reesei) hypercellulolytic mutant RUT-C30 lacks a 85 kb (29 gene-encoding) region of the wild-type genome. BMC Genomics. 2008;9:327.
Seidl V, Seibel C, Kubicek CP, Schmoll M. Sexual development in the industrial workhorse Trichoderma reesei. Proc Natl Acad Sci U S A. 2009;106:13909–14.
Strich R, Khakhina S, Mallory MJ. Ume6p is required for germination and early colony development of yeast ascospores. FEMS Yeast Res. 2011;11:104–13.
Vitikainen M, Arvas M, Pakula T, Oja M, Penttila M, Saloheimo M. Array comparative genomic hybridization analysis of Trichoderma reesei strains with enhanced cellulase production properties. BMC Genomics. 2010;11:441.
White MJD. Modes of speciation, a series of books in biology. San Francisco: W.H. Freeman; 1978. p. 455.
Zanders SE, Eickbush MT, Yu JS, Kang JW, Fowler KR, Smith GR, et al. Genome rearrangements and pervasive meiotic drive cause hybrid infertility in fission yeast. Elife. 2014;3:e02630.
Acknowledgements
This work was supported by Academia Sinica and the Ministry of Science and Technology, Taiwan (ROC) for T.F.W. We thank Monika Schmoll for her generosity in sharing the T. reesei strains.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Li, WC., Chuang, YC., Chen, CL., Wang, TF. (2016). Hybrid Infertility: The Dilemma or Opportunity of Applying Sexual Development to Improve Trichoderma reesei Industrial Strains. In: Schmoll, M., Dattenböck, C. (eds) Gene Expression Systems in Fungi: Advancements and Applications. Fungal Biology. Springer, Cham. https://doi.org/10.1007/978-3-319-27951-0_16
Download citation
DOI: https://doi.org/10.1007/978-3-319-27951-0_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-27949-7
Online ISBN: 978-3-319-27951-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)