Abstract
The effect of paternal aging on fertility, embryo quality, and offspring health is an important area of study that has received far less attention than the age effect in women. This is, in part, due to the fact that in females there are dramatic alterations to fertility and pregnancy outcomes that abruptly occur as a female ages. Such abrupt alterations to pregnancy success and/or embryonic and offspring health are not seen in males. Instead, there are subtle alterations to pregnancy success and offspring phenotypes that occur as a man ages. It is believed that, at least in part, these alterations can be explained by perturbations to the sperm epigenome that occur over time. This chapter will explore the effect of aging on the sperm epigenome and the potential impacts these perturbations may have on embryonic development and ultimately offspring health.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
4.1 Introduction
The human sperm is a highly specialized cell, elegantly equipped with the minimum necessary to deliver a haploid genome through the female reproductive tract to the oocyte. Upon fusion with the oolemma, the sperm deposits not only half of the genetic material into the oocyte but also initiates signal transduction cascades responsible for completion of meiosis in the egg and the initiation of embryogenesis. The role of the sperm in delivering DNA and activating the oocyte has long been appreciated. In addition, a growing body of data indicates that the epigenetic, as well as the genetic, landscape of the sperm has direct effects on embryogenesis and offspring phenotypes and that paternal epigenetic contributions can, in some cases, confer transgenerational effects (Milekic et al. 2014; Govorko et al. 2012; Carone et al. 2010; Hammoud et al. 2009).
A variety of natural and extraneous influences can impact the sperm epigenome with potential downstream consequences (Guerrero-Bosagna et al. 2012; Hare and Moran 1979; Hemminki et al. 1999; Marczylo et al. 2012). This chapter will focus on the effects of male age on sperm epigenetics. Age has been shown to consistently and predictably affect the epigenetic profiles of numerous cell types (Richardson 2003; Christensen et al. 2009; Day et al. 2013). Remarkably, the age-induced epigenetic changes observed in sperm appear to be greater in magnitude and often more consistent than changes reported in other cell types (Jenkins et al. 2013). While much remains to be learned about the epigenetic contributions of the sperm to the early embryo, a growing body of evidence suggests that some alterations in the sperm epigenome escape the early waves of epigenetic reprogramming. These changes may explain some of the increased risks of certain diseases that are observed more frequently in the offspring of older fathers (Hemminki et al. 1999; Frans et al. 2008, 2013).
4.2 The Sperm Epigenome
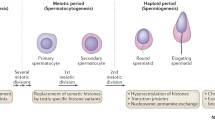
The sperm is morphologically and functionally distinct from any other cell type. Perhaps the greatest distinction between the sperm cell and other cell types is the nuclear structure. While the DNA of somatic cells is packaged around histones, the majority of sperm histones are displaced in a two-step process during spermiogenesis, first by transition proteins, which are subsequently replaced by protamines 1 and 2 (P1 and P2) to form a tight toroidal structure that compresses the nucleus 6–20 times tighter than the somatic cell nucleus (Fig. 4.1) (Balhorn 2007; Ward and Coffey 1991). In normal fertile men, the ratio of P1:P2 is approximately 1:1. Importantly, infertile men often display an altered P1:P2 ratio, and deviations from the normal ratio are associated with abnormal semen parameters, increased DNA damage, and reduced fertilization and implantation rates (Aoki et al. 2005, 2006).
Illustration of epigenetic structure in the mature sperm and the dramatic organization that occurs in the early embryo immediately following fertilization. The top panel shows the protamine-bound mature sperm, undergoing chromatin decondensation marked by the removal of protamine proteins. The bottom panel shows the active demethylation that occurs in the paternal pronucleus, as well as the passive, replication-dependent demethylation that occurs in the maternal pronucleus
These observations were the first to suggest that the epigenetic status of sperm might be important for early development. While the mature sperm nucleus is comprised primarily of protamine-bound DNA, about 5 % of the DNA remains bound to histones (Hammoud et al. 2009). Until recently, it was unclear whether the persistent histones were the result of incomplete histone replacement or whether they served a functional purpose. Several years ago, our lab demonstrated that histones are consistently retained at specific loci including developmental gene promoters, genes encoding microRNAs, and imprinted loci. In addition, it was found that the retained histones often display bivalency, the presence of both activating and silencing modifications within the same region, which is reminiscent of stem cell signatures (Hammoud et al. 2009). These findings suggest that the epigenetic status of sperm is tightly regulated and likely mechanistically important for embryogenesis and early development. Following fertilization, the sperm nucleus undergoes decondensation and pronuclear development, and the protamines are replaced by oocyte-derived histones. During this process, the majority of DNA methylation marks are removed to restore totipotency to the sperm and oocyte genomes (Fig. 4.1), which clearly raises questions regarding the importance of pre-fertilization epigenetic marks; however, two important considerations are warranted. First, the identity of unmodified loci remains uncertain, raising the possibility that key sperm loci remain unchanged and functionally important during embryogenesis. Second, data suggest that epigenetic abnormalities in male gametes may affect embryo development, and there is evidence to suggest that these abnormalities can affect offspring phenotype. Even less data are available on the impacts of age-associated sperm epigenetic alterations and their impacts on the embryo. Despite this, there are many indications that age-associated epigenetic alterations may play a role in both embryogenesis and offspring health.
4.3 Delayed Parenthood
Advanced paternal age has recently become a heavily investigated topic as a result of multiple studies demonstrating ties between advanced paternal age and various offspring abnormalities. Additional trends contributing to the increasing interest in the role of advanced paternal age in reproduction is the trend in delayed parentage (Mills et al. 2011). Though this trend is justified by increasing life expectancies in both sexes, advanced paternal age may affect general semen parameters and sperm quality ultimately altering fecundity and offspring health. While many couples consider the risks associated with advanced maternal age in family planning decisions, very little thought has been given to the age of male partners. In recent history, paternal age has steadily increased, particularly in developed countries. This trend is believed to be associated with increased life expectancy, socioeconomic pressures, and divorce rates with subsequent remarriage at older ages (Kuhnert and Nieschlag 2004). During a 10-year span (1993–2003) in Great Britain, the percent of fathers within the age range of 35–54 increased from 25 % of total births to 40 %. Associated with this trend was a decrease in the number of births to fathers less than 35 years of age from 74 % of total births to only 60 % (Bray et al. 2006). Over two decades in Australia (1988–2008), the average age of fathers has increased by approximately 3 years (Australian Bureau of Statistics 2009). Similarly, the average age of fathers in Germany increased by 2 years over a 10-year period (Kuhnert and Nieschlag 2004). Congruent trends can be found in the United States and many other developed countries. As average paternal age continues to increase, it is becoming increasingly important to characterize the potential consequences of advanced paternal age on embryonic development and offspring health.
4.4 Heritability of Epigenetic Alteration Through the Paternal Germline
Though poorly understood, there is clear evidence that demonstrates a unique mechanism of heritability through the paternal germline. This idea initially became of great interest to many different scientific fields as a result of findings from growing catalogs of epidemiologic data coupled with landmark studies in mouse models. Specifically, data collected during and following massive crop failures in Sweden in the late 1800s and early 1900s was used to perform large retrospective studies in human populations. From these studies, it was found that paternal diet, independent of other factors, contributes to offspring disease susceptibility and general health in ways never before identified (Kaati et al. 2007; Pembrey et al. 2006). Though the nature of the data set made it impossible to understand any biological mechanisms that underlie these alterations, many believe it plausible that alterations of heritable epigenetic marks in gametes play some role in the process. In support of this idea is the work on the agouti viable yellow gene in mouse models, which demonstrated that nutrition can affect offspring phenotype through heritable altered epigenetic marks (Waterland and Jirtle 2003). This and other work have stimulated the study of transgenerational inheritance as we see it today.
Many intriguing studies have come as a result of the increased emphasis on transgenerational inheritance in the literature. One recent study found that male mice fed a low-protein diet, when mated with a normal female, sire offspring with altered expression of many genes important in metabolism and cholesterol synthesis (Carone et al. 2010). Similarly, metabolic alterations, specifically changes in insulin sensitivity, were also seen in the female offspring of male rats fed high-fat diets (Ng et al. 2010).
Although no concrete mechanism for inheritance of altered metabolic activity has been identified or any other nongenetic inheritance from the male germline, there are intriguing candidates including epigenetic inheritance through altered sperm DNA methylation. A recent study strongly supports the idea of transgenerational inheritance. Govorko et al. demonstrated that the offspring of male mice exposed in utero to alcohol had altered hypothalamic proopiomelanocortin (POMC) gene activity as a result of hypermethylation at the POMC promoter and that these deficits were passed down through the F3 generation (Govorko et al. 2012). Interestingly, although the methylation marks at the POMC promoter were similar in the F1 female and male (both exposed to prenatal alcohol), the alterations were not inherited via the maternal germline, suggesting a unique mechanism of epigenetic inheritance through the paternal germline (Govorko et al. 2012). Taken together, these data demonstrate the likelihood that the sperm epigenome plays an essential role in embryogenesis and is capable of contributing to offspring health.
4.5 Age-Associated Sperm Epigenetic Alterations
An important consideration in the role of paternal aging on embryo quality and offspring health is the degree to which the sperm is susceptible to genomic or epigenomic perturbation that could lead to embryonic or offspring dysfunction and disease. Because of the plastic nature of epigenetic marks in the sperm and the potential heritability of any perturbations, sperm epigenetics, in particular DNA methylation, has become one of the main candidates on which studies have focused. Only recently has data become available to describe the epigenetic landscape of the aged sperm, and these have focused primarily on DNA methylation in both human and mouse models. It is informative to describe this in context of somatic cell alteration associated with age where it is known that DNA methylation is altered in many somatic cell types with age in relatively consistent patterns (Wilson and Jones 1983; Oakes et al. 2003). From the few studies that have been performed, it appears that sperm methylation patterns resultant from aging are far different and of greater magnitude than what is seen in somatic cells (Jenkins et al. 2013, 2014). In fact, these cells display a virtually opposite profile of epigenetic change with age (Fig. 4.2). Although this may appear counterintuitive, it is important to note that other genomic alterations, such as telomere length, follow similar trends between these two tissue types (Eisenberg 2011). Furthermore, the idea that the magnitude of methylation alteration is greater in sperm as compared to somatic cells with aging is not without precedence. Work in support of this idea demonstrates that frequently dividing cells have more striking methylation changes associated with age than do cells that divide less frequently. As sperm undergo large amounts of division over the lifespan of an individual, it is not surprising that the magnitude of epigenetic change is greater in sperm over time than in other human tissues.
General tissue-specific age-associated alterations that occur in sperm and other somatic tissues. Sperm tend to have slight increases in global methylation with age, while regionally there is a bias toward methylation loss. In somatic cells the opposite is true, as global methylation decreases and regional methylation increases with age
Two recent studies on human sperm from anonymous donors have revealed distinct patterns of methylation alteration with age (Jenkins et al. 2013, 2014). These studies utilized sperm donors who collected two samples many years apart (between 10 and 20 years approximately). This allowed the authors to analyze paired data to determine the intraindividual impact of aging on the sperm methylome. It was discovered that there is an increase in the global level of methylation in human sperm, a surprising finding based on the baseline hypermethylation in the mature sperm and the contrasting global hypomethylation that occurs with age in somatic cells (Jenkins et al. 2013, 2014). A number of regional alterations (approximately 1,000 bps in length) were also significantly altered with age and displayed a strong bias toward demethylation. This finding is, again, in opposition to what has been described in somatic cells where there is a bias toward regional hypermethylation (Jenkins et al. 2014). Alterations at these sites were confirmed with the use of targeted bisulfite sequencing in an independent cohort of unpaired general population sperm samples. These findings were remarkably consistent at the identified regions of alteration (Fig. 4.3). Intriguingly, it appeared that the age-associated regional alterations identified were enriched at genes known to be associated with neuropsychiatric disease. Similar results were identified in mice where regional hypomethylation was common in the sperm of aged mice though no global changes were identified (Milekic et al. 2014). Interestingly, all offspring of older males had similar alterations to methylation patterns in brain tissue coupled with alterations in social behaviors. Taken together, age-associated methylation perturbations represent a plausible mechanism by which the increased incidence of disease in the offspring of older fathers may be transmitted.
An example of sperm-specific regional methylation. At this relatively small genomic window (approximate 250 bps), there is a significant decrease in fraction methylation (y axis) at each CpG (x axis) that occurs within men over 50 (n = 9) when compared to men under 40 (n = 12). These data represent an example of one of many loci significantly affected by age in (Jenkins et al. 2014)
4.6 Embryo Quality, Pregnancy Outcomes, and Offspring Health
The effects of paternal age on pregnancy outcome and embryo quality are controversial. This controversy is mainly a result of the scant data available on the subject. Some reports suggest that there is a significant decline in fertility (as measured by time to pregnancy) with age, while others report no such associations (Hassan and Killick 2003; Begueria et al. 2014). Additional data does suggest that paternal age is a significant factor when compounded with maternal age (de la Rochebrochard and Thonneau 2002). Other studies support these data by suggesting an increased frequency of fetal loss, increased time to pregnancy, and decreased probability of conception in older men (Selvin and Garfinkel 1976; Ford et al. 2000; Dunson et al. 2002). However, there are conflicting data which suggest little to no effect of paternal age on fertility in natural conception or with the use of assisted reproductive technologies (ART) (Begueria et al. 2014; Olsen 1990; Bellver et al. 2008). Similar controversy exists in the effect of paternal age on embryo quality with the use of ART with some studies showing no effect (Bellver et al. 2008; Ferreira et al. 2010) and some suggesting decreased quality of embryos sired by older fathers on day 3, 4, and 5 (Luna et al. 2009; Frattarelli et al. 2008). The most compelling indication that paternal age may affect embryo quality is data on miscarriage. In general, the consensus from the available data is that advanced paternal age is a risk factor for miscarriage though no real mechanisms for this finding have been elucidated (Kleinhaus et al. 2006; Slama et al. 2005). Other studies evaluating ART with donor eggs (to completely remove the influence of maternal factors) found no associations between paternal age and risk of miscarriage (Begueria et al. 2014). Taken together, much work remains to determine what, if any, effect advanced paternal age has on male fertility and embryo quality.
The subtlety of the effect of age on male fertility, and particularly pregnancy outcomes, is in striking contrast to the dramatic decline seen in female fertility. In fact, even men of advanced age are able to sire offspring with little difficulty, though possibly with slightly reduced efficiency which is why paternal age has largely been ignored and has received far less attention in the clinical setting than the age of the female partner. The fact that males are still fertile at advanced ages may present, and potentially complicate, another issue that, while subtle, is far more consistent, namely, the effect of paternal age on offspring health and disease susceptibility. It has been shown that the offspring of older fathers have increased incidence of various forms of cancer, including hematological and central nervous system tumors (Hemminki et al. 1999; Oksuzyan et al. 2012; Murray et al. 2002; Yip et al. 2006), though the data remains somewhat controversial. Furthermore, it has long been suggested that advanced paternal age is a risk factor for schizophrenia (Hare and Moran 1979; Miller et al. 2011; Matheson et al. 2011; Wohl and Gorwood 2007). More recently, it has been suggested that advanced paternal age is significantly associated with many forms of neuropsychiatric or neurocognitive diseases including autism spectrum disorders (ASD) (Gardener et al. 2009; Hultman et al. 2011), bipolar disorder (Frans et al. 2008; Menezes et al. 2010), and general increases in behavioral issues (Kuja-Halkola et al. 2012; Saha et al. 2009a) in children of older fathers though some controversy exists. In addition, some studies indicate that children of older fathers display slightly reduced IQ compared with children of younger fathers (Malaspina et al. 2005; Saha et al. 2009b), although the differences are small, and conflicting reports exist (Svensson et al. 2011).
Taken together, it is clear that advanced paternal age does not have a dramatic affect on pregnancy outcomes, embryo quality, or fertility in general, but it may impact offspring health and disease susceptibility. While the lack of striking age-associated fertility declines in males has garnered it little attention in the study of fertility, it is this same maintenance of fertility that might require more study in the field of transgenerational inheritance. Age-associated alterations to sperm, which appear to affect offspring health, do not seem to be catastrophic to spermatogenesis or cause declines in fertility. This, in turn, means that aged sperm are entirely competent to fertilize an oocyte and produce viable offspring, while harboring alterations that may potentially affect offspring health.
4.7 Future Directions
To gain a more complete understanding of the epigenetic alterations in sperm that are capable of embryonic or offspring phenotype alterations, much work is still needed. A number of genomic regions have been identified that have both methylation alterations with age and are important in various cell processes and diseases known to have increased occurrence in the offspring of older males. To determine if these marks can contribute to disease susceptibility in the offspring or affect events in the embryo, a number of unanswered questions must be addressed.
What is the impact of altered methylation profiles at our regions of interest? To completely understand the alterations which have been identified and their impact on offspring phenotype or embryo development, it must be determined if these alterations are associated with transcriptional changes. Future work can target genomic sites that are known to be altered with age in mouse models to determine if (1) there are changes to transcription in the embryo and (2) determine if there are altered transcript levels in various tissues in the offspring (should the sperm be competent to generate viable offspring).
Do the altered methylation marks seen in sperm escape, or impact in any way, embryonic epigenetic reprogramming? This is an essential question to fully understand the impact of an altered epigenetic profile. It is feasible that an alteration could affect embryogenesis in one of two ways. First, it could directly affect transcription of an important developmental factor. Second, the epigenetic abnormality may result in not a targeted perturbation but in the global alteration of epigenetic reprogramming, effectively altering an important aspect of embryogenesis, likely to the point of embryonic arrest.
Do methylation perturbations contribute to neuropsychiatric disorders in the offspring or perturbations to embryogenesis? To date, there are many intriguing studies that have provided some small degree of insight into the effect of aging on the sperm epigenome. However, much of the potential impacts are simply extrapolation of the available data without any real targeted studies to prove causative relationships. While the data is exciting, future targeted work is still required to enable us to reach these further conclusions.
4.8 Conclusions
The role of the paternal epigenome in embryogenesis should not be downplayed. It appears from a growing body of evidence that the sperm epigenetic landscape is essential in facilitating gene poising and general transcription regulation at genes important in embryonic development (Hammoud et al. 2009). However, with our current understanding, we are unable to definitively determine that sperm epigenetic alterations associated with age are causative of any poor pregnancy outcomes or decreased embryo quality declines. In fact, the aged male remains remarkably fertile with, at most, only modest declines in fecundity. When we consider this fact coupled with the data regarding known and consistent age-associated alterations to the paternal epigenome, it is easy to contemplate the implications of these alterations beyond embryogenesis. Specifically, a great deal of focus has now been given to the increased incidence of diseases seen in the offspring of older fathers and the transgenerational impacts that they impose. This is of particular concern in developed countries where the age of paternity is steadily increasing. While the questions regarding paternal age and epigenetic alterations that may affect embryogenesis are essential and must be addressed further, the impact of these alterations on the offspring appears to be a more relevant question due to the fact that the alterations identified with aging do not appear to affect (at least in a great degree) the competency of sperm to yield viable offspring.
Many important questions must still be addressed in regard to the epigenetic findings associated with advanced paternal age. While we know that there are real alterations that occur with remarkable consistency, the impact of these alterations is unknown. Mouse data suggesting similar methylation patterns in the brain of offspring sired by older fathers is intriguing (particularly when coupled with the identified behavioral abnormalities), but this also requires a great degree of further study. We currently have a great deal of genomic targets that are known to be altered in the sperm of men with advanced age, and these can be used to analyze potential implications in the embryo and the offspring. Taken together, while having learned much about the impacts of advanced age in the recent past, there is still a great deal of work that needs to be performed to truly elucidate the impact of age-associated sperm epigenetic alterations on the embryo and beyond.
References
Aoki VW et al (2005) DNA integrity is compromised in protamine-deficient human sperm. J Androl 26:741–748. doi:10.2164/jandrol.05063
Aoki VW, Emery BR, Liu L, Carrell DT (2006) Protamine levels vary between individual sperm cells of infertile human males and correlate with viability and DNA integrity. J Androl 27:890–898. doi:10.2164/jandrol.106.000703
Balhorn R (2007) The protamine family of sperm nuclear proteins. Genome Biol 8:227. doi:10.1186/gb-2007-8-9-227, gb-2007-8-9-227 [pii]
Begueria R et al (2014) Paternal age and assisted reproductive outcomes in ICSI donor oocytes: is there an effect of older fathers? Hum Reprod 29:2114–2122. doi:10.1093/humrep/deu189
Bellver J, Garrido N, Remohi J, Pellicer A, Meseguer M (2008) Influence of paternal age on assisted reproduction outcome. Reprod Biomed Online 17:595–604
Australian Bureau of Statistics (2009) Births, Australia (2008) Summary of findings. Births
Bray I, Gunnell D, Davey Smith G (2006) Advanced paternal age: how old is too old? J Epidemiol Community Health 60:851–853. doi:10.1136/jech.2005.045179, 60/10/851 [pii]
Carone BR et al (2010) Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143:1084–1096. doi:10.1016/j.cell.2010.12.008
Christensen BC et al (2009) Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 5, e1000602. doi:10.1371/journal.pgen.1000602
Day K et al (2013) Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome Biol 14:R102. doi:10.1186/gb-2013-14-9-r102
de la Rochebrochard E, Thonneau P (2002) Paternal age and maternal age are risk factors for miscarriage; results of a multicentre European study. Hum Reprod 17:1649–1656
Dunson DB, Colombo B, Baird DD (2002) Changes with age in the level and duration of fertility in the menstrual cycle. Hum Reprod 17:1399–1403
Eisenberg DT (2011) An evolutionary review of human telomere biology: the thrifty telomere hypothesis and notes on potential adaptive paternal effects. Am J Hum Biol 23:149–167. doi:10.1002/ajhb.21127
Ferreira RC et al (2010) Negative influence of paternal age on clinical intracytoplasmic sperm injection cycle outcomes in oligozoospermic patients. Fertil Steril 93:1870–1874. doi:10.1016/j.fertnstert.2008.12.043
Ford WC et al (2000) Increasing paternal age is associated with delayed conception in a large population of fertile couples: evidence for declining fecundity in older men. The ALSPAC study team (Avon longitudinal study of pregnancy and childhood). Hum Reprod 15:1703–1708
Frans EM et al (2008) Advancing paternal age and bipolar disorder. Arch Gen Psychiatry 65:1034–1040. doi:10.1001/archpsyc.65.9.1034
Frans EM et al (2013) Autism risk across generations: a population-based study of advancing grandpaternal and paternal age. JAMA Psychiatry 70:516–521. doi:10.1001/jamapsychiatry.2013.1180
Frattarelli JL, Miller KA, Miller BT, Elkind-Hirsch K, Scott RT Jr (2008) Male age negatively impacts embryo development and reproductive outcome in donor oocyte assisted reproductive technology cycles. Fertil Steril 90:97–103. doi:10.1016/j.fertnstert.2007.06.009
Gardener H, Spiegelman D, Buka SL (2009) Prenatal risk factors for autism: comprehensive meta-analysis. Br J Psychiatry 195:7–14. doi:10.1192/bjp.bp.108.051672
Govorko D, Bekdash RA, Zhang C, Sarkar DK (2012) Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biol Psychiatry 72:378–388. doi:10.1016/j.biopsych.2012.04.006
Guerrero-Bosagna C et al (2012) Epigenetic transgenerational inheritance of vinclozolin induced mouse adult onset disease and associated sperm epigenome biomarkers. Reprod Toxicol 34:694–707. doi:10.1016/j.reprotox.2012.09.005
Hammoud SS et al (2009) Distinctive chromatin in human sperm packages genes for embryo development. Nature 460:473–478. doi:10.1038/nature08162, nature08162 [pii]
Hare EH, Moran PA (1979) Raised parental age in psychiatric patients: evidence for the constitutional hypothesis. Br J Psychiatry 134:169–177
Hassan MA, Killick SR (2003) Effect of male age on fertility: evidence for the decline in male fertility with increasing age. Fertil Steril 79(Suppl 3):1520–1527, S0015028203003662 [pii]
Hemminki K, Kyyronen P, Vaittinen P (1999) Parental age as a risk factor of childhood leukemia and brain cancer in offspring. Epidemiology 10:271–275
Hultman CM, Sandin S, Levine SZ, Lichtenstein P, Reichenberg A (2011) Advancing paternal age and risk of autism: new evidence from a population-based study and a meta-analysis of epidemiological studies. Mol Psychiatry 16:1203–1212. doi:10.1038/mp.2010.121
Jenkins TG, Aston KI, Cairns BR, Carrell DT (2013) Paternal aging and associated intraindividual alterations of global sperm 5-methylcytosine and 5-hydroxymethylcytosine levels. Fertil Steril 100:945–951. doi:10.1016/j.fertnstert.2013.05.039
Jenkins TG, Aston KI, Pflueger C, Cairns BR, Carrell DT (2014) Age-associated sperm DNA methylation alterations: possible implications in offspring disease susceptibility. PLoS Genet 10, e1004458. doi:10.1371/journal.pgen.1004458
Kaati G, Bygren LO, Pembrey M, Sjostrom M (2007) Transgenerational response to nutrition, early life circumstances and longevity. Eur J Hum Genet 15:784–790. doi:10.1038/sj.ejhg.5201832
Kleinhaus K et al (2006) Paternal age and spontaneous abortion. Obstet Gynecol 108:369–377. doi:10.1097/01.AOG.0000224606.26514.3a
Kuhnert B, Nieschlag E (2004) Reproductive functions of the ageing male. Hum Reprod Update 10:327–339. doi:10.1093/humupd/dmh030, dmh030 [pii]
Kuja-Halkola R, Pawitan Y, D’Onofrio BM, Langstrom N, Lichtenstein P (2012) Advancing paternal age and offspring violent offending: a sibling-comparison study. Dev Psychopathol 24:739–753. doi:10.1017/S095457941200034X
Luna M et al (2009) Paternal age and assisted reproductive technology outcome in ovum recipients. Fertil Steril 92:1772–1775. doi:10.1016/j.fertnstert.2009.05.036
Malaspina D et al (2005) Paternal age and intelligence: implications for age-related genomic changes in male germ cells. Psychiatr Genet 15:117–125
Marczylo EL, Amoako AA, Konje JC, Gant TW, Marczylo TH (2012) Smoking induces differential miRNA expression in human spermatozoa: a potential transgenerational epigenetic concern? Epigenetics 7:432–439. doi:10.4161/epi.19794
Matheson SL, Shepherd AM, Laurens KR, Carr VJ (2011) A systematic meta-review grading the evidence for non-genetic risk factors and putative antecedents of schizophrenia. Schizophr Res 133:133–142. doi:10.1016/j.schres.2011.09.020
Menezes PR et al (2010) Paternal and maternal ages at conception and risk of bipolar affective disorder in their offspring. Psychol Med 40:477–485. doi:10.1017/S003329170999064X
Milekic MH et al (2014) Age-related sperm DNA methylation changes are transmitted to offspring and associated with abnormal behavior and dysregulated gene expression. Mol Psychiatry. doi:10.1038/mp.2014.84
Miller B et al (2011) Meta-analysis of paternal age and schizophrenia risk in male versus female offspring. Schizophr Bull 37:1039–1047. doi:10.1093/schbul/sbq011
Mills M, Rindfuss RR, McDonald P, te Velde E (2011) Why do people postpone parenthood? Reasons and social policy incentives. Hum Reprod Update 17:848–860. doi:10.1093/humupd/dmr026, dmr026 [pii]
Murray L et al (2002) Association of early life factors and acute lymphoblastic leukaemia in childhood: historical cohort study. Br J Cancer 86:356–361. doi:10.1038/sj.bjc.6600012
Ng SF et al (2010) Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature 467:963–966. doi:10.1038/nature09491, nature09491 [pii]
Oakes CC, Smiraglia DJ, Plass C, Trasler JM, Robaire B (2003) Aging results in hypermethylation of ribosomal DNA in sperm and liver of male rats. Proc Natl Acad Sci USA 100:1775–1780. doi:10.1073/pnas.0437971100, 0437971100 [pii]
Oksuzyan S, Crespi CM, Cockburn M, Mezei G, Kheifets L (2012) Birth weight and other perinatal characteristics and childhood leukemia in California. Cancer Epidemiol 36:e359–e365. doi:10.1016/j.canep.2012.08.002
Olsen J (1990) Subfecundity according to the age of the mother and the father. Dan Med Bull 37:281–282
Pembrey ME et al (2006) Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet 14:159–166. doi:10.1038/sj.ejhg.5201538
Richardson B (2003) Impact of aging on DNA methylation. Ageing Res Rev 2:245–261
Saha S, Barnett AG, Buka SL, McGrath JJ (2009a) Maternal age and paternal age are associated with distinct childhood behavioural outcomes in a general population birth cohort. Schizophr Res 115:130–135. doi:10.1016/j.schres.2009.09.012
Saha S et al (2009b) Advanced paternal age is associated with impaired neurocognitive outcomes during infancy and childhood. PLoS Med 6:e40. doi:10.1371/journal.pmed.1000040
Selvin S, Garfinkel J (1976) Paternal age, maternal age and birth order and the risk of a fetal loss. Hum Biol 48:223–230
Slama R et al (2005) Influence of paternal age on the risk of spontaneous abortion. Am J Epidemiol 161:816–823. doi:10.1093/aje/kwi097
Svensson AC, Abel K, Dalman C, Magnusson C (2011) Implications of advancing paternal age: does it affect offspring school performance? PLoS One 6:e24771. doi:10.1371/journal.pone.0024771
Ward WS, Coffey DS (1991) DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod 44:569–574
Waterland RA, Jirtle RL (2003) Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23:5293–5300
Wilson VL, Jones PA (1983) DNA methylation decreases in aging but not in immortal cells. Science 220:1055–1057
Wohl M, Gorwood P (2007) Paternal ages below or above 35 years old are associated with a different risk of schizophrenia in the offspring. Eur Psychiatry 22:22–26. doi:10.1016/j.eurpsy.2006.08.007
Yip BH, Pawitan Y, Czene K (2006) Parental age and risk of childhood cancers: a population-based cohort study from Sweden. Int J Epidemiol 35:1495–1503. doi:10.1093/ije/dyl177
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Jenkins, T.G., Aston, K.I., Meyer, T., Carrell, D.T. (2015). The Sperm Epigenome, Male Aging, and Potential Effects on the Embryo. In: Bronson, R. (eds) The Male Role in Pregnancy Loss and Embryo Implantation Failure. Advances in Experimental Medicine and Biology, vol 868. Springer, Cham. https://doi.org/10.1007/978-3-319-18881-2_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-18881-2_4
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-18880-5
Online ISBN: 978-3-319-18881-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)