Abstract
Lung cancer is one of the most common malignant tumors and is the leading cause of cancer mortality worldwide. However, drug resistance induced by chemotherapeutants to lung cancer cells is the primary issue during the chemotherapy of lung cancer. Many mechanisms such as the changes of drug metabolism related genes and signal pathways are involved in the chemoresistance. MicroRNAs (miRNAs) are a class of endogenetic, non-coding, short-chain and small RNAs that regulate cell growth, apoptosis and signal transduction. miRNA polymorphisms associate with drug metabolism and drug resistance formation. Furthermore, differentially expressed miRNAs play critical roles in prediction of the sensitivity to chemotherapeutic agents in lung cancer. Regulation of specific miRNA expression will break a new path for overcoming lung cancer resistance and the personalized therapy. Together, in this chapter we have discussed the current understanding of the role of miRNA on drug resistance, and the potential implications of miRNA in lung cancer targeted therapy.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Based on the report in Cancer Statistics 2013 in USA, lung and bronchus cancers in men and in women continue to be the leading common causes of cancer death, although cancer death rates have declined 20 % from their peak in 1991 (215.1 per 100,000 population) to 2009 (173.1 per 100,000 population) [1]. Chemotherapy and targeted therapy are also the mainstream in lung cancer treatment. However, the major problem in lung cancer therapy is the emergence of inherent and acquired drug resistance of the cancer cells [2]. MicroRNAs (miRNAs) are a new class of small, non-coding RNAs that range in size from 19 to 25 nucleotides (nt) and have important roles in a variety of biologic processes [3]. miRNAs are predicted to regulate the expression of up to one-third of human protein-coding genes, and they are involved pathogenesis, diagnosis, treatment and prognosis [4–7]. The accumulating evidences demonstrate that miRNAs regulate drug sensitivity and/or resistance to chemotherapeutic agents [8–11]. Therefore, to explore the role of miRNAs in drug resistance may accelerate novel therapeutic strategies for lung cancer treatment [12].
2 Drug Resistance in Lung Cancer
Chemotherapy is a major treatment modality in both primary and palliative care for lung cancer patients. However, some patients do not respond to such therapy, or they respond well initially and then gradually relapse. This may lead to an increase in the drug dosage, which generally increases the adverse affects, yet fails to improve the clinical prognosis or outcome.
2.1 Current Opinion of MDR in Lung Cancer
Non-small cell lung cancer (NSCLC) cells are often intrinsically resistant to certain anticancer drugs, whereas small-cell lung cancer (SCLC) cells can acquire resistance with continued administration of the drug. Moreover, at the time of diagnosis, the majority of patients with lung cancer most often already have metastatic disease, making it difficult to use other therapeutic options, such as surgery or radiation [13]. Thus, a better understanding of the different mechanisms underlying multiple drug resistance (MDR) is of utmost importance if we are to develop strategies to overcome it. Although numerous mechanisms are associated with MDR in lung cancer, we are a long way from fully understanding how to overcome drug resistance.
2.2 Representative Features of MDR
There are three separate forms of MDR have been characterized in more detail [14]: (a) atypical MDR, (b) classical MDR and (c) non-Pgp MDR. Although all three MDR phenotypes have much in common with respect to cross-resistance patterns, the underlying mechanisms certainly differ. Atypical MDR is associated with quantitative and qualitative alterations in topoisomerase II alpha, a nuclear enzyme that actively participates in the lethal action of cytotoxic drugs [15]. Moreover, atypical MDR cells do not overexpress P-glycoprotein, and are unaltered in their ability to accumulate drugs. Given that classical and non-Pgp MDR is transcriptional activation of membrane-bound transport proteins, these transport proteins belong to the ATP-binding cassette (ABC) superfamily of transport systems. The classical MDR phenotype is characterized by a reduced ability to accumulate drugs, due to activity of an energy-dependent uni-directional, membrane-bound, drug-efflux pump with broad substrate specificity [16]. The classical MDR drug pump is composed of a transmembrane glycoprotein (P-glyco-protein, Pgp) with a molecular weight of 170 kDa, and is, in man, encoded by the so-called multidrug resistance (MDR1) gene. Typically, non-Pgp MDR has no P-glycoprotein expression, yet has about the same cross-resistance pattern as classical MDR. This non-Pgp MDR phenotype is caused by overexpression of the multidrug resistance-associated protein (MRP) gene, which encodes a 190 kDa membrane-bound glycoprotein. MRP probably works by direct extrusion of cytotoxic drugs from the cell and/or by mediating sequestration of the drugs into intracellular compartments, both leading to a reduction in effective intracellular drug concentrations. Together, all these three types of MDR have much similar mechanism in drug resistance induction.
2.3 Molecular Mechanisms of MDR
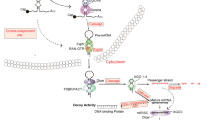
Many molecular mechanisms have been identified, such as (a) drug transporters involved in the efflux of chemotherapeutic drugs, (b) drug inactivation by sulfur-containing macromolecules and role of antioxidants, (c) DNA repair pathways inducing resistance to chemotherapy, (d) modifications or alterations of drug target sites, (e) loss of intracellular death mechanisms, (f) resistance to small molecule inhibitors and (g) alteration of drug metabolism pathway (Fig. 3.1) [17–22]. Additional contributing factors include ineffective drug delivery to the tumor, increased metabolism and therefore a shortened half-life of the drug, lack of drug specificity to the tumor, and tumor vasculature [23, 24]. These factors make it even harder to pinpoint the exact mechanism underlying resistance to a particular drug. It is hoped that identification of new targets and understanding their contribution to lung cancer drug resistance will provide opportunities for innovative therapies in overcoming drug resistance.
The typical molecular cellular mechanisms for drug resistance. Drug resistance is often seen through: ① failure of the drug to enter the cell by loss of the cell surface receptors or transporters; ② alteration of drug target site; ③ change or inactivation of drug; ④ alteration of drug metabolism pathway; ⑤ increased repair of damaged DNA; ⑥ active transport out of the cell
2.4 miRNA Analysis – A Novel Screening Tool in Lung Cancer Chemotherapy
There is a need for the development of new tools to screen patients prior to beginning chemotherapy. More recently a tool known as lung metagene score was developed in an attempt to individualize treatment for lung cancer patients [25]. The lung metagene score (formerly known as the lung metagene predictor) is a screening tool developed to estimate the risk of recurrence in early stage NSCLC [26, 27]. By comparing microarray data of untreated and drug-treated NSCLC cells, Heller et al. identify 33 miRNAs whose expression is upregulated after drug treatment and which are associated with a CpG island [28]. Moreover, resveratrol, a plant phenolic phytoalexin that has been reported to have antitumor properties in several types of cancers, alters miRNA expression profiles in NSCLC cells [29]. Specifically, miR-21 acts a biomarker predictive for platinum-based adjuvant chemotherapy response in NSCLC patients [30]. Although mRNA analysis is a novel screening tool for lung cancer chemotherapy, this technique is still under development and needs extensive clinical trials before it can be used as standard tool in the management of patients with lung cancer. This will also allow initiation of early and aggressive chemotherapy in patients with a high risk of recurrence in early stage NSCLC. Moreover, such screening tools will also allow us to choose the most appropriate chemotherapy drug that will be most effective in the treatment of such patients, based on their miRNA profiling.
3 miRNAs in Lung Cancer: Tiny Molecules with Huge Impacts
miRNAs play important roles in the regulation of a wide class of cellular processes by sequestering target mRNAs and inhibiting translation of the proteins that they encode.
3.1 miRNAs Altered in Lung Carcinogenesis and Development
A large number of human miRNA genes are frequently located at fragile sites, as well as in minimal regions of loss of heterozygosity, minimal regions of amplification (minimal amplicons), or common breakpoint regions. Overall, more than one half of miR genes are in cancer-associated genomic regions or in fragile sites [31]. This phenomenon indicates miRNAs may act as oncogenes (oncomiRs) or tumor suppressors (tumor suppressor miRs) in lung carcinogenesis. For example, the mutations of tumor suppressor miRs result in carcinogenesis.
3.1.1 miRNAs as Oncogenes-OncomiRs
Aberrations within the cancer miRome were first described in B-cell chronic lymphocytic leukemia, where the locus for the tumor suppressor miR-15 is deleted in 68 % of patients [32]. miRNA-specific phenotypes have since been described for a number of other malignancies. Several ‘oncomiRs’-miRNAs behaving as oncogenes have now been identified and validated to play different roles in lung cell transformation and carcinogenesis (Table 3.1). Specially, miR-21 is a prominent oncogenic driver in a variety of cancers [33] and a number of studies have now shown miR-21 to be up-regulated in NSCLC of various stages and histologies [34–38]. Three targets have been validated for this miRNA: phosphatase and tensin homolog (PTEN), human mutS homolog 2 (hMSH2) (in NSCLC) and programmed cell death 4 (PDCD4) (in breast and colon cancers). Among these, both PTEN and PDCD4 are tumor suppressors that control cell growth and proliferation [35, 39, 40]. PTEN is also shown to be a target of miR-221/222 [41]. Overexpression of these two miRNAs in NSCLC cell lines plays a role in resistance to tumor necrosis factor (TNF)-related apoptosis-inducing ligand [42] and enhances migration through activation of the serine/threonine-specific protein kinase-Akt signaling pathway [41]. Although these findings require in vivo validation, miR-221/222 are known to be frequently upregulated in several solid tumors, including lung cancer [43]. Overexpression of miR-155 in adenocarcinoma and squamous cell lung cancer has also been shown across multiple studies [36, 44]. Its prognostic value and to its potential targets are now being explored [45]. Oncogenic properties of the miR-17-92 cluster, which is housed in the intronic region of chromosome 13q31.3, enhance tumorigenesis by regulating the translational product of proto-oncogene c-Myc (c-Myc) [46, 47]. The miRNAs belonging to this cluster (miR-17, -18a, -19a, -20a, -19b-1 and -92-1) are frequently present in SCLC, and their potential therapeutic applications in SCLC are now undergoing study [48]. Inhibition of miR-17-5p and miR-20a with antisense oligonucleotides can induce apoptosis selectively in lung cancer cells overexpressing miR-17-92, suggesting the possibility of ‘oncomiR addiction’ to expression of these miRNAs in a subset of lung cancers [49]. Collectively, the present findings contribute towards better understanding of the oncomiRs, which might ultimately lead to the future translation into clinical applications by inhibition of oncomiRs.
3.1.2 miRNAs as Tumor Suppressors-Tumor Suppressor miRs
Downregulation of certain miRNAs in cancer is indicative of their tumor-suppressor characteristics. The miRNA let-7 is involved in cell cycle progression [50] and is a classic example of a miRNA behaving as a tumor suppressor in human cancers. Decreased expression of let-7 family members is often observed across many cancers, including lung cancer, and is often associated with poor prognosis [43, 51]. Putative mechanisms of let-7 down-regulation in cancer include genetic alterations [31], regulation of K-RAS, c-Myc and HMGA2 oncogenes [52–54], direct targeting of Dicer mRNA [55] and cell proliferation control in a cyclin-dependent manner [56]. Increased levels of K-RAS and its hyperactivity can occasionally be attributed to a polymorphism in the K-RAS 3′ untranslated region (3′UTR) binding site for let-7 (let-7 complementarity site 6 or LCS6). The allelic frequency of LCS6 in a large population is reported to be 5.8 % (n = 2,433) and as much as 20 % in NSCLC [57]. Underexpression of another miRNA family, the miR-34 cluster, is correlated with poor survival in male smokers with squamous cell carcinoma (SCC) stages I–IIIa [58, 59]. Induction of miR-34 results in apoptosis of lung cancer cells [60]. Expression levels of miR-34a/b/c are shown to be directly correlated with expression of the p53 tumor-suppressor protein, whereas ectopic expression of these miRNAs induces cell-cycle arrest in a mouse fibroblast model [61]. Another study indicates miR-34 expression is inversely correlated with Axl, a receptor that induces proliferation, migration and invasion in cancer, in a panel of NSCLC cell lines and 44 NSCLC resection specimens [62]. Exogenous delivery (local and systemic) of lipid-formulated miR-34 is also shown to reduce tumor size in a mouse model of NSCLC. The expression of miR-29 family is inversely correlated to DNA methyltransferase (DNMT) 3A and DNMT3B in lung cancer tissues and that miR-29 directly targets both DNMT3A and -3B. The enforced expression of miR-29 in lung cancer cell lines restores normal patterns of DNA methylation, induces reexpression of methylation-silenced tumor suppressor genes, such as fragile histidine triad (FHIT) and WW domain-containing oxidoreductase (WWOX), and inhibits tumorigenicity in vitro and in vivo [63]. Overexpression of miR-126 in a lung cancer cell line results in a decrease in Crk protein without any alteration in the associated mRNA. These lung cancer cells exhibit a decrease in adhesion, migration, and invasion [64]. Both miR-15a and miR-16 are frequently deleted or down-regulated in squamous cell carcinomas and adenocarcinomas of the lung. In these tumors, expression of miR-15a/miR-16 inversely correlates with the expression of cyclin D1. These two miRs are implicated in cell cycle regulation in an Rb-dependent manner [65]. Furthermore, miR-1 is downregulated in human primary lung cancer tissues and cell lines. Expression of miR-1 in nonexpressing A549 and H1299 cells reverses their tumorigenic properties, such as growth, replication potential, motility/migration, clonogenic survival, and tumor formation in nude mice. Exogenous miR-1 significantly reduces expression of oncogenic targets, such as MET, a receptor tyrosine kinase, and Pim-1, a Ser/Thr kinase, frequently upregulated in lung cancer [66]. Taken together, these findings suggest a possible therapeutic potential for artificially enhancing cellular levels of tumor suppressor miRs.
3.2 miRNAs as Biomarkers for Early Diagnosis in Lung Cancer
Recently the report from the National Lung Cancer Screening Trial demonstrates an improved survival attributable to early detection of lung cancer by screening [67]. This finding underscores the importance of lung cancer screening research. Current efforts at screening primarily involve clinical history such as smoking and asbestos exposure, imaging in the form of low-dose computed tomography (CT), and either white light or autofluorescence bronchoscopy. The presence of miRNAs in body fluids offers an avenue to improve on these methods by quantitation of miRNAs in these fluids to enhance these efforts.
The first significant effort in this direction comes from Chen and colleagues [68]. By deep sequencing of pooled sera from patients with and without lung cancer, 8 miRNAs are identified as differentially expressed in comparison populations. These miRNAs then are validated in a few clinical samples by RT-PCR. A convincing external validation of these results is still pending. Another exploratory study looks at the potential for microarray profiling of serum-derived miRNAs [69]. In this study, the authors demonstrate the technical feasibility of performing such profiling and demonstrate the high accuracy of predicting cancer presence versus absence by cross-validation analysis. In fact, screening for circulating miRNAs in the peripheral blood can be used as a potential diagnostic tool in lung cancer [70]. Although no independent validation of such a signature is performed and cancers of different histology are grouped into one class, this is an important proof-of-principle study that serum miRNA profiling can be used to classify patients with and without cancer. A different approach to this problem is to perform miRNA microarray profiling of exosomes isolated from serum. Such a study demonstrates a higher exosome content in patients with lung cancer compared with controls [71]. The miRNA expression content of exosomes parallels that of the tumor using a panel of 12 miRNAs, supporting the hypothesis that these are tumor-derived exosomes. A third approach is to perform miRNA profiling of whole blood with the hypothesis that the body’s response to the tumor may be used to detect the presence of the tumor. Such an approach in a small pilot study demonstrates promising results worthy of future investigation [72]. In this study, the investigators used Paxgenetubes (Becton, Dickinson and Company, NJ) to collect whole blood. These tubes have a lysis agent as well as an RNA stabilizer that limits the changes in RNA induced by specimen processing. The miRNA profile of whole blood of 17 patients with lung cancer is compared with those of 19 controls without lung cancer. A 24-miRNA signature using a support vector machine algorithm has a classification accuracy of 95.4 %. Problems common to all of these studies include the absence of suitable controls and the lack of strong external validation data. The first is particularly important as tobacco exposure leads to alteration of miRNAs [73], and if nonsmoking controls are chosen, then the differences noticed may be a result of smoking exposure and not necessarily the presence of lung cancer. The approach of using whole blood is limited by the fact the miRNAs are expressed robustly in nonnucleated cells such as red blood cells (RBCs) [74], and the effect of the presence of cancer on the miRNA expression of RBCs is largely unknown; therefore, the biologic basis of the signature is difficult to delineate. However, these are useful data worthy of future study. Extensive validation of such signatures is essential to be convincing.
As well as other body fluids, sputum is abundant in miRNAs. Endogenous miRNAs are present in sputum in a remarkably stable form and sensitively and specifically detected by real-time RT-PCR. Detection of mir-21 expression produces 69.66 % sensitivity and 100.00 % specificity in diagnosis of NSCLC, as compared with 47.82 % sensitivity and 100.00 % specificity by sputum cytology [75]. Another study evaluates the utility of miRNA biomarkers of squamous lung cancer in human sputa. A directed RT-PCR study using 6 miRNAs demonstrates reasonable sensitivity (76 %) and specificity (96 %) of a 3-miRNA signature (miR-205, miR-210, and miR-708) for the detection of squamous cell lung cancer [76]. A similar study by the same group indicates a 4-miRNA signature (miR-21, miR-486, miR-375, and miR-200b) has a sensitivity of 80.6 % and a specificity of 91.7 % [77]. The optimized five miRNAs panel (miR-21, miR-143, miR-155, miR-210 and miR-372) detects NSCLC with 83.3 % sensitivity and 100 % specificity in 30 prospectively accrued study patients [78]. Therefore, these findings suggest sputum miRNA profiling using cluster analysis is a promising approach for the early detection of lung cancer. For better diagnosis, further larger follow-up studies using this approach are warranted.
3.3 miRNAs in Lung Cancer Prognosis
Many studies have attempted to use miRNA expression patterns to predict the prognosis of lung cancer patients. For the most part, these experiments have been performed with NSCLC. The clinical question is an important one as a significant proportion of patients with even early, adequately resected lung cancer recur. It would be useful to generate a prediction algorithm to identify those that will recur; if such a prediction can be made accurately, and then this subset of high-risk patients can be potentially treated by adjuvant chemotherapy to decrease their disease recurrence rate.
The miRNA profiling of lung cancers can predict outcome is first reported by Yanaihara et al. [36]. In this early study, the investigators compare the miRNA expression profiles of numerous lung cancers with their corresponding normal counterparts. As a secondary analysis, they demonstrate that miRNAs let-7 and miR-155 can predict disease outcome. The second major study to address this issue is published by Yu et al. [79]. In this well-designed study, RT-PCR-based miRNA profiling is performed on training and test sets to identify a 5-miRNA-based prognostic classifier for lung cancer. This finding also is validated in an independent validation set. After these early studies, two microarray-based studies [51, 80], also demonstrate the prognostic potential of miRNAs. The first study is in squamous cell lung cancer and finds miR-146b to be a significant prognostic miRNA. A similar study by Patnaik et al. also demonstrates miR-146b-3p to have prognostic value along with several other miRNAs previously not associated with lung cancer prognosis. The prognostic value of miR-21 and miR-205 overexpression is also validated in NSCLC [37]. Genetic variants of miRNA sequences are associated with NSCLC survival [81]. Additional two studies have validated this concept [58, 82]. Interestingly, one large study has looked at the ability of serum miRNAs to prognosticate lung cancer [83]. Although the ability to predict prognosis reaches the statistical significance, the prediction accuracy is modest. A more recent study demonstrates no ability of measurement of expression of a limited set of miRNAs to predict the prognosis of lung cancer, adding a cautionary note to such investigations [84]. In summary, several additional steps are necessary before the concept of miRNA-based prognostication can be used clinically. First, additional validation studies are crucial; preferably these should be conducted in a prospective fashion. Second, the studies should focus on prediction accuracy and not solely on time-to-recurrence or time-to-survival end-points. Third, the day-to-day variability of the assays used as well as the variability across several samples from the same tumor should be assessed; this has not yet been performed in a systematic manner. Fourth, it will be useful to assess the ability of these assays to prognosticate based on CT-guided biopsies as this may spur neoadjuvant trials for patients at a high risk for recurrence. As is shown in the list of issues, these assays are several years away from clinical application.
3.4 miRNAs in Lung Cancer Therapy
Since miRNAs act as oncomiRs or tumor suppressor miRs in lung cancer, they can be used as potential agents in cancer treatment [85]. In NSCLC cells, anti-miRNA oligonucleotides (AMOs) show a inhibitory effect on cell growth [86]. A recent paradigm shift in the field of cancer is the concept of personalized therapy. This shift is based on the assumption that one can predict the response of an individual tumor to a particular drug. As with gene expression profiling, miRNA profiling is being considered for the prediction of response to therapy. Conceptually, miRNA expression has been demonstrated to be associated with cancer cell line sensitivity. Thus far, three different groups have published miRNA expression profiles of the NCI-60 cancer cell lines [87–89]. These expression profiles have a moderate correlation with cell line sensitivity to many drugs. An example of such a correlation is demonstrated in Fig. 3.2. In this figure, expression profiles performed using the Affymetrix (Affymetrix Inc., Calif) miRNA microarray profiling of the NCI-60 cancer cell lines (unpublished data) are used to predict sensitivity to oxaliplatin. Other investigators have demonstrated that altering the levels of specific miRNAs changes the sensitivity of cancer cell lines to therapeutic agents [8, 90]. A small body of literature also associates miRNA expression to chemosensitivity prediction in other cancer systems [91–93]. Of particular interest is the potential of such prediction in SCLC demonstrated by a preliminary study [94]. However, this field is still in its infancy and is faced with many hurdles. For one, in vitro sensitivity may not correlate with in vivo sensitivity, as cancer behavior is the result of interplay between cancer cells and their environment. Also, most treatment modalities use multiple agents and confounding is an issue. Some attempts at tackling these issues involve bioinformatics algorithms such as the COXEN algorithm [95]. Clinically, an evolution of platforms and bioinformatics is required to overcome these issues.
4 miRNAs in Lung Cancer Drug Resistance
Although the studies that miRNAs are involved in special signal transduction pathways and regulatory mechanisms in lung cancer drug resistance just begins, there are a large number of experimental and clinical studies have shown that miRNAs play critical roles in chemotherapy resistance.
4.1 miR-Polymorphisms and miR-Targeted mRNA Polymorphisms Alter Drug Metabolism
Polymorphisms in the miRNA regulatory pathway (miR-polymorphisms or single nucleotide polymorphisms [SNPs] that interfere with mRNA function [miRSNPs]) are a novel class of functional polymorphisms present in the human genome [96]. They include not only miR-polymorphisms, but also polymorphisms in miR-targeted mRNAs.
In general, miRSNPs reside at or near a miRNA-binding site of a functional gene, influencing its expression by interfering with miRNA function [97–99]. In the clinical researches, it is found that the expression and polymorphisms in miR-targeted mRNAs are involved in drug responses. The expressions of multidrug resistance protein-1 (MRP1), breast cancer resistance protein (BCRP), lung resistance-related protein (LRP), and excision repair cross-complementing group-1 (ERCC1) in patients with advanced NSCLC are correlated with response to cisplatin-based chemotherapy [100]. These DNA repair gene polymorphisms are useful as predictors of clinical outcome to the platinum-based chemotherapy. Epidermal growth factor receptor (EGFR) kinase inhibitors induce dramatic clinical responses in NSCLC patients with advanced disease. EGFR gene polymorphism in intron 1 contains a polymorphic single sequence dinucleotide repeat (CA-SSR) shows a statistically significant correlation with the gefitinib response and is appeared to be a useful predictive marker of the development of clinical outcome containing skin rashes with gefitinib treatment. The other polymorphisms of EGFR are also associated with increased EGFR promoter activity. EGFR gene mutations and polymorphisms are also associated with EGFR kinase inhibitors response and toxicity [101]. Recent studies reveal that miRNAs affect drug metabolism by targeting ATP-binding cassette (ABC) genes, a class of drug transporters on cell membrane. Adenovirus-mediated RNA interference on endogenous miRNAs silences the ABC multidrug resistance protein 2 (ABCC2) expression in a mouse model [102]. Meanwhile, Zhu et al. reports that miR-27a and miR-451 are involved in activating the expression of P-glycoprotein, the MDR1 gene product that confers cancer cell resistance to a broad range of chemotherapeutics [103]. Expressions of miR-27a and miR-451 are upregulated in MDR cancer cell lines A2780DX5 and KB-V1, as compared with their parental lines A2780 and KB-3-1. Treatment of A2780DX5 cells with the antagomirs of miR-27a or miR-451 decreases the expression of P-glycoprotein and MDR1 mRNA. In contrast, the mimics of miR-27a and miR-451 increase MDR1 expression in the parental cells A2780. The sensitivity to and intracellular accumulation of cytotoxic drugs that are transported by P-glycoprotein are enhanced by the treatment with the antagomirs of miR-27a or miR-451. Meanwhile, To et al. shows that miR-519c regulates ABCG2 expression at the 3′UTR of its mRNA through modulation of transcript stability and protein translation and then leads to drug resistance [104]. Similarly, miR-328-directed downregulation of ABCG2 expression in MCF-7/MX100 cells results in an increased mitoxantrone sensitivity [105]. Interestingly, our recent data indicates miR-31 targets ABCB9, an important member of ABC family, and inhibits cisplatin-induced cell apoptosis in NSCLC (accepted data by Cancer Letters). Using a bioinformatical tool, some miRNAs interact with drug transporter, nuclear receptor (NR) and human cytochrome P450 [106–108]. More and more studies show that miRNAs are involved in drug metabolism and distribution by regulating drug-metabolizing enzymes, drug transporters and/or NR genes (Fig. 3.3). These results demonstrate the dysregulated miRNA expression plays an important role in an abnormal drug metabolism.
4.2 miRNA Polymorphisms and Drug Resistance
MiR-polymorphisms potentially influence drug uptake, metabolism and distribution by regulating multiple gene expressions. As a result, polymorphisms affect the treatment efficiency or induce resistance to the special anticancer drug during this process.
Analysis of the publicly available SNP database reveals the presence of a relatively high level of variations in the 3′UTRs of miRNA target genes [109]. However, relatively low levels of variation are observed in the miRNA seed region of a functional miRNA. Approximately 250 SNPs are found to potentially create target sites for miRNAs [110]. Functional polymorphisms in the 3′UTRs of several genes have been reported to be associated with diseases by affecting gene expression [111]. Some of these polymorphisms may interfere with the function of miRNA and are potential miR-polymorphisms able to affect the expression of miRNA targets [112]. Therefore, miR-polymorphisms may result in gain or loss of a miRNA function. Based on the current knowledge of this field, miR-polymorphisms can be classified in the following three major categories [96]: (a) polymorphisms affecting miRNA biogenesis, (b) miR-polymorphisms in miRNA target sites and (c) miR-polymorphisms altering epigenetic regulation of miRNA genes. Recently, miR-polymorphisms have been shown to affect drug response and have the potential to confer drug resistance [96]. It is demonstrated that a C to T SNP, identified in a case–control study of childhood leukemia patients, occurring with 14.2 % allelic frequency in the Japanese population, is present near the miR-24 binding site in the 3′UTR of the dihydrofolate reductase (DHFR) gene. The T allele of the SNP results in loss of miR-24-mediated regulation of DHFR, high DHFR protein levels and confers methotrexate (MTX) resistance [99]. This finding may also be useful in predicting the clinical outcome of MTX treatment.
4.3 miRNAs Altered in Drug Resistance
Recently, increasing studies have indicated that aberrant miRNA expression is strongly implicated in anticancer drug resistance phenotype. Their involvements in tumor cells response to chemotherapeutic agents are being confirmed by more and more reports.
4.3.1 miRNAs Affect Drug Sensitivity to EGFR Mutants
DNA sequencing of 623 genes with known or potential relationships to lung adenocarcinoma reveals more than 1,000 somatic mutations across the samples. It has been identified 26 genes that are mutated at significantly high frequencies and thus are probably involved in carcinogenesis. Notably, EGFR is one of the frequently mutated genes [113]. Interestingly, miRNAs may regulate EGFR mutation in NSCLC. Comparing paired lung cancer tissue with adjacent normal lung parenchyma, miR-126*, miR-145, miR-21, miR-182, miR-183 and miR-210 are found to be the most differentially expressed miRNAs. Most interestingly, an obvious inhibition of cell growth is observed in the EGFR mutant lung adenocarcinoma after transfection of pre-miR-145. These results also show that restoration of tumor suppressor miR-145 inhibits cancer cell growth in EGFR mutant lung adenocarcinoma [114]. Further study on these specific differentially expressed miRNAs may provide important information on peculiar tumorigenetic pathways and may identify useful biomarkers.
EGFR gene mutations, which are correlated with sensitivity to EGFR-tyrosine kinase inhibitors (EGFR-TKIs), are more frequent in never-smoker lung cancers. A miRNA expression profiling of 28 cases of never-smoker lung cancer identifies aberrantly expressed miRNAs, which are much fewer than in lung cancers of smokers and includes miRNAs previously identified (e.g., upregulated miR-21) and unidentified (e.g., downregulated miR-138) in those smoker cases. The changes in expression of some of these miRNAs, including miR-21, are more remarkable in cases with EGFR mutations than in those without these mutations. A significant correlation between phosphorylated-EGFR (p-EGFR) and miR-21 levels in lung carcinoma cell lines and the suppression of miR-21 by an EGFR-TKI, AG1478, suggests that the EGFR signaling is a pathway positively regulating miR-21 expression. In the never-smoker-derived lung adenocarcinoma cell line H3255 with mutant EGFR and high levels of p-EGFR and miR-21, antisense inhibition of miR-21 enhances AG1478-induced apoptosis. In a never-smoker-derived adenocarcinoma cell line H441 with wild-type EGFR, the antisense miR-21 not only shows the additive effect with AG1478 but also induces apoptosis by itself [115]. These results suggest that aberrantly increased expression of miR-21, which is enhanced further by the activated EGFR signaling pathway, plays a significant role in lung carcinogenesis in never-smokers, as well as in smokers, and is a potential therapeutic target in both EGFR-mutant and wild-type cases.
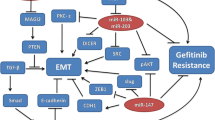
To understand the role of miRNA in EGFR-TKI-resistant NSCLCs, Garofalo et al. examines the changes in miRNA that are mediated by tyrosine kinase receptors. They report that miR-30b/c, miR-221 and miR-222 are modulated by both EGF and MET receptors, whereas miR-103 and miR-203 are controlled only by MET. Importantly, they show that these miRNAs have important roles in gefitinib-induced apoptosis and epithelial-mesenchymal transition (EMT) of NSCLC cells in vitro and in vivo by inhibiting the expression of the genes encoding BCL2-like 11 (BIM), apoptotic peptidase activating factor 1 (APAF-1), protein kinase Cε (PKC-ε) and sarcoma viral oncogene homolog (SRC) [116]. These findings suggest that modulation of specific miRNAs may provide a therapeutic approach for the treatment of NSCLC.
A recent study in NSCLC cell lines demonstrates that the tumor microenvironment elicits transforming growth factor β1 (TGFβ1) and stimulates a miRNA gene expression program that induces resistance to anti-EGFR therapy and drives lung tumor cells to EMT, invasion, and metastasis [117]. Moreover, miR-214 regulates the acquired resistance to gefitinib via the PTEN/Akt pathway in EGFR-mutant cell lines [118]. To overcome this kind of resistance, Rai et al. focuses on EGFR suppression using miR-7, targeting multiple sites in the 3′UTR of EGFR mRNA. In this study, two EGFR-TKI-sensitive cell lines (PC-9 and H3255) and two EGFR-TKI-resistant cell lines harboring T790M (RPC-9 and H1975) are used. They construct miR-7-2 containing miR-7-expressing plasmid and the miR-7 expression level of the transfectants is approximately 30-fold higher, and the luciferase activity is ablated by 92 %. The results show miR-7 significantly inhibits cell growth not only in PC-9 and H3255 but also in RPC-9 and H1975. Expressions of insulin receptor substrate-1 (IRS-1), RAF-1, and EGFR are suppressed in these four cell lines. Injection of the miR-7-expressing plasmid reveals a remarkable tumor regression in a mouse xenograft model using RPC-9 and H1975. Moreover, EGFR, RAF-1, and IRS-1 are suppressed in the residual tumors [119]. These findings indicate promising therapeutic applications of miR-7-expressing plasmids against EGFR oncogene-addicted lung cancers including T790M resistance by liposomal delivery.
4.3.2 miRNAs Affect TRAIL Resistance
The TNF-related apoptosis inducing ligand (TRAIL) has gained much attention due to its specific anti-tumor potential without toxic side effects. Thus TRAIL is a promising new anticancer biotherapeutic [120]. As shown by many preclinical studies, TRAIL efficiently induces apoptosis in numerous tumor cell lines but not in the majority of normal cells. However, an increasing number of publications report on a predominance of TRAIL resistance in primary human tumor cells, which require sensitization for TRAIL-induced apoptosis. Sensitization of cancer cells by treatment with chemotherapeutic drugs and irradiation has been shown to restore TRAIL sensitivity in many TRAIL-resistant tumor cells [121]. How miRNAs regulate TRAIL resistance needs to be fully explored.
To define novel pathways that regulate susceptibility to TRAIL in NSCLC, a genome-wide expression profiling of miRNAs is performed by Garofalo and colleagues. They show that in TRAIL-resistant NSCLC cells, levels of different miRNAs are increased and in particular, miR-221 and -222. Then they demonstrate that these miRNAs impair TRAIL-dependent apoptosis by inhibiting the expression of key functional proteins. Indeed, transfection with anti-miR-221 and -222 renders CALU-1-resistant cells sensitive to TRAIL. Conversely, H460-sensitive cells treated with -221 and -222 pre-miRNAs become resistant to TRAIL. Both miR-221 and -222 target the 3′UTR of Kit and p27(kip1) mRNAs, but interfere with TRAIL signaling mainly through p27(kip1) [42]. The results show that high expression levels of miR-221 and -222 are needed to maintain the TRAIL-resistant phenotype, thus making these miRNAs as promising therapeutic targets or diagnostic tool for TRAIL resistance in NSCLC. Interestingly, these two miRNAs are upregulated by the MET proto-oncogene. A recent study shows that miR-130a, expresses at low level in lung cancer cell lines, by targeting MET was able to reduce TRAIL resistance in NSCLC cells through the c-Jun-mediated down-regulation of miR-221 and miR-222 [122]. Together, a better understanding of miR-221/-222 regulation in drug resistance is the key in developing new strategies in NSCLC therapy.
PED/PEA-15 (phosphoprotein enriched in diabetes, PED) is a death effector domain family member of 15 kDa with a broad antiapoptotic function found overexpressed in a number of different human tumors, including lung cancer. Incoronato et al. identifies miR-212 as a negative regulator of PED expression. Furthermore, they also show that ectopic expression of miR-212 increases TRAIL-induced cell death in NSCLC cells. In contrast, inhibition of endogenous miR-212 by use of antago-miR results in increase of PED protein expression and resistance to TRAIL treatment. Besides, in NSCLC, they show both in vitro and in vivo that PED and miR-212 expressions are inversely correlated, that is, PED is upregulated and miR-212 is rarely expressed [123]. These findings suggest that miR-212 should be considered as a tumor suppressor because it negatively regulates the antiapoptotic protein PED and regulates TRAIL sensitivity.
4.3.3 Ectopic Expressions of miRNAs Reverse Therapeutic Effects of Anti-cancer Drugs
It’s well known that miRNAs are strongly implicated in drug resistance, cell survival and apoptosis. Therefore, it is likely that they can also modulate sensitivity and resistance to anticancer drugs in substantial ways. To test this hypothesis, Blower and colleagues investigate the pharmacologic roles of three microRNAs previously implicated in cancer biology (let-7i, miR-16, and miR-21) and also use in silico methods to test pharmacologic miRNA effects more broadly. In the experimental system, they increase the expression of individual miRNAs by transfecting their precursors (which are active) or suppress the expression by transfection of antisense oligomers. In three NCI-60 human cancer cell lines, a panel of 60 lines is used for anticancer drug discovery. They assess the growth-inhibitory potencies of 14 structurally diverse compounds with known anticancer activities. Changing the cellular levels of let-7i, miR-16, and miR-21 affect the potencies of a number of the anticancer agents by up to fourfold. The effect is the most prominent with mir-21, with 10 of 28 cell-compound pairs showing significant shifts in growth-inhibitory activity. Varying mir-21 levels change potencies in opposite directions depending on compound class; indicating that different mechanisms determine toxic and protective effects. In silico comparison of drug potencies with miRNA expression profiles across the entire NCI-60 panel reveal that approximately 30 miRNAs, including mir-21, show highly significant correlations with numerous anticancer agents [8]. There results support a substantial role for miRNAs in anticancer drug response, suggesting novel potential approaches to the improvement of chemotherapy.
The primary researches show miRNAs as biomarkers are associated with TKI resistance, such as up-regulations of miR-21 and miR-23b predict an increase of anticancer drug-sunitinib resistance, while down-regulation of miR-424 indicates an increase of resistance of erlotinib and vandetanib (data not published, but released on AACR 2010). The data from our own group demonstrate miR-31 is significantly upregulated in cisplatin-resistant cell line, as compared to that in cisplatin-sensitive cell line. As a result, miR-31 overexpression induces drug resistance in cisplatin-sensitive cell line and miR-31 knockdown rescues drug sensitivity in cisplatin-resistant cell line. MiR-31 exerts an anti-apoptotic effect probably through inhibition of ABCB9 and provides a novel strategy using miR-31 as a potential target in NSCLC chemotherapy (data accepted by Cancer Letters). Similarly, in A549 cell line, miR-1 is downregulated and exogenous miR-1 enhances their sensitization to doxorubicin-induced apoptosis [66].
Therefore, lung cancer resistance emerged in clinical treatments is closely related to some upregulated or downregulated miRNAs. To regulate the expressions of these miRNAs is an ideal approach to control therapeutic effects (Table 3.2). It is clear that more than one target or mechanism of drug resistance is activated in certain drugs. The targets or mechanisms that can be activated with more than one drug are more attractive for the broad therapeutic potential. miRNAs or antagomiRNAs might be more efficient in avoiding resistance or increasing the effectiveness of malignant tumors to chemotherapy. Furthermore, miRNAs might be used as a biomarker to predict the response to chemotherapy and the survival in patients with malignant tumors. In addition, miRNAs combined with traditional chemotherapy agents might provide a new strategy to treat malignant tumors in the future.
5 miRNAs and Lung Cancer Targeted Therapy: Small Molecules with Unlimited Potentials
As we have discussed above, miRNAs have fast become a field of interest, particularly for their discovered involvement in a number of different oncogenic pathways and the potential they bring for a deeper look into the mechanisms of chemoresistance and future therapies that can be used to circumvent this resistance. miRNAs are already showing their potential as biomarkers to predict treatment response in a number of different cancers, and continued clinical validation is needed to hone in on the most promising of these predictions. There has also been preliminary research showing the involvement of miRNAs in key pathways regulating cancer cell growth, proliferation, invasion, and so on, and this pushes them toward use as novel targets for new anticancer treatments. Again, further research is warranted to validate involvement in these and other pathways, and to continue pursuing miRNA delivery systems that can be translated to therapeutic treatments.
5.1 miRNAs as New Therapeutic Targets
Preclinical models have consistently underlined the feasibility and efficacy of miRNA-based therapies, either alone or in combination with current targeted therapies. The appealing strength of such therapeutic option dwells in miRNAs ability to concurrently target multiple genes, frequently in the context of a specific network/pathway, making miRNA-based therapy extremely efficient in regulating distinct biological processes relevant to normal and pathological cell homeostasis [124]. There are two main therapeutic strategies to target miRNA expression: miRNA reduction and miRNA replacement (Fig. 3.4) [6].
Potential miRNA-based therapeutic strategies. The function of oncogenic miRNAs could be stopped by small-molecule inhibitors (regulation of miRNAs expression at the transcriptional level), antisense oligonucleotides (binding by complementarity miRNAs and inducing either duplex formation or miRNA degradation), miRNAs masking (molecules complementary to the 3′UTR of the target miRNA, resulting in competitive inhibition of the downstream target effects) or miRNAs sponges (oligonucleotide constructs with multiple complementary miRNA binding sites to the target miRNA). Tumor suppressor miRNAs function can be restored by introducing systemic miRNAs (miRNA mimics) or inserting genes coding for miRNAs into viral constructs
The use of oligonucleotides or virus-based constructs can either block the expression of an oncogenic miRNA or reintroduce the loss of expression of a tumor suppressor miRNA. A different approach is the use of drugs to modulate miRNA expression by targeting their transcription and their processing [124]. There are some fundamental issues, which have impeded development of miRNA-based treatments. First, we need to clearly demonstrate a tissue-specific delivery and develop a more efficient cellular uptake of synthetic oligonucleotides to achieve sustained target inhibition. This should result in significantly enhanced patient benefits and reduced drug toxicity. In fact, the second and even more challenging problem to overcome is the biological instability of miRNAs in bodily fluids or tissues, as unmodified oligonucleotides are rapidly degraded by cellular and serum nucleases, requiring huge doses of drugs. As a result, various chemical modifications in oligonucleotides have been investigated, such as morpholinos, peptide nucleic acids, cholesterol conjugation and phosphorothioate backbone modifications. Among others, the locked nucleic acid (LNA) constructs provide the most promising results. LNA nucleosides are a class of nucleic acid analogues in which the ribose ring is “locked” by a methylene bridge connecting the 2′-O atom to the 4′-C atom. This feature confers to LNA oligonucleotides great advantages including: (a) High hybridization affinity towards complementary single-stranded RNA and complementary single-stranded or double-stranded DNA; (b) Excellent mismatch discrimination, and (c) High aqueous solubility. The so-called “LNA anti-miR” constructs have been successfully used in several in vitro and in vivo studies to knockdown the expression of specific miRNAs [125, 126]. This success has culminated in the first two miRNA-based clinical trials for the treatment of hepatitis C virus (HCV) infection by targeting miR-122 with an LNA-antimiR (miravirsen or SPC3649; Santaris Pharma, Denmark) [125, 126]. The phase IIa clinical trial [126] has shown a dose-dependent, long reduction in HCV RNA that continues to fall after completion of treatment without any recorded serious adverse effects.
The discovery of exosome-specific miRNA circulation among bodily fluids provided the “Trojan horse” for the forthcoming development of miRNA delivery vehicles for systemic gene therapy: exosomes, as natural cell-derived nano-carriers, are immunologically inert and possess an intrinsic ability to cross biological barriers [127]. On the other hand, exosome-released miRNAs represent a novel mechanism of cross-talk and genetic exchange between cells. Interestingly, cancer-released exosomes have been shown to carry oncogenic miRNAs, and the inhibition of cancer-related exosome secretion has been demonstrated to significantly reduce the metastatic potential of lung cancer cell lines [127]. The LNA and exosome data drew attention to the potential of miRNAs for cancer treatment. The development of safe and specific methods of delivery of miRNA-based treatments will allow modulation of miRNAs to become in the next few years a central feature of cancer treatment and management.
5.2 miRNA Therapeutics in Lung Cancer
Almost two decades have passed since the initial discovery of miRNAs, and many important biological roles have been attributed to this class of RNAs. Currently, the scientific community is pursuing different strategies to employ miRNA therapeutics [124]. Emerging miRNA antisense and mimic technologies, as well as other novel mechanisms of delivery, are being explored. In the very near future, we will be seeing more published studies focusing on assessing the effectiveness, pharmacokinetics and toxicity of miRNA mimics and inhibitors. One major obstacle in advancing miRNA-based therapeutics lies in the very property of these complex molecules-their ability to act on multiple cellular targets could realize diverse side effects [128]. Evaluating off-target effects will be a necessity in early-phase human studies when arriving at the recommended dose and schedule for clinical efficacy trials. One example demonstrated a therapeutic use of miRNAs in a rodent model of NSCLC. Artificial let-7 is directly injected into already-established tumor mass, leading to tumor regression [129]. Similarly, in vivo application of miR-17-5p antagomir results in a reduction of therapy-resistant neuroblastoma in mice [130]. Various routes of delivery as well as formulations are also being investigated. Systemic delivery of the tumor suppressors let-7 and miR-34a complexed with a neutral lipid emulsion are shown to preferentially target lung tumors, resulting in up to 60 % tumor burden reduction in mouse models of lung cancer [131]. Intranasal delivery of let-7 also leads to tumor growth reduction in a mouse xenograft model [132]. Meanwhile, Wiggins et al. [10] develops a therapeutic formulation using chemically synthesized tumor suppressor miR-34a and a lipid-based delivery vehicle that blocks tumor growth in mouse models of NSCLC. This formulation is effective when administered locally or systemically, it is well tolerated and does not induce an immune response [133]. Taken together, these studies demonstrate the therapeutic potential of miRNAs in lung cancer.
5.3 MiRNA Based-Therapeutic Targets in Invasive and Metastatic Lung Cancer
The most deadly aspect of cancer is its ability to invade and metastasize. Garofalo et al. [41] shows that overexpressed miR-221 and miR-222, by targeting PTEN and TIMP3 tumor suppressors, induce TRAIL resistance and enhance cellular migration through the activation of the Akt pathway and metallopeptidases in aggressive NSCLC cells. They further demonstrate that the MET oncogene is involved in miR-221 and miR-222 activation through the c-Jun transcription factor. Muniyappa et al. [134] identifies that miR-29a has a significant anti-invasive and anti-proliferative effect on lung cancer cells in vitro. miR-29a functions as an antioncomir and this function is likely mediated through the post-transcriptional fine tuning of the cellular levels of several proteins, including RAN (a member of the RAS oncogene family). Gibbons et al. [135] demonstrates that forced expression of miR-200 abrogates the capacity of metastatic lung adenocarcinoma cells to undergo EMT, invade, and metastasize. Tumor cell metastasis is regulated by miR-200 expression that changes in response to contextual extracellular cues. Ma et al. [136] reports that silencing of miR-10b with antagomirs to mice bearing highly metastatic cells significantly increases the levels of Hoxd10 and markedly suppresses formation of lung metastases. miR-10b antagomir is well tolerated by normal animals and it appears to be a promising candidate for the development of new anti-metastasis agents. These results indicate miRNAs can be used as therapeutic targets in invasive and metastatic lung cancers.
5.4 Therapeutic Potential of miRNAs in Lung Cancer Chemotherapy
Emerging evidence has also shown that some miRNAs could target genes related to drug sensitivity, resulting in the altered sensitivity of cancer cells to anti-cancer drugs [137]. Guo et al. [138] indicates that transfection of the drug resistant SCLC cells with the mimics of miR-134 greatly increases the sensitivity to anti-cancer drugs cisplatin, etoposide, and doxorubicin. miR-134 increases the cell survival by inducing G1 arrest and downregulates MRP1/ABCC1 protein in drug-resistant SCLC cells. Zhu et al. [139] demonstrates that enforced miR-181b expression reduces BCL2 protein level and sensitizes multidrug resistant lung cancer cells to cisplatin-induced apoptosis. Galluzzi et al. [10] also reports that pre-miR-181a and pre-miR-630 enhances and reduces cisplatin-triggered cell death in NSCLC cells, respectively. Pre-miR-181a and pre-miR-630 consistently modulated mitochondrial/postmitochondrial steps of the intrinsic pathway of apoptosis, including Bax oligomerization, mitochondrial transmembrane potential dissipation, and the proteolytic maturation of caspase-9 and caspase-3. Another two different groups show that both miR-98 and miR-34a regulates cisplatin-induced A549 cell death by inhibiting TP53 pathway [140, 141]. Similarly, miR-622 maybe function as a tumor suppressor by targeting K-RAS and enhancing the anticarcinogenic effect of resveratrol [142]. Therefore, miR-622 is potentially useful as a clinical therapy. miR-100 resensitizes docetaxel-resistant human lung adenocarcinoma cells (SPC-A1) to docetaxel by targeting Plk1 [143]. Thus, this suggests that downregulation of miR-100 could lead to Plk1 over-expression and eventually to docetaxel chemoresistance of human lung adenocarcinoma. miR-200b reverses chemoresistance of docetaxel-resistant human lung adenocarcinoma cells by targeting E2F3 [144]. The results suggest that downregulation of miR-200b could lead to E2F3 overexpression and in turn contribute to chemoresistance of lung adenocarcinoma cells to docetaxel. miR-126 enhances the sensitivity of NSCLC cells to anticancer agents LY294002, an inhibitor of the phosphoinositidyl-3 kinase (PI3K)/Akt signaling pathway, by targeting vascular endothelial growth factor (VEGF) A [145]. The identification of a miR-337-3p as a modulator of cellular response to taxanes, and STAT3 and RAP1A as regulatory targets which mediate that response, defines a novel regulatory pathway modulating paclitaxel sensitivity in lung cancer cells, which may provide novel adjuvant strategies along with paclitaxel in the treatment of lung cancer and may also provide biomarkers for predicting paclitaxel response in NSCLC [9]. miR-513a-3p sensitizes human lung adenocarcinoma cells to chemotherapy by targeting GSTP1 [146]. Interestingly, serum miR-125b can be used as a diagnostic or prognostic biomarker for advanced NSCLC patients receiving cisplatin-based chemotherapy [147]. Recently, Franchina et al. showed that circulating miR-22, miR-24 and miR-34a act as novel predictive biomarkers to pemetrexed-based chemotherapy in advanced NSCLC patients [148].
5.5 Therapeutic Potential of miRNAs in Lung Cancer Radiotherapy
Another role for miRNAs that deserves mention is that of sensitizers to radiotherapy. This is of particular importance given that many tumors require combinations of chemotherapy and radiotherapy as optimal modes of treatment. miRNAs may modulate the DNA damage response, thus sensitizing tumor cells to both chemotherapy and radiotherapy.
The miRNA regulatory network may also be a potentially useful therapeutic target for overcoming the radioresistance of lung cancer. The overexpression of let-7a decreases expression of K-RAS and radiosensitizes A549 cells. Inhibition of Lin28, a repressor of let-7, attenuates K-RAS expression and radiosensitizes A549 cells. The Lin28-let7 regulatory network may be a potentially useful therapeutic target for overcoming the radioresistance of human cancers having activated K-RAS signaling [149]. Comparing with resistant NSCLC patients, five miRNAs (miR-126, miR-let-7a, miR-495, miR-451 and miR-128b) are significantly upregulated and seven miRNAs (miR-130a, miR-106b, miR-19b, miR-22, miR-15b, miR-17-5p and miR-21) are greatly downregulated in radiotherapy sensitive group. Overexpression of miR-126 inhibits the growth of SK-MES-1 cells and promotes its apoptosis induced by irradiation. The expression level of p-Akt decreases in the miR-126 overexpression group. After treating with PI3K constitutively activator (IGF-1) and inhibitor (LY294002), miR-126 overexpression has no significant effects on the apoptosis of SK-MES-1 cells. These results show miR-126 promotes NSCLC cells apoptosis induced by irradiation through the PI3K-Akt pathway [150]. The expression of miR-9 and let-7 g could enhance the efficiency of radiotherapy for lung cancer treatment through the inhibition of NF-κB1[151]. A small number of NSCLC cell lines have a high level of endogenous miR-101. The ectopic miR-101 is able to radiosensitize most NSCLC cells, except for the NSCLC cell lines that had a much higher endogenous miR-101 level [152]. In the p53 wild type, K-RAS mutated NSCLC cells, the overexpression of miR-34b increases radiosensitivity at low doses of radiation [153]. miR-214 is upregulated in radiotherapy-resistant NSCLC cells relative to radiosensitive counterparts and miR-214 modulates radiotherapy response of NSCLC cells through regulation of p38MAPK, apoptosis and senescence [154]. Overexpression of miR-449a in CL1-0 cells effectively increases irradiation-induced DNA damage and apoptosis, alters the cell cycle distribution and eventually leads to sensitization of CL1-0 to irradiation [155]. Therefore, continued investigations will benefit the understanding of miRNA-based radiotherapy in lung cancer.
6 Conclusion
The discovery that noncoding regions of the genome harbored regulatory molecules that could regulate basic cellular functions is one of the most important discoveries in recent history. This discovery alone has provided insight into the mechanisms of tumor initiation and progression, identified potential biomarkers, and, finally, created the possibilities for novel small-molecule therapies. Each of these complex areas continues to be investigated. Among these noncoding RNAs, miRNAs are a short, noncoding class of RNAs that are involved in global regulation of protein expression, act on multiple targets and serve as physiological buffers in biological responses to internal and external stimuli.
It is becoming apparent that therapeutic inhibition or mimicking of a single miRNA can simultaneously target multiple genes within similar pathways and signaling cascades. miRNAs are now being studied for their potential as a new generation of therapeutics. The main challenges for miRNA therapeutics are stability, safety, and delivery to appropriate cells within a tissue or organ [156]. Technological advances have enabled important discoveries of molecular, cellular, clinical, and therapeutic cancer research findings in recent years [157]. There are various therapeutic tools that are currently being investigated to manipulate upregulated miRNAs, such as antagomir, small molecule, and miRNA sponge. Antagomir is the most widely used approach to regulate miRNA levels in vivo, including LNA antisense oligonucleotides. One study group finds that small molecule can also be used to modulate the functionality of a specific miRNA [158]. miRNA sponge is another technique that uses a vector expressing miRNA target sites to scavenge a miRNA and prevent it from regulating its natural targets [159]. On the other hand, there are also approaches to mimic or reexpress the downregulated miRNAs, such as lipid-formulated mimic and adeno-associated viruses’ vector.
Nevertheless, considerable obstacles should be overcome before miRNA therapy becomes a real option for the management of cancer. The exciting therapeutic potential of miRNA is accompanied by the recognition of off-target effects. Integrative vectors allow life-long gene supplementation, but involve the risk of potential tumorigenicity resulting from activation of oncogenes located in the vicinity of the integration site [160]. The long terminal repeat enhancer element is removed and an internal promoter is added in lentiviral vector to solve this problem [161]. Although the development of these therapeutic tools are still in infancy, interdisciplinary helps from nanobiotechnology making research on miRNA therapeutics rapidly moving forward [162]. Several organizations already have active preclinical or clinical trials engaged in developing miRNA therapeutics in various cancers, including lung cancer, prostate cancer, liver cancer, esophageal cancer, leukemia, skin cancer, and renal cell carcinoma (Fig. 3.5) [163, 164]. Recently, the therapeutic applicability of LNA-antimir technology has been reported in non-human primates. Treatment of chronically infected chimpanzees with a LNA-modified oligonucleotide complementary to miR-122 leads to long-lasting suppression of hepatitis C virus viremia, with no evidence of viral resistance or side effects in the treated animals [125]. This encouraging result highlights the scaling up of miRNAs from laboratory to translational research [165, 166]. Although there are still numerous hurdles in the development of miRNAs as a novel class of therapeutics, the available findings indicate the great potential of miRNAs in lung cancer therapy.
To date, a compelling body of evidence points to the direct involvement of miRNAs in lung carcinogenesis. Combined with high tissue specificity and stability in formalin-fixed paraffin-embedded (FFPE) tissues and bodily fluids, miRNA use in diagnosis, classification and prediction of disease course has become an appealing area of biomarker and therapeutic research. With more discoveries on the horizon, miRNA-based signatures could become prominent clinical tools for patient management and care.
7 Future Directions
As next-generation sequencing approaches become more affordable, they will begin to dominate the discovery field in miRNA research, uncovering novel miRNAs and their associations with lung oncopathology. Linking individual miRNAs to their respective targets within the context of complex pathway networks that are commonly deregulated in cancer would be crucial in painting the ‘big picture’ underlying these cellular processes. While the miRNA signatures reported thus far provide evidence for the translational value of miRNAs and their utility as theranostics, much validation is still needed. In the future, we can expect to see standardization of sample collection techniques, discovery platforms and data analysis techniques to aid in cross-study comparison of results. Overall, miRNAs harbor immense potential in various areas of cancer biology, and realization of their potential as therapeutic targets is simply a matter of time.
References
Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63:11–30
Nishio K, Nakamura T, Koh Y, Suzuki T, Fukumoto H, Saijo N (1999) Drug resistance in lung cancer. Curr Opin Oncol 11:109–115
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Boeri M, Pastorino U, Sozzi G (2012) Role of microRNAs in lung cancer: microRNA signatures in cancer prognosis. Cancer J 18:268–274
Di Lisio L, Martinez N, Montes-Moreno S, Piris-Villaespesa M, Sanchez-Beato M, Piris MA (2012) The role of miRNAs in the pathogenesis and diagnosis of B-cell lymphomas. Blood 120:1782–1790
Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M (2012) MicroRNAs in cancer management. Lancet Oncol 13:e249–e258
Melo SA, Kalluri R (2012) Molecular pathways: microRNAs as cancer therapeutics. Clin Cancer Res 18:4234–4239
Blower PE, Chung JH, Verducci JS et al (2008) MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther 7:1–9
Du L, Subauste MC, DeSevo C (2012) miR-337-3p and its targets STAT3 and RAP1A modulate taxane sensitivity in non-small cell lung cancers. PLoS One 7:e39167
Galluzzi L, Morselli E, Vitale I et al (2010) miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res 70:1793–1803
Kim J, Kang Y, Kojima Y et al (2013) An endothelial apelin-FGF link mediated by miR-424 and miR-503 is disrupted in pulmonary arterial hypertension. Nat Med 19:74–82
Maftouh M, Avan A, Galvani E, Peters GJ, Giovannetti E (2013) Molecular mechanisms underlying the role of microRNAs in resistance to epidermal growth factor receptor-targeted agents and novel therapeutic strategies for treatment of non-small-cell lung cancer. Crit Rev Oncog 18:317–326
Shanker M, Willcutts D, Roth JA, Ramesh R (2010) Drug resistance in lung cancer. Lung Cancer Targets Ther 1:4
Nooter K, Stoter G (1996) Molecular mechanisms of multidrug resistance in cancer chemotherapy. Pathol Res Pract 192:768–780
Krishna R, Mayer LD (2000) Multidrug resistance (MDR) in cancer. Mechanisms, reversal using modulators of MDR and the role of MDR modulators in influencing the pharmacokinetics of anticancer drugs. Eur J Pharm Sci 11:265–283
Liu FS (2009) Mechanisms of chemotherapeutic drug resistance in cancer therapy – a quick review. Taiwan J Obstet Gynecol 48:239–244
Gottesman MM (2002) Mechanisms of cancer drug resistance. Annu Rev Med 53:615–627
Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM (1999) Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu Rev Pharmacol Toxicol 39:361–398
Townsend DM, Tew KD (2003) The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene 22:7369–7375
Eastman A, Schulte N (1988) Enhanced DNA repair as a mechanism of resistance to cis-diamminedichloroplatinum(II). Biochemistry 27:4730–4734
Kavallaris M, Kuo DY, Burkhart CA et al (1997) Taxol-resistant epithelial ovarian tumors are associated with altered expression of specific beta-tubulin isotypes. J Clin Invest 100:1282–1293
Sethi T, Rintoul RC, Moore SM et al (1999) Extracellular matrix proteins protect small cell lung cancer cells against apoptosis: a mechanism for small cell lung cancer growth and drug resistance in vivo. Nat Med 5:662–668
Morin PJ (2003) Drug resistance and the microenvironment: nature and nurture. Drug Resist Updat 6:169–172
Green SK, Frankel A, Kerbel RS (1999) Adhesion-dependent multicellular drug resistance. Anticancer Drug Des 14:153–168
Urgard E, Vooder T, Vosa U et al (2011) Metagenes associated with survival in non-small cell lung cancer. Cancer Inform 10:175–183
Chen HY, Yu SL, Chen CH et al (2007) A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med 356:11–20
Shedden K, Taylor JM, Enkemann SA et al (2008) Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 14:822–827
Heller G, Weinzierl M, Noll C et al (2012) Genome-wide miRNA expression profiling identifies miR-9-3 and miR-193a as targets for DNA methylation in non-small cell lung cancers. Clin Cancer Res 18:1619–1629
Bae S, Lee EM, Cha HJ et al (2011) Resveratrol alters microRNA expression profiles in A549 human non-small cell lung cancer cells. Mol Cells 32:243–249
Gao W, Lu X, Liu L, Xu J, Feng D, Shu Y (2012) MiRNA-21: a biomarker predictive for platinum-based adjuvant chemotherapy response in patients with non-small cell lung cancer. Cancer Biol Ther 13:330–340
Calin GA, Sevignani C, Dumitru CD et al (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 101:2999–3004
Calin GA, Dumitru CD, Shimizu M et al (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 99:15524–15529
Jazbutyte V, Thum T (2010) MicroRNA-21: from cancer to cardiovascular disease. Curr Drug Targets 11:926–935
Zhong Z, Dong Z, Yang L, Gong Z (2012) miR-21 induces cell cycle at S phase and modulates cell proliferation by down-regulating hMSH2 in lung cancer. J Cancer Res Clin Oncol 138:1781–1788
Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH (2010) MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta 411:846–852
Yanaihara N, Caplen N, Bowman E et al (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9:189–198
Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES (2008) Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem 54:1696–1704
Rotunno M, Zhao Y, Bergen AW et al (2010) Inherited polymorphisms in the RNA-mediated interference machinery affect microRNA expression and lung cancer survival. Br J Cancer 103:1870–1874
Lu Z, Liu M, Stribinskis V et al (2008) MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 27:4373–4379
Fassan M, Pizzi M, Giacomelli L et al (2011) PDCD4 nuclear loss inversely correlates with miR-21 levels in colon carcinogenesis. Virchows Arch 458:413–419
Garofalo M, Di Leva G, Romano G et al (2009) miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 16:498–509
Garofalo M, Quintavalle C, Di Leva G et al (2008) MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene 27:3845–3855
Navarro A, Marrades RM, Vinolas N et al (2009) MicroRNAs expressed during lung cancer development are expressed in human pseudoglandular lung embryogenesis. Oncology 76:162–169
Volinia S, Calin GA, Liu CG et al (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 103:2257–2261
Jiang S, Zhang HW, Lu MH et al (2010) MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res 70:3119–3127
O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435:839–843
Hayashita Y, Osada H, Tatematsu Y et al (2005) A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 65:9628–9632
Ebi H, Sato T, Sugito N et al (2009) Counterbalance between RB inactivation and miR-17-92 overexpression in reactive oxygen species and DNA damage induction in lung cancers. Oncogene 28:3371–3379
Matsubara H, Takeuchi T, Nishikawa E et al (2007) Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene 26:6099–6105
Reinhart BJ, Slack FJ, Basson M et al (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403:901–906
Raponi M, Dossey L, Jatkoe T et al (2009) MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res 69:5776–5783
Lee YS, Dutta A (2007) The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21:1025–1030
Kumar MS, Erkeland SJ, Pester RE et al (2008) Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A 105:3903–3908
Johnson SM, Grosshans H, Shingara J et al (2005) RAS is regulated by the let-7 microRNA family. Cell 120:635–647
Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T (2008) let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis 29:2073–2077
Johnson CD, Esquela-Kerscher A, Stefani G et al (2007) The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67:7713–7722
Chin LJ, Ratner E, Leng S et al (2008) A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res 68:8535–8540
Landi MT, Zhao Y, Rotunno M et al (2010) MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res 16:430–441
Gallardo E, Navarro A, Vinolas N et al (2009) miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis 30:1903–1909
Tarasov V, Jung P, Verdoodt B et al (2007) Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 6:1586–1593
He L, He X, Lim LP et al (2007) A microRNA component of the p53 tumour suppressor network. Nature 447:1130–1134
Mudduluru G, Ceppi P, Kumarswamy R, Scagliotti GV, Papotti M, Allgayer H (2011) Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer. Oncogene 30:2888–2899
Fabbri M, Garzon R, Cimmino A et al (2007) MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A 104:15805–15810
Crawford M, Brawner E, Batte K et al (2008) MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun 373:607–612
Bandi N, Zbinden S, Gugger M et al (2009) miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res 69:5553–5559
Nasser MW, Datta J, Nuovo G et al (2008) Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem 283:33394–33405
Patz EF Jr, Caporaso NE, Dubinett SM et al (2010) National Lung Cancer Screening Trial American College of Radiology Imaging Network Specimen Biorepository originating from the Contemporary Screening for the Detection of Lung Cancer Trial (NLST, ACRIN 6654): design, intent, and availability of specimens for validation of lung cancer biomarkers. J Thorac Oncol 5:1502–1506
Chen X, Ba Y, Ma L et al (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18:997–1006
Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B (2009) Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One 4:e6229
Roth C, Kasimir-Bauer S, Pantel K, Schwarzenbach H (2011) Screening for circulating nucleic acids and caspase activity in the peripheral blood as potential diagnostic tools in lung cancer. Mol Oncol 5:281–291
Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH (2009) Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 10:42–46
Keller A, Leidinger P, Borries A (2009) miRNAs in lung cancer – studying complex fingerprints in patient’s blood cells by microarray experiments. BMC Cancer 9:353
Pottelberge GR, Mestdagh P, Bracke KR et al (2011) MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 183:898–906
Chen SY, Wang Y, Telen MJ, Chi JT (2008) The genomic analysis of erythrocyte microRNA expression in sickle cell diseases. PLoS One 3:e2360
Xie Y, Todd NW, Liu Z et al (2010) Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer 67:170–176
Xing L, Todd NW, Yu L, Fang H, Jiang F (2010) Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol 23:1157–1164
Yu L, Todd NW, Xing L et al (2010) Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer 127:2870–2878
Roa WH, Kim JO, Razzak R et al (2012) Sputum microRNA profiling: a novel approach for the early detection of non-small cell lung cancer. Clin Invest Med 35:E271
Yu SL, Chen HY, Chang GC et al (2008) MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 13:48–57
Patnaik SK, Kannisto E, Knudsen S, Yendamuri S (2010) Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res 70:36–45
Hu Z, Chen J, Tian T et al (2008) Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest 118:2600–2608
Gao W, Yu Y, Cao H et al (2010) Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother 64:399–408
Hu Z, Chen X, Zhao Y et al (2010) Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol 28:1721–1726
Voortman J, Goto A, Mendiboure J et al (2010) MicroRNA expression and clinical outcomes in patients treated with adjuvant chemotherapy after complete resection of non-small cell lung carcinoma. Cancer Res 70:8288–8298
Weidhaas JB, Babar I, Nallur SM et al (2007) MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res 67:11111–11116
Fei J, Lan F, Guo M, Li Y, Liu Y (2008) Inhibitory effects of anti-miRNA oligonucleotides (AMOs) on A549 cell growth. J Drug Target 16:688–693
Blower PE, Verducci JS, Lin S et al (2007) MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther 6:1483–1491
Gaur A, Jewell DA, Liang Y et al (2007) Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res 67:2456–2468
Liu H, D’Andrade P, Fulmer-Smentek S et al (2010) mRNA and microRNA expression profiles of the NCI-60 integrated with drug activities. Mol Cancer Ther 9:1080–1091
Kong W, He L, Coppola M et al (2010) MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem 285:17869–17879
Hwang JH, Voortman J, Giovannetti E et al (2010) Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One 5:e10630
Xie L, Chen X, Wang L et al (2010) Cell-free miRNAs may indicate diagnosis and docetaxel sensitivity of tumor cells in malignant effusions. BMC Cancer 10:591
Yu ZW, Zhong LP, Ji T, Zhang P, Chen WT, Zhang CP (2010) MicroRNAs contribute to the chemoresistance of cisplatin in tongue squamous cell carcinoma lines. Oral Oncol 46:317–322
Ranade AR, Cherba D, Sridhar S et al (2010) MicroRNA 92a-2*: a biomarker predictive for chemoresistance and prognostic for survival in patients with small cell lung cancer. J Thorac Oncol 5:1273–1278
Lee JK, Havaleshko DM, Cho H et al (2007) A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci U S A 104:13086–13091
Mishra PJ, Bertino JR (2009) MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics 10:399–416
Bertino JR, Banerjee D, Mishra PJ (2007) Pharmacogenomics of microRNA: a miRSNP towards individualized therapy. Pharmacogenomics 8:1625–1627
Mishra PJ, Banerjee D, Bertino JR (2008) MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: introducing microRNA pharmacogenomics. Cell Cycle 7:853–858
Mishra PJ, Humeniuk R, Longo-Sorbello GS, Banerjee D, Bertino JR (2007) A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci U S A 104:13513–13518
Li J, Li ZN, Du YJ, Li XQ, Bao QL, Chen P (2009) Expression of MRP1, BCRP, LRP, and ERCC1 in advanced non-small-cell lung cancer: correlation with response to chemotherapy and survival. Clin Lung Cancer 10:414–421
Osawa K (2009) Gene polymorphisms and chemotherapy in non-small cell lung cancer. Zhongguo Fei Ai Za Zhi 12:837–840
Narvaiza I, Aparicio O, Vera M et al (2006) Effect of adenovirus-mediated RNA interference on endogenous microRNAs in a mouse model of multidrug resistance protein 2 gene silencing. J Virol 80:12236–12247
Zhu H, Wu H, Liu X et al (2008) Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol 76:582–588
To KK, Zhan Z, Litman T, Bates SE (2008) Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol Cell Biol 28:5147–5161
Pan YZ, Morris ME, Yu AM (2009) MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol 75:1374–1379
Takagi S, Nakajima M, Mohri T, Yokoi T (2008) Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem 283:9674–9680
Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T (2006) MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res 66:9090–9098
Yu AM (2007) Small interfering RNA in drug metabolism and transport. Curr Drug Metab 8:700–708
Chen K, Rajewsky N (2006) Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet 38:1452–1456
Saunders MA, Liang H, Li WH (2007) Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A 104:3300–3305
Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39:1278–1284
Barnes MR, Deharo S, Grocock RJ, Brown JR, Sanseau P (2007) The micro RNA target paradigm: a fundamental and polymorphic control layer of cellular expression. Expert Opin Biol Ther 7:1387–1399
Ding L, Getz G, Wheeler DA et al (2008) Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455:1069–1075
Cho WC, Chow AS, Au JS (2009) Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer 45:2197–2206
Seike M, Goto A, Okano T et al (2009) MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A 106:12085–12090
Garofalo M, Romano G, Di Leva G et al (2012) EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med 18:74–82
Bryant JL, Britson J, Balko JM et al (2012) A microRNA gene expression signature predicts response to erlotinib in epithelial cancer cell lines and targets EMT. Br J Cancer 106:148–156
Wang YS, Wang YH, Xia HP, Zhou SW, Schmid-Bindert G, Zhou CC (2012) MicroRNA-214 regulates the acquired resistance to gefitinib via the PTEN/AKT pathway in EGFR-mutant cell lines. Asian Pac J Cancer Prev 13:255–260
Rai K, Takigawa N, Ito S et al (2011) Liposomal delivery of MicroRNA-7-expressing plasmid overcomes epidermal growth factor receptor tyrosine kinase inhibitor-resistance in lung cancer cells. Mol Cancer Ther 10:1720–1727
Schaefer U, Voloshanenko O, Willen D, Walczak H (2007) TRAIL: a multifunctional cytokine. Front Biosci 12:3813–3824
Koschny R, Walczak H, Ganten TM (2007) The promise of TRAIL–potential and risks of a novel anticancer therapy. J Mol Med (Berl) 85:923–935
Acunzo M, Visone R, Romano G et al (2012) miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene 31:634–642
Incoronato M, Garofalo M, Urso L et al (2010) miR-212 increases tumor necrosis factor-related apoptosis-inducing ligand sensitivity in non-small cell lung cancer by targeting the antiapoptotic protein PED. Cancer Res 70:3638–3646
Garzon R, Marcucci G, Croce CM (2010) Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov 9:775–789
Lanford RE, Hildebrandt-Eriksen ES, Petri A et al (2010) Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327:198–201
Lindow M, Kauppinen S (2012) Discovering the first microRNA-targeted drug. J Cell Biol 199:407–412
Fabbri M (2012) TLRs as miRNA receptors. Cancer Res 72:6333–6337
Allen KE, Weiss GJ (2010) Resistance may not be futile: microRNA biomarkers for chemoresistance and potential therapeutics. Mol Cancer Ther 9:3126–3136
Trang P, Medina PP, Wiggins JF et al (2010) Regression of murine lung tumors by the let-7 microRNA. Oncogene 29:1580–1587
Fontana L, Fiori ME, Albini S et al (2008) Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One 3:e2236
Trang P, Wiggins JF, Daige CL et al (2011) Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther 19:1116–1122
Esquela-Kerscher A, Trang P, Wiggins JF et al (2008) The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 7:759–764
Wiggins JF, Ruffino L, Kelnar K et al (2010) Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res 70:5923–5930
Muniyappa MK, Dowling P, Henry M et al (2009) MiRNA-29a regulates the expression of numerous proteins and reduces the invasiveness and proliferation of human carcinoma cell lines. Eur J Cancer 45:3104–3118
Gibbons DL, Lin W, Creighton CJ et al (2009) Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev 23:2140–2151
Ma L, Reinhardt F, Pan E et al (2010) Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol 28:341–347
Sarkar FH, Li Y, Wang Z, Kong D, Ali S (2010) Implication of microRNAs in drug resistance for designing novel cancer therapy. Drug Resist Updat 13:57–66
Guo L, Liu Y, Bai Y, Sun Y, Xiao F, Guo Y (2010) Gene expression profiling of drug-resistant small cell lung cancer cells by combining microRNA and cDNA expression analysis. Eur J Cancer 46:1692–1702
Zhu W, Shan X, Wang T, Shu Y, Liu P (2010) miR-181b modulates multidrug resistance by targeting BCL2 in human cancer cell lines. Int J Cancer 127:2520–2529
Wang X, Dong K, Gao P et al (2013) MicroRNA-34a sensitizes lung cancer cell lines to DDP treatment independent of p53 status. Cancer Biother Radiopharm 28:45–50
Zhang S, Zhang C, Li Y, Wang P, Yue Z, Xie S (2011) miR-98 regulates cisplatin-induced A549 cell death by inhibiting TP53 pathway. Biomed Pharmacother 65:436–442
Han Z, Yang Q, Liu B et al (2012) MicroRNA-622 functions as a tumor suppressor by targeting K-Ras and enhancing the anticarcinogenic effect of resveratrol. Carcinogenesis 33:131–139
Feng B, Wang R, Chen LB (2012) MiR-100 resensitizes docetaxel-resistant human lung adenocarcinoma cells (SPC-A1) to docetaxel by targeting Plk1. Cancer Lett 317:184–191
Feng B, Wang R, Song HZ, Chen LB (2012) MicroRNA-200b reverses chemoresistance of docetaxel-resistant human lung adenocarcinoma cells by targeting E2F3. Cancer 118:3365–3376
Zhu X, Li H, Long L (2012) miR-126 enhances the sensitivity of non-small cell lung cancer cells to anticancer agents by targeting vascular endothelial growth factor A. Acta Biochim Biophys Sin (Shanghai) 44:519–526
Zhang X, Zhu J, Xing R et al (2012) miR-513a-3p sensitizes human lung adenocarcinoma cells to chemotherapy by targeting GSTP1. Lung Cancer 77:488–494
Cui EH, Li HJ, Hua F et al (2013) Serum microRNA 125b as a diagnostic or prognostic biomarker for advanced NSCLC patients receiving cisplatin-based chemotherapy. Acta Pharmacol Sin 34:309–313
Franchina T, Amodeo V, Bronte G et al (2014) Circulating miR-22, miR-24 and miR-34a as novel predictive biomarkers to pemetrexed-based chemotherapy in advanced non small cell lung cancer. J Cell Physiol 229:97–99
Oh JS, Kim JJ, Byun JY, Kim IA (2010) Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras. Int J Radiat Oncol Biol Phys 76:5–8
Wang XC, Du LQ, Tian LL et al (2011) Expression and function of miRNA in postoperative radiotherapy sensitive and resistant patients of non-small cell lung cancer. Lung Cancer 72:92–99
Arora H, Qureshi R, Jin S, Park AK, Park WY (2011) miR-9 and let-7g enhance the sensitivity to ionizing radiation by suppression of NFkappaB1. Exp Mol Med 43:298–304
Chen S, Wang H, Ng WL, Curran WJ, Wang Y (2011) Radiosensitizing effects of ectopic miR-101 on non-small-cell lung cancer cells depend on the endogenous miR-101 level. Int J Radiat Oncol Biol Phys 81:1524–1529
Balca-Silva J, Sousa Neves S, Goncalves AC et al (2012) Effect of miR-34b overexpression on the radiosensitivity of non-small cell lung cancer cell lines. Anticancer Res 32:1603–1609
Salim H, Akbar NS, Zong D et al (2012) miRNA-214 modulates radiotherapy response of non-small cell lung cancer cells through regulation of p38MAPK, apoptosis and senescence. Br J Cancer 107:1361–1373
Liu YJ, Lin YF, Chen YF et al (2013) MicroRNA-449a enhances radiosensitivity in CL1-0 lung adenocarcinoma cells. PLoS One 8:e62383
Cho WC (2010) MicroRNAs as therapeutic targets for lung cancer. Expert Opin Ther Targets 14:1005–1008
Cho WC (2010) Conquering cancer through discovery research. IUBMB Life 62:655–659
Young DD, Connelly CM, Grohmann C, Deiters A (2010) Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J Am Chem Soc 132:7976–7981
Ebert MS, Neilson JR, Sharp PA (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4:721–726
Gonzalez-Aseguinolaza G, Prieto J (2010) Durable correction of inherited metabolic liver disorders requires preventing transgene off-targeting from gene therapy vectors: the value of microRNAs. Gastroenterology 139:726–729
Fischer A, Hacein-Bey-Abina S, Cavazzana-Calvo M (2010) 20 years of gene therapy for SCID. Nat Immunol 11:457–460
Rossbach M (2010) Small non-coding RNAs as novel therapeutics. Curr Mol Med 10:361–368
Seto AG (2010) The road toward microRNA therapeutics. Int J Biochem Cell Biol 42:1298–1305
Wahid F, Shehzad A, Khan T, Kim YY (2010) MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta 1803:1231–1243
Nana-Sinkam SP, Croce CM (2013) Clinical applications for microRNAs in cancer. Clin Pharmacol Ther 93:98–104
Uchino K, Ochiya T, Takeshita F (2013) RNAi Therapeutics and Applications of MicroRNAs in Cancer Treatment. Jpn J Clin Oncol 43:596–607
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Gong, Z. et al. (2014). The Role of MicroRNA in Lung Cancer Drug Resistance and Targeted Therapy. In: Sarkar, F. (eds) MicroRNA Targeted Cancer Therapy. Springer, Cham. https://doi.org/10.1007/978-3-319-05134-5_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-05134-5_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-05133-8
Online ISBN: 978-3-319-05134-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)