Abstract
There is mounting evidence that the placenta can be considered as a programming agent of adult health and diseases. Placental weight and shape at term are correlated with the development of metabolic diseases in adulthood in humans. Maternal obesity and malnutrition predispose the offspring to developing metabolic syndrome, a vicious cycle leading to transmission to subsequent generation(s), with differences in response and susceptibility according to the sex of the individual. Adaptations in placental phenotype in response to maternal diet and body composition alter fetal nutrient provision. This finding implies important epigenetic changes. However, the epigenetics of placental development in studies of developmental origins of health and disease (DOHaD) is still poorly documented, particularly concerning overnutrition. We used histology, microarray analyses and epigenetic techniques to investigate the effects of a high fat diet (HFD) or low protein diet on mouse placental development, respectively. We showed for the first time that not only the gene sets but also their biological functions affected by the HFD differed markedly between the two sexes. Remarkably, genes of the epigenetic machinery as well as global DNA methylation level showed sexual dimorphism. Imprinted gene expression was altered, with locus-specific changes in DNA methylation. Thus, these findings demonstrate a striking sexual dimorphism of programming trajectories in response to the same environmental challenge, implicating sex chromosome genes, not just hormones. Explaining the sex-specific causal variables and how males versus females respond and adapt, and to what extent, to environmental perturbations should help physicians and patients anticipate disease susceptibility.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction to the Sexually Dimorphic Effects of Gestational Obesity and Suboptimal Nutrition
According to the Developmental Origins of Adult Health and Disease (DOHaD) concept, environmental conditions during specific windows of mammalian development can have lasting effects on cell fate, organogenesis, metabolic pathways and physiology, thereby influencing life-long physical health and the susceptibility to lifestyle-induced diseases in adulthood (Barker and Osmond 1988; McMillen and Robinson 2005). There is evidence to suggest that maternal overnutrition, gestational diabetes and obesity are deleterious to the health of offspring, inducing the same range of defects as maternal mal- or undernutrition and leading to the development of metabolic syndrome (Armitage et al. 2005; Boloker et al. 2002; Dabelea et al. 2000; Levin and Govek 1998; Nathanielsz et al. 2007) in offspring, with a striking sex specificity (Dunn et al. 2010; Gabory et al. 2009, 2012, 2013; Gallou-Kabani et al. 2007). The number of overweight or obese women of child-bearing age is growing, reaching 25 % in Europe and 50 % in the US (WHO), and potentially triggering a vicious cycle, with transmission to subsequent generations and increasing prevalence of these lifestyle diet-induced disorders. Interestingly, the adverse metabolic consequences of dietary manipulations can be improved or prevented by applying mild food restriction or a normal control diet to the mother (Giraudo et al. 2010; Srinivasan et al. 2006), by reducing maternal obesity by bariatric surgery (Kral et al. 2006; Smith et al. 2009) or by the addition to the maternal diet of specific nutrients involved at different levels of carbon metabolism essential for DNA methylation (Boujendar et al. 2003; Burdge and Lillycrop 2010; Dolinoy et al. 2006; Lillycrop et al. 2005; Torrens et al. 2006; Waterland et al. 2007).
The principal modifiers of disease risk, presentation and treatment include sex and age. Differences between the sexes have been reported for most non-communicable diseases (NCDs), including metabolic diseases, hypertension, cardiovascular disease, cancer and psychiatric and neurological disorders. Sex differences in responses to drugs and diets have also been described, and many DOHaD reports have indicated that exposure to an adverse environment during specific windows of developmental programming may affect the long-term health and susceptibility to NCDs of the offspring in a sex-specific manner (Bale 2011; Barker 1992; Gabory et al. 2009; van Abeelen et al. 2011; Waddell and McCarthy 2012). Large sex differences in the incidence and progression of diseases suggest that sex-biased factors are an untapped source of factors providing protection against disease (Arnold and Lusis 2012).
Sociocultural considerations have largely obscured divergences in the biology of the sexes, which have been studied essentially only in terms of the organizational and activational effects of sex hormones after sexual differentiation. However, unequal gene expression by the sex chromosomes has an impact much earlier, beginning at conception, and may set the backdrop for events in later life (reviewed in Al-Khan et al. 2011; Clifton 2010; Gabory et al. 2009). The sex chromosomes have a disproportionately large impact on human health and disease. The unique evolutionary pathway of the X and Y chromosomes has resulted in these chromosomes having highly atypical gene contents and activities (Graves 2010), resulting in differences between the sexes even before the initiation of adrenal and gonad development (Penaloza et al. 2009).
In all adult tissues examined to date, including the gonads and brain, the expression of different genes with different functions from different networks is modulated in a sex-specific manner. Gene expression analyses, either for candidate genes or at the genome-wide level, show that both the trajectories under basal conditions and those modulating responses differ between the sexes (Gabory et al. 2009). The mechanisms regulating the differences between the sexes in adults have been investigated intensively in the liver, but dimorphic gene expression has also been reported in several mouse tissues, including the kidneys, lacrimal gland and brain (reviewed in Gabory et al. 2009). A microarray analysis revealed that 14 % (brain) to 70 % (liver) of active genes display sexual dimorphism. Interestingly, these genes cluster together not only on the sex chromosomes but also on several autosomes (Yang et al. 2006). van Nas et al. (2009) identified sexually dimorphic modules of networks of genes with highly correlated patterns of expression implicated in genetic and metabolic traits and genes affected by gonadal hormones in the adipose tissue, brain, liver and muscle. In adult liver, the genes mapping to the sex chromosomes play a lesser role in the modulation of sex-specific gene expression than gonadal hormones (Wauthier et al. 2010). Statistically significant sex differences have been reported for 78 % of the metabolites in the human plasma metabolome (Mittelstrass et al. 2011). For this reason, molecular investigations of the similarities and differences between the responses of male and female conceptuses to various maternal exposures are of considerable interest.
Sex differences in the rate of fetal growth have long been recognized (Lubchenco et al. 1963). The sex of the embryo affects the size of both the fetus and the placenta, together with the ability of the placenta to respond to adverse stimuli (Clifton 2010; Clifton et al. 2010; Mao et al. 2010). In mice and cattle, accelerated development is already evident in XY blastocysts; cell division among male embryos occurs more rapidly than in female embryos (Mittwoch 1993) and, in humans, boys grow more rapidly than girls from the earliest stages of gestation (Eriksson et al. 2009). These differences may start as early as the blastocyst stage in bovines: one third of the genes actively expressed in bovine blastocysts display sexually dimorphic expression (Bermejo-Alvarez et al. 2008, 2010). Analysis of genes involved in amino acid transport and metabolism identified sex differences both in average placental gene expression between male and female and in the relationships between placental gene expression and maternal factors (Sturmey et al. 2010). Ontological analysis of such data suggests a higher global transcriptional level in females and greater protein metabolism levels in males. Specifically, global glucose metabolism and pentose-phosphate pathway activity are twice and four times greater in bovine male vs. female blastocysts, respectively, with similar metabolic differences being seen for human embryos at the same stages (for review Bermejo-Alvarez et al. 2011). At birth, placental weights and FPI (fetus-to-placenta weight ratio index, reflecting placental efficiency) tend to be greater in boys than girls (Lampl et al. 2010). These observations suggest that males may be both more responsive to growth-promoting influences and more susceptible to supply disturbances (Lampl et al. 2010; Wallace et al. 2012).
As a critical messenger between the maternal environment and the fetus, the placenta may play a key role not only in buffering environmental effects transmitted by the mother but also in expressing and modulating effects due to preconceptional exposure of both the mother and the father to stressful conditions. Sexually dimorphic patterns of expression have recently been reported for individual genes or pathways in placentas from humans and rodents, potentially accounting for differences in the sensitivity of male and female fetuses to maternal diet (reviewed in Gabory et al. 2009). However, few groups have studied global sexual dimorphism in the placenta with microarrays, focusing in particular on the impact of maternal diet, asthma or stress on placental gene expression through systematic investigations of the relationship between diet and the expression of sexually dimorphic genes (Mao et al. 2010; Sood et al. 2006). Even fewer studies have investigated the associated epigenetic changes (Gabory et al. 2012; Gallou-Kabani et al. 2010).
Figure 1 shows how such influences may operate on the transmission of environmental influences to subsequent generation(s) and illustrates the central role of the placenta in the sex specificity of these parent-of-origin effects. Support for the possibility of inter- and transgenerational effects is also emerging, making it important to know the role played by the placenta and the possible maternal and or paternal epigenetic imprints carried by the gametes forming the zygote. Indeed, maternally or paternally transmitted, non-erased epigenetic alterations of key developmental genes may perturb early trophoblast development in a sex-specific manner (Fig. 1).
Sex-specific transmission of exposure to environment to subsequent generations. Environmental factors—including nutrition, psychosocial stress, toxins, endocrine disruptors, tobacco, alcohol, and microbiota—impact individual (F0) epigenetic landscapes, and hence gene pathways and networks, in ways that differ between the sexes. For example, maternal and paternal preconceptional exposures can alter gamete epigenetic marks, some of which can be transmitted to the subsequent (F1) generation. Additionally consequences of maternal F0 exposure during pregnancy (stress, metabolism, diet, hormonal changes…) can be transmitted from the maternal to the fetal compartment via the placenta in a sex-specific manner and can affect F1 tissue development. Programming of somatic tissues can lead to changes in long-term health outcomes in the first generation. Moreover, primordial germ cells, which develop and undergo reprogramming during fetal development, can also be affected by F0 maternal environment and contribute genetic and epigenetic information to the F2 generation. Maternal and paternal lineages affect the transmission of such influences differently. In particular, intergenerational effects of F0 exposure on the maternal lineage can be seen in the F1 and F2 generations, and transgenerational to F3, whereas the paternal lineage can transmit intergenerational effects to the F1 and transgenerational effects to the F2 and F3 generations
Placenta Is a Sexually Dimorphic Organ
During pregnancy, the placenta maintains fetal homeostasis by regulating nutrient transfer from the mother to the fetus. This ‘gateway’ to the fetus is affected by numerous environmental factors, each of which may modifies epigenetic marks and gene expression within the placenta (Novakovic and Saffery 2012). Various environmental factors, including nutrient status and tissue oxygenation, may modify placental development and function (Cross and Mickelson 2006; Liang et al. 2010). Studies of rodents and large animals have shown placental development to be highly adaptable, with many means of compensating for poor nutritional conditions (Coan et al. 2010; Constância et al. 2005; Gheorghe et al. 2010; Mao et al. 2010; Reynolds et al. 2010).
The placenta has traditionally been considered an asexual organ, and many studies focusing on the placenta have not taken the sex of the embryo into account (Clifton 2010). Predisposition to various diseases in adulthood, including type 2 diabetes (T2D), hypertension and cardiovascular disease (CVD), is affected by the food consumed by the mother during pregnancy. It thus appears likely that sex-specific placental functions contribute to the differences in frequency of these diseases between the sexes. The sex of the embryo affects the size of both the fetus and the placenta, together with morbidity and the ability of the placenta to respond to adverse stimuli (Clifton 2010; Clifton et al. 2010; Mao et al. 2010). Moreover, Wang et al. (2011) showed that the maternal decidual renin-angiotensin system (RAS) in humans is regulated in a sex-specific manner at term, both before and after labor. Thus, the sex of the fetus affects gene expression, indicating possible functional differences as a function of fetal sex.
The fact that boys tend to be longer at birth than girls suggests that the placenta may be more efficient for male than for female fetuses, although it may have a smaller reserve capacity. From the earliest stages of gestation, boys grow more rapidly than girls and are therefore at greater risk of undernutrition (Eriksson et al. 2009). Unfavorable programming, whether immediately before conception or during gestation, may result in various defects potentially translated into differences in susceptibility to disease between boys and girls (Clifton 2010; Dunn et al. 2010; Eriksson et al. 2009; Gabory et al. 2009; Gallou-Kabani et al. 2010; Ng et al. 2010). Thus, the placenta constitutes an ideal organ in which to study the sensing, by the fetus, of stresses, starvation, endocrine disruptors and obesity-prone diets or lifestyles, in a sex-specific manner.
Neither Sex Nor Maternal HFD Triggers Gross Morphological Changes in Placental Layers
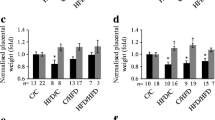
We investigated the impact of a high-fat diet (HFD) during gestation on fetoplacental development in a mouse model. We collected placentas in the middle of the fetal period, when the morphological development of the placenta is complete and fetal growth is maximal (Gallou-Kabani et al. 2010). We observed a major effect of sex on placental weight, with male placentas being larger than female placentas. Under the HFD, the placental weight was increased in both males and females. There was no effect of sex or diet on fetal weight. Therefore the FPI was greater in females than in males and was reduced under a HFD.
Experimental and epidemiological studies in humans and animal models have demonstrated that predisposition to impaired glucose tolerance, blood pressure and coronary heart disease is associated with either a low or high fetus-to-placenta weight ratio and a decrease in FPI under maternal HFD, for both male and female placentas (Gallou-Kabani et al. 2010). This finding indicates a change in placental efficiency, which could be due to either a change in placental development or to gross morphological changes in the different layers. Using in situ hybridization with several probes, we could not detect any changes in the respective proportion of the two layers (Gallou-Kabani et al. 2010). To assess whether these weight changes were associated with morphological changes, we analyzed the placenta with classical histological approaches. No obvious changes were observed in the structure of the labyrinth or spongiotrophoblast. The proportion of the placenta occupied by the labyrinth was not affected by diet or sex of the offspring. However, as shown by the dysregulation of important genes involved in placental development, we cannot exclude subtle changes (Cross et al. 2003; Rawn and Cross 2008). Altogether this finding suggests that the observed increase in placental weight was distributed evenly between the layers. Moreover, overall placental and labyrinth shapes, estimated from minor and major lengths, did not differ between the four groups. Thus, neither the sex of the offspring nor HFD in the mother triggered gross morphological changes in the respective structure and size of the placental layers (Gabory et al. 2012).
Sexual Dimorphism and Sensitivity to Diet for 9/20 Genes from Four Clusters of Imprinted Genes
It has been suggested that changes in imprinted gene dosage in the placentas may compromise the prenatal control of nutritional resources (Charalambous et al. 2007). Indeed, monoallelic behavior and sensitivity to changes in regional epigenetic state render imprinted genes both vulnerable and adaptable. However, the underlying mechanisms remain unclear.
We investigated whether a HFD during pregnancy modified the expression of imprinted genes in the placenta. We compared gene expression patterns in total placenta homogenates, for male and female offspring, by the RT-qPCR analysis of 20 imprinted genes. Sexual dimorphism and/or sensitivity to diet were observed for nine genes from four clusters on chromosomes 6, 7, 12 and 17. Six genes (Slc22a1, Slc22a2, Slc22a3, Rtl1, Dlk1 and Dio3) displayed changes in expression pattern when the mother was fed the HFD. We observed sex-specific sensitivity to the HFD, with effects limited to or more pronounced in the female placenta for Dlk1, Dio3, Slc22a1 or in the male placenta for Slc22a2 only. Our results are therefore consistent with previous findings that female placentas display more striking changes in gene expression in response to maternal diet than do male placentas. As suggested by Penaloza et al. (2009), this difference in cell behavior and sensitivity appears to be driven by the genetic sex of the cells, with the effects of factors such as hormones subsequently being superimposed on this difference.
Concerning the chromosome 17 cluster (Slc22a2, Slc22a1 and Slc22a3), the sex steroid hormone estrogen down-regulates renal organic cation transport in animals and may contribute to sex-related differences in xenobiotic accumulation and excretion (Asaka et al. 2006; Pelis et al. 2007; Urakami et al. 2000). However, caution is required when extrapolating transport-related sex differences between species and organs. These data on sexual dimorphism in organic cation transport are nonetheless potentially interesting when trying to understand the differences between the sexes in terms of the response in the placenta. For the chromosome 12 cluster, an effect of diet was observed for the paternally expressed Dlk1, Rtl1 and Dio3 genes but not for the maternally expressed Gtl2/meg3 genes, with female placentas again being more sensitive than male placentas to the effects of the HFD.
This study was the first to demonstrate that the placentas of male and female fetuses from mothers fed a HFD displayed changes in the expression of selected imprinted genes from different clusters, with these changes differing between the sexes.
Differential CpG Methylation of the Igf2r DMR
The transporter genes Slc22a2 and Slc22a3 are imprinted specifically in mouse placenta (Wagschal and Feil 2006). The promoters of the repressed paternal alleles of these genes do not display DNA methylation (Sleutels et al. 2002). The Igf2r imprint control element (ICE), which is a differentially methylated région (DMR) containing 30 CpG, plays a crucial role in regulating many imprinted genes in this cluster. We therefore investigated whether adaptation of the nutrient supply to fetal demand in pregnant mice fed a HFD involved the ICE/DMR regulating these important placental transporter systems (Gallou-Kabani et al. 2010). In a bisulphite-sequencing analysis of all 30 CpG within the DMR of the chromosome 17 cluster, we found no statistically significant difference between the sexes or the two diets. However, a CpG by CpG analysis revealed sex- and diet-specific differential methylation of individual CpGs in two conspicuous subregions of the DMR. Significantly different levels of methylation between the sexes were found for the first four CpGs in fetuses from mothers fed the HFD. Similarly, different levels of methylation between the sexes were found for the next five CpG and for CpG 20 in fetuses from mothers fed the control diet (CD). CpG 2 was the only CpG displaying both diet response and sexual dimorphism (Gallou-Kabani et al. 2010).
Bioinformatic analysis suggested that the CpGs displaying sex- and/or diet-specific differential methylation in the DMR might lie within recognition elements or binding sites for transcription factors or factors involved in chromatin remodelling, or within a higher-order chromatin architecture including Pax4, Smarca3, Nrf2/Arp, Ppar/Rxr, Egr3, Rxr, Stat6 (Gallou-Kabani et al. 2010).
Placental Gene Expression Is Sexually Dimorphic Under CD and Under HFD
Using Affymetrix exon microarrays, we analyzed gene expression in our placenta samples. We obtained expression data for four groups: female (F) or male (M) placentas from mothers under the control (CD) or high-fat (HFD) diet (F CD, F HFD, M CD and M HFD). First, we analyzed the data to detect sexual dimorphism in gene expression by comparing M CD vs F CD and M HFD vs F HFD. Second, to describe the placental response to a maternal HFD challenge, we compared F HFD vs F CD and M HFD vs M CD. Altogether, for these four comparisons, a bioinformatic analysis was carried out using the Ingenuity Pathway Analysis (IPA) software.
We first investigated whether there was a basal sexual dimorphism between female and male placentas from mothers fed a CD during pregnancy. We detected 104 genes displaying sexual dimorphism, of which 97 were mapped in IPA: 54 with higher expression in female than in male placentas and 43 with higher expression in male than in female placentas (Fig. 2). Among the genes mapped in IPA, 97 genes were sexually dimorphic in CD conditions and 93 genes in HFD conditions. Interestingly, only 11 genes displayed sex-dimorphic expression in both CD and HFD placentas. Among these 11 genes, Adra1a, Eif2s3x, Kdm5c (Jarid1c) and Ogt were more strongly expressed in female placentas (the last three are located on the X-chromosome) and Cst6, Ppp2r2c, Zfp36l2, Ddx3y, Eif2s3y and Kdm5d (Jarid1d) were more strongly expressed in male placentas. Interestingly, four Y-linked genes were represented on the microarray: Ddx3y, Eif2s3y, Kdm5d and Sry. The expression of this last gene is undetectable in placenta. Therefore, we can observe male-specific expression for three of four Y-linked genes in our microarray data. Notably, the amplitude of the differences in gene expression levels observed between males and females was very close under a CD and under a HFD. Interestingly, concerning the known classical sex hormone receptor genes, only the expressions of the Estrogen Receptor Related beta (Esrrb) or Nr3b2 and the hormone nuclear receptor Retinoid X receptor gamma (Rxrg) or Nr2b3 genes were sexually dimorphic under a CD (more expressed in male and more expressed in female, respectively).
Sexual dimorphism in placental gene expression under control diet (CD) or high fat diet (HFD). The Venn diagram illustrates that, among the genes mapped by Ingenuity Pathway Analysis (IPA), 97 were sexually dimorphic in CD conditions and 93 in HFD conditions. Eighty-six genes were dimorphic in basal conditions (CD) but not in HFD placentas, 81 were not dimorphic in basal conditions but became dimorphic under HFD conditions and only 11 genes displayed sex-dimorphic expression in both CD and HFD placentas. The tables corresponds to the list of biological functions and their annotations associated with the gene sets, as analyzed by IPA
The Transcriptional Response of Placenta to Maternal HFD Is Female- and Male-Specific
To characterize the effect of the maternal HFD on gene expression and whether a HFD triggers different changes in males and females, we analyzed this effect in the two sexes separately. We compared the female HFD with the female CD microarrays and the male HFD with the male CD microarrays. We found that 168 genes were affected by maternal HFD in female placentas, with 164 genes mapped in IPA, whereas 190 genes were affected in male placentas, with 187 mapped in IPA. About half of these genes were upregulated and half downregulated in both female and male placentas (Fig. 3). Interestingly, only 16 genes were dysregulated in both sexes, with the same amplitude, namely CcdC56, Sh3bgrl3 and Sumo1, which were upregulated, and Cxcl2, GcGr, Klk8, Mpg, Pcca, Pdgfß, Pla2g15, Ppp2r3c, Rhbdf2, Slc7a2, Tmem62 and Zfp37, which were downregulated. Among dysregulated genes and gene families, some have been reported to be important for placental development: Adra2a, Ceacam10, Crabp2, Gata1, Gcm1, Mmp1a, Mmp 16, Pdgfß and Rarb (Cross et al. 2003; Rawn and Cross 2008; Watson and Cross 2005).
Effect of maternal HFD on gene expression in female or in male placentas. The Venn diagram illustrates that, among the genes mapped by Ingenuity Pathway Analysis (IPA), 164 were affected by maternal HFD in female placentas, whereas 187 were affected in male placentas. A total of 148 genes are specific to the female response and 171 are specific to the male response. Only 16 genes are affected by maternal HFD in both sexes. The tables corresponds to the list of biological functions and their annotations associated with the gene sets, as analyzed by IPA
Several studies have reported differences in gene expression between male and female placentas in humans and mice (Clifton et al. 2009; Mao et al. 2010; Sood et al. 2006; Suter et al. 2010). In humans, Clifton et al. showed that the placenta adapts in a sexually dimorphic manner to chronic maternal asthma. The growth of female fetuses is reduced, increasing chances of survival, whereas normal growth of male fetuses is associated with a poor outcome in the case of acute asthma exacerbation (Clifton 2010; Clifton et al. 2009). In mice, Mao et al. (2010) reported that, in E12.5 mice fed a low-fat diet (10 %) or a very HFD (60 %), similar to the CD and HFD used in this study, respectively, or an intermediate chow diet (25 %), there were more changes in gene expression with both diets in female than male placentas. In the present report, but at a later stage, E15.5, we confirmed that the affected gene sets differed between females and males. However our transcriptomic analysis provides no evidence for a greater reactivity to maternal HFD in either male or female mouse placentas in terms of placental and fetal growth or the numbers of dysregulated genes. Moreover, using the Ingenuity Pathway Analysis, we show for the first time that the gene sets change not only from a quantitative point of view but also, more strikingly, from a qualitative point of view. Indeed, the associated biological functions affected by the HFD differed markedly between the two sexes. In the absence of a greater reactivity but with clearly different trajectories, it is difficult to say whether one sex copes better than the other under a HFD, as previously suggested in other contexts (Clifton 2010; Eriksson et al. 2009). Differences in adaptation between males and females may therefore be context-, species- and stage-specific.
Diet Effect and Sexual Dimorphism in Global DNA Methylation in Mouse Tissues and Placenta
Global DNA methylation was assessed by the LUMA technique, in which the ratio of genomic DNA digested by the methylation-sensitive enzyme HpaII to that digested with the methylation-insensitive enzyme MspI indicates the level of cytosine demethylation (Karimi et al. 2006). An effect of sex was observed under the CD. Male placentas displayed lower levels (3.3 %) of methylation than female placentas. Diet had an effect on global % methylation but was statistically significant only in females (2.4 %). Female placentas from mothers fed the HFD displayed lower levels of methylation. Such a global hypomethylation was also observed in brains of offspring of HFD-fed mouse mothers (Vucetic et al. 2010). However, it remains difficult to speculate about the potential role of the high fat/low carbohydrate composition of the diet on the one carbon metabolism, in the absence of relevant mechanisms to account for a potential link.
This dimorphism may be due to the presence of an inactive X (Xi) chromosome in the female. However, Weber et al. (2005) overturned previous views by showing that Xi was hypermethylated at only a subset of gene-rich regions and, unexpectedly, displayed overall hypomethylation with respect to its active counterpart. Hellman and Chess (2007) have shown that the active X (Xa) chromosome in females has levels of allele-specific methylation that are twice those of Xi. A bipartite methylation-demethylation program results in Xa-specific hypomethylation at gene promoters and hypermethylation at gene bodies in both male and female active Xa chromosomes (Hellman and Chess 2007).
We investigated this difference in methylation further by assessing the methylation levels of the two major repetitive elements containing most of the genomic 5-methylcytosine bases: LINE-1 (long interspersed nucleotide element-1) and SINE-1 (short interspersed nucleotide element-1), represented by human Alu elements and the homologous mouse B1 elements. The methylation levels of both LINE-1 and SINE-1 have been reported to be good indicators of cellular 5-methylcytosine level (i.e., global DNA methylation level; Fryer et al. 2009; Jeong and Lee 2005). In our placenta model, no difference in the level of LINE-1 or B1 repetitive element methylation was observed between the sexes or between the diets (CD and HFD). These differences are therefore probably located in non-genic regions, gene bodies and centromeric heterochromatin.
Effect of Maternal Diet and Sexual Dimorphism on the Epigenetic Machinery Enzymes
The mouse extraembryonic membranes, yolk sac and placenta are characterized by global undermethylation of DNA with respect to embryonic somatic lineages. The major differences in methylation between the two lineages probably affect non-gene regions such as, for example, centromeric heterochromatin (Ng et al. 2008). Indeed, in both chorionic villi and placental fibroblasts, large differences have been observed both globally and between various chromosome structures within individual metaphases (Kokalj-Vokac et al. 1998). As previously mentioned, female placentas displayed significantly higher levels of methylation than male placentas under control conditions (Gallou-Kabani et al. 2010). We also showed an effect of maternal HFD, with hypomethylation in female placentas only (Gallou-Kabani et al. 2010). However, the levels of expression of neither Dnmt3a, Dnmt3b nor Dnmt1 differed between the sexes or between diets. In contrast, this hypomethylation was consistent with the observation of Dnmt3l downregulation only in female HFD placentas. Dnmt3l has no DNA methylase activity but is a cofactor of the de novo DNA methyltransferases (Kobayashi et al. 2012; Smallwood et al. 2011). Our data do not exclude a role for Dnmt3a, Dnmt3b and Dnmt1 in global methylation differences. Indeed the expression of these genes and the levels of the corresponding proteins may have varied before the E15.5 stage. Time course studies and studies of the cell type distribution of DNA methylation during development are therefore of interest.
Six other epigenetic machinery genes, Kmt1a, Kmt1b, Kmt2f, Kdm5c, Kdm5d, and Prmt7, were dysregulated under maternal HFD as an effect of diet, sex of the fetus, or both. Kmt1a and Kmt1b encode methyltransferases involved in trimethylation of the lysine 9 residue of histone H3 (H3K9me3 is mostly associated with a repressed chromatin state). The two paralogues, Kdm5c and Kdm5d, demethylases of lysine 4 of histone H3 (H3K4me3 is mostly associated with an active chromatin state), map to the X and Y chromosomes, respectively. Kdm5c escapes X-inactivation and is more strongly expressed in females than in males (Li and Carrel 2008). Kdm5c and Kdm5d are highly similar in nucleotide and amino acid sequence, but whether the role and targets of these two enzymes are identical, divergent or with partial compensation in their functions is still unclear (Xu et al. 2008). Prmt7 is an arginine methyl transferase (H3R2) and Setd1a (Kmt2f) is a H3K4 methyltransferase. Little is known about these two enzymes and their role is not described in placenta.
The proteins encoded by Y-linked genes may or may not have the same functions, the same target sequences or the same patterns of expression, according to age or tissue, as their X paralogues. In our study, in placentas of HFD-fed mouse mothers, the Y- and X-linked histone demethylase paralogue genes Kdm5c and Kdm5d were sexually dimorphic. In another report, in mouse brain, expression of the Y version of the gene in male mice did not compensate for the dosage imbalance between the two sexes in the expression of their X homologs escaping X-inactivation. Figure 4 shows that, in placentas from mothers on a CD or HFD, the Y-linked Kdm5d gene expression in males was not able to compensate for the expression of Kdm5c, its X-linked paralogue escaping XIC, in females (Gabory et al. 2012). Thus the epigenetic enzymes produced by these two genes could mark the epigenome in a sex-specific manner, both at the quantitative and qualitative levels (Xu et al. 2002).
Analysis of the mRNA expression of the Kdm5c and Kdm5d paralogues. Three PCR primer pairs have been designed for recognizing specifically either Kdm5c or Kdm5d cDNA and for recognizing both Kdm5c/5d cDNA. Their expression was studied in male and female placentas in pregnant female mice fed either a control diet (CD) or a high-fat diet (HFD) from E0.5 to sacrifice at E15.5 stage. Kdm5c expression is higher in females (pink bars) than males (blue bars), and Kdm5d is expressed only in males, regardless of maternal diet. The Kdm5c/5d PCR shows that the combined expression of Kdm5d and Kdm5c expression in males is not of equivalent magnitude as the expression of Kdm5c from both alleles in females
The involvement of all these enzymes in an important network is consistent with (1) documented crosstalk between the H3K4 and H3K9 methylation marks, with H3K9 methylation partly controlled by the Kmt1a and Kmt1b enzymes, and (2) crosstalks between histone methylation and acetylation and DNA methylation. Dnmt3l has been shown to recruit histone deacetylases (Deplus et al. 2002) and to interact with H3K4me3 (Ooi et al. 2007). Moreover, the H3K4 demethylase Kdm5c interacts with H3K9me3, which can be methylated by Kmt1a and 1b (Iwase et al. 2007). It will therefore be important to determine how the crosstalk between key repressive or activating marks and their modifying enzymes can be disturbed by environmental challenges, leading to developmental and metabolic malprogramming.
Effects of Maternal Diet and Sexual Dimorphism on Relevant Histone Marks
Since Kdm5c, Kdm5d, Kmt1a and Kmt1b enzymes are responsible for histone modifications of H3K4 and H3K9, respectively, we investigated whether the differences in gene expression led to differences in the global levels of pertinent histone methylation marks. In line with the changes in expression of histone modifying enzymes, we analyzed global histone modifications. The H3K4me3 signal was not sexually dimorphic, as might have been expected given the dimorphic expression of Kdm5c and Kdm5d, and was equivalent in both diets despite the dysregulation of Kmt2f. Similarly, H3K9me3 was not affected by diet despite the downregulation of Kmt1a and Kmt1b. However, Western blotting provides a global evaluation of histone marks and cannot exclude subtle changes in particular target sequences of these enzymes. Thus, diet itself or the mechanisms used by cells to compensate for dietary imbalances for adaptation must have a major impact on the epigenetic machinery. As previously highlighted for DNA methylation, time course studies and studies of the cell type distribution of histone marks during development are required.
Not Just Hormones: Role of X and Y Chromosome-Linked Genes
Increasing numbers of reports are challenging the traditional view of the influences of gonadal hormones and highlighting other roles for sex chromosomes (reviewed in Davies and Wilkinson 2006; Dunn et al. 2010; Gabory et al. 2009, 2013; Howerton and Bale 2012). In bovine blastocysts, sex determines the expression levels of one third of all actively expressed genes (Bermejo-Alvarez et al. 2010). Sexual dimorphism has also been observed in total embryonic cells isolated from mice at E10.5, before the occurrence of sexual differentiation. These cells responded differently to the dietary stressors applied, even before the production of fetal sex hormones (Penaloza et al. 2009). The genes present on the sex chromosomes (which are asymmetrically inherited between males and females) may influence sexually dimorphic gene expression (Davies and Wilkinson 2006). In most placental mammals, X inactivation is random. However, extraembryonic X inactivation is thought to be paternally imprinted in rodents. At least 150 loci are known to escape inactivation and may therefore be expressed from both X chromosomes (Carrel and Willard 2005). Studies in mice and rats demonstrating sex differences in placental responses to changes in the maternal environment may thus indicate a role for these escaped genes, as the placentas of female fetuses may produce twice as much of the corresponding proteins as those of male fetuses (Howerton and Bale 2012). Alternatively, the small number of expressed genes present on the Y chromosome may be involved. Many X-homologous regions are found on the Y chromosome, but most do not recombine and are referred to as male-specific regions. These regions contain 78 single and multicopy genes encoding about 27 different proteins in humans. In total, 29 genes are conserved in the pseudo-autosomal regions (PARs) of the X and Y chromosomes. In human term placentas, Sood et al. (2006) showed that many of the sex-correlated genes are located on the sex chromosomes, but that some are autosomal. Thus X- and Y-linked genes may modulate the expression of different sets of autosomal genes, leading to physiological differences between males and females (Gabory et al. 2013).
Conclusions: Why Sex Matters
These findings highlight the importance of studying both sexes in epidemiological protocols or dietary interventions in both humans and experimental animal models. Our results pave the way for explorations concerning the possible targeting, by fatty acids and other nutrients, of conspicuous regions in the genome harboring binding sites for the recruitment of diet- and tissue-specific chromatin remodeling complexes. Elucidation of the ways in which epigenetic modifications fix the effects of early environmental events, in a sex-specific manner, ensuring sustained responses to transient stimuli resulting in modified gene expression patterns and phenotypes later in life, remains a key challenge (Attig et al. 2010).
References
Al-Khan A, Aye IL, Barsoum I, Borbely A, Cebral E, Cerchi G, Clifton VL, Collins S, Cotechini T, Davey A, Flores-Martin J, Fournier T, Franchi AM, Fretes RE, Graham CH, Godbole G, Hansson SR, Headley PL, Ibarra C, Jawerbaum A, Kemmerling U, Kudo Y, Lala PK, Lassance L, Lewis RM, Menkhorst E, Morris C, Nobuzane T, Ramos G, Rote N, Saffery R, Salafia C, Sarr D, Schneider H, Sibley C, Singh AT, Sivasubramaniyam TS, Soares MJ, Vaughan O, Zamudio S, Lash GE (2011) IFPA Meeting 2010 Workshops Report II: placental pathology; trophoblast invasion; fetal sex; parasites and the placenta; decidua and embryonic or fetal loss; trophoblast differentiation and syncytialisation. Placenta 32(Suppl 2):S90–S99
Armitage JA, Taylor PD, Poston L (2005) Experimental models of developmental programming: consequences of exposure to an energy rich diet during development. J Physiol 565:3–8
Arnold AP, Lusis AJ (2012) Understanding the sexome: measuring and reporting sex differences in gene systems. Endocrinology 153:2551–2555
Asaka J, Terada T, Okuda M, Katsura T, Inui K (2006) Androgen receptor is responsible for rat organic cation transporter 2 gene regulation but not for rOCT1 and rOCT3. Pharm Res 23:697–704
Attig L, Gabory A, Junien C (2010) Early nutrition and epigenetic programming: chasing shadows. Curr Opin Clin Nutr Metab Care 13:284–293
Bale TL (2011) Sex differences in prenatal epigenetic programming of stress pathways. Stress 14:348–356
Barker DJ (1992) The fetal origins of diseases of old age. Eur J Clin Nutr 46(Suppl 3):S3–S9
Barker DJ, Osmond C (1988) Low birth weight and hypertension. BMJ 297:134–135
Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A (2008) Epigenetic differences between male and female bovine blastocysts produced in vitro. Physiol Genom 32:264–272
Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A (2010) Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc Natl Acad Sci U S A 107:3394–3399
Bermejo-Alvarez P, Rizos D, Lonergan P, Gutierrez-Adan A (2011) Transcriptional sexual dimorphism during preimplantation embryo development and its consequences for developmental competence and adult health and disease. Reproduction 141:563–570
Boloker J, Gertz SJ, Simmons RA (2002) Gestational diabetes leads to the development of diabetes in adulthood in the rat. Diabetes 51:1499–1506
Boujendar S, Arany E, Hill D, Remacle C, Reusens B (2003) Taurine supplementation of a low protein diet fed to rat dams normalizes the vascularization of the fetal endocrine pancreas. J Nutr 133:2820–2825
Burdge GC, Lillycrop KA (2010) Nutrition, epigenetics, and developmental plasticity: implications for understanding human disease. Ann Rev Nutr 30:1–7
Carrel L, Willard HF (2005) X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434:400–404
Charalambous M, da Rocha ST, Ferguson-Smith AC (2007) Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr Opin Endocrinol Diabetes Obes 14:3–12
Clifton VL (2010) Review: sex and the human placenta: mediating differential strategies of fetal growth and survival. Placenta 31(Suppl):S33–S39
Clifton VL, Engel P, Smith R, Gibson P, Brinsmead M, Giles WB (2009) Maternal and neonatal outcomes of pregnancies complicated by asthma in an Australian population. Aust N Z J Obstet Gynaecol 49:619–626
Clifton VL, Hodyl NA, Murphy VE, Giles WB, Baxter RC, Smith R (2010) Effect of maternal asthma, inhaled glucocorticoids and cigarette use during pregnancy on the newborn insulin-like growth factor axis. Growth Horm IGF Res 20:39–48
Coan PM, Vaughan OR, Sekita Y, Finn SL, Burton GJ, Constancia M, Fowden AL (2010) Adaptations in placental phenotype support fetal growth during undernutrition of pregnant mice. J Physiol 588:527–538
Constância M, Angiolini E, Sandovici I, Smith P, Smith R, Kelsey G, Dean W, Ferguson-Smith A, Sibley CP, Reik W, Fowden A (2005) Adaptation of nutrient supply to fetal demand in the mouse involves interaction between the Igf2 gene and placental transporter systems. Proc Natl Acad Sci U S A 102:19219–19224
Cross JC, Mickelson L (2006) Nutritional influences on implantation and placental development. Nutr Rev 64:S12–S18, discussion S72–91
Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, Simmons DG, Yamamoto H, Kingdom JCP (2003) Genes, development and evolution of the placenta. Placenta 24:123–130
Dabelea D, Hanson RL, Lindsay RS, Pettitt DJ, Imperatore G, Gabir MM, Roumain J, Bennett PH, Knowler WC (2000) Intrauterine exposure to diabetes conveys risks for type 2 diabetes and obesity: a study of discordant sibships. Diabetes 49:2208–2211
Davies W, Wilkinson LS (2006) It is not all hormones: alternative explanations for sexual differentiation of the brain. Brain Res 1126:36–45
Deplus R, Brenner C, Burgers WA, Putmans P, Kouzarides T, de Launoit Y, Fuks F (2002) Dnmt3L is a transcriptional repressor that recruits histone deacetylase. Nucleic Acids Res 30:3831–3838
Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL (2006) Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect 114:567–572
Dunn GA, Morgan CP, Bale TL (2010) Sex-specificity in transgenerational epigenetic programming. Horm Behav 59:290–295
Eriksson JG, Kajantie E, Osmond C, Thornburg K, Barker DJ (2009) Boys live dangerously in the womb. Am J Hum Biol 22:330–335
Fryer AA, Nafee TM, Ismail KM, Carroll WD, Emes RD, Farrell WE (2009) LINE-1 DNA methylation is inversely correlated with cord plasma homocysteine in man: a preliminary study. Epigenetics 4:394–398
Gabory A, Attig L, Junien C (2009) Sexual dimorphism in environmental epigenetic programming. Mol Cell Endocrinol 25:8–18
Gabory A, Ferry L, Fajardy I, Jouneau L, Gothie JD, Vige A, Fleur C, Mayeur S, Gallou-Kabani C, Gross MS, Attig L, Vambergue A, Lesage J, Reusens B, Vieau D, Remacle C, Jais JP, Junien C (2012) Maternal diets trigger sex-specific divergent trajectories of gene expression and epigenetic systems in mouse placenta. PLoS One 7:e47986
Gabory A, Roseboom TJ, Moore T, Moore LG, Junien C (2013) Placental contribution to the origins of sexual dimorphism in health and diseases: sex chromosomes and epigenetics. Biol Sex Diff 2013:4–5
Gallou-Kabani C, Vige A, Gross MS, Rabes JP, Boileau C, Larue-Achagiotis C, Tome D, Jais JP, Junien C (2007) C57BL/6J and A/J mice fed a high-fat diet delineate components of metabolic syndrome. Obesity (Silver Spring) 15:1996–2005
Gallou-Kabani C, Gabory A, Tost J, Karimi M, Mayeur S, Lesage J, Boudadi E, Gross MS, Taurelle J, Vige A, Breton C, Reusens B, Remacle C, Vieau D, Ekström TJ, Jais JP, Junien C (2010) Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS One 5:e14398
Gheorghe CP, Goyal R, Mittal A, Longo LD (2010) Gene expression in the placenta: maternal stress and epigenetic responses. Int J Dev Biol 54:507–523
Giraudo SQ, Della-Fera MA, Proctor L, Wickwire K, Ambati S, Baile CA (2010) Maternal high fat feeding and gestational dietary restriction: effects on offspring body weight, food intake and hypothalamic gene expression over three generations in mice. Pharmacol Biochem Behav 97:121–129
Graves JA (2010) Review: sex chromosome evolution and the expression of sex-specific genes in the placenta. Placenta 31(Suppl):S27–S32
Hellman A, Chess A (2007) Gene body-specific methylation on the active X chromosome. Science 315:1141–1143
Howerton CL, Bale TL (2012) Prenatal programing: at the intersection of maternal stress and immune activation. Horm Behav 62:237–242
Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y (2007) The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128:1077–1088
Jeong KS, Lee S (2005) Estimating the total mouse DNA methylation according to the B1 repetitive elements. Biochem Biophys Res Commun 335:1211–1216
Karimi M, Johansson S, Stach D, Corcoran M, Grander D, Schalling M, Bakalkin G, Lyko F, Larsson C, Ekstrom TJ (2006) LUMA (LUminometric Methylation Assay) – a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res 312:1989–1995
Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, Suzuki Y, Kono T (2012) Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet 8:e1002440
Kokalj-Vokac N, Zagorac A, Pristovnik M, Bourgeois CA, Dutrillaux B (1998) DNA methylation of the extraembryonic tissues: an in situ study on human metaphase chromosomes. Chromosome Res 6:161–166
Kral JG, Biron S, Simard S, Hould FS, Lebel S, Marceau S, Marceau P (2006) Large maternal weight loss from obesity surgery prevents transmission of obesity to children who were followed for 2 to 18 years. Pediatrics 118:e1644–e1649
Lampl M, Gotsch F, Kusanovic JP, Gomez R, Nien JK, Frongillo EA, Romero R (2010) Sex differences in fetal growth responses to maternal height and weight. Am J Hum Biol 22:431–443
Levin BE, Govek E (1998) Gestational obesity accentuates obesity in obesity-prone progeny. Am J Physiol 275:R1374–R1379
Li N, Carrel L (2008) Escape from X chromosome inactivation is an intrinsic property of the Jarid1c locus. Proc Natl Acad Sci U S A 105:17055–17060
Liang C, Decourcy K, Prater MR (2010) High-saturated-fat diet induces gestational diabetes and placental vasculopathy in C57BL/6 mice. Metabolism 59:943–950
Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC (2005) Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr 135:1382–1386
Lubchenco LO, Hansman C, Dressler M, Boyd E (1963) Intrauterine growth as estimated from liveborn birth-weight data at 24 to 42 weeks of gestation. Pediatrics 32:793–800
Mao J, Zhang X, Sieli PT, Falduto MT, Torres KE, Rosenfeld CS (2010) Contrasting effects of different maternal diets on sexually dimorphic gene expression in the murine placenta. Proc Natl Acad Sci U S A 107:5557–5562
McMillen IC, Robinson JS (2005) Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev 85:571–633
Mittelstrass K, Ried JS, Yu Z, Krumsiek J, Gieger C, Prehn C, Roemisch-Margl W, Polonikov A, Peters A, Theis FJ, Meitinger T, Kronenberg F, Weidinger S, Wichmann HE, Suhre K, Wang-Sattler R, Adamski J, Illig T (2011) Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS Genet 7:e1002215
Mittwoch U (1993) Blastocysts prepare for the race to be male. Hum Reprod 8:1550–1555
Nathanielsz PW, Poston L, Taylor PD (2007) In utero exposure to maternal obesity and diabetes: animal models that identify and characterize implications for future health. Obstet Gynecol Clin North Am 34:201–212, vii–viii
Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M (2008) Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol 10:1280–1290
Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ (2010) Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature 467:963–966
Novakovic B, Saffery R (2012) The ever growing complexity of placental epigenetics - role in adverse pregnancy outcomes and fetal programming. Placenta 33:959–970
Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH (2007) DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448:714–717
Pelis RM, Hartman RC, Wright SH, Wunz TM, Groves CE (2007) Influence of estrogen and xenoestrogens on basolateral uptake of tetraethylammonium by opossum kidney cells in culture. J Pharmacol Exp Ther 323:555–561
Penaloza C, Estevez B, Orlanski S, Sikorska M, Walker R, Smith C, Smith B, Lockshin RA, Zakeri Z (2009) Sex of the cell dictates its response: differential gene expression and sensitivity to cell death inducing stress in male and female cells. FASEB J 23:1869–1879
Rawn SM, Cross JC (2008) The evolution, regulation, and function of placenta-specific genes. Annu Rev Cell Dev Biol 24:159–181
Reynolds LP, Borowicz PP, Caton JS, Vonnahme KA, Luther JS, Hammer CJ, Maddock Carlin KR, Grazul-Bilska AT, Redmer DA (2010) Developmental programming: the concept, large animal models, and the key role of uteroplacental vascular development. J Anim Sci 88:E61–E72
Sleutels F, Zwart R, Barlow DP (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415:810–813
Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G (2011) Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet 43:811–814
Smith J, Cianflone K, Biron S, Hould FS, Lebel S, Marceau S, Lescelleur O, Biertho L, Simard S, Kral JG, Marceau P (2009) Effects of maternal surgical weight loss in mothers on intergenerational transmission of obesity. J Clin Endocrinol Metab 94:4275–4283
Sood R, Zehnder JL, Druzin ML, Brown PO (2006) Gene expression patterns in human placenta. Proc Natl Acad Sci U S A 103:5478–5483
Srinivasan M, Katewa SD, Palaniyappan A, Pandya JD, Patel MS (2006) Maternal high-fat diet consumption results in fetal malprogramming predisposing to the onset of metabolic syndrome-like phenotype in adulthood. Am J Physiol Endocrinol Metab 291:E792–E799
Sturmey RG, Bermejo-Alvarez P, Gutierrez-Adan A, Rizos D, Leese HJ, Lonergan P (2010) Amino acid metabolism of bovine blastocysts: a biomarker of sex and viability. Mol Reprod Dev 77:285–296
Suter M, Abramovici A, Showalter L, Hu M, Shope CD, Varner M, Aagaard-Tillery K (2010) In utero tobacco exposure epigenetically modifies placental CYP1A1 expression. Metabolism 59:1481–1490
Torrens C, Brawley L, Anthony FW, Dance CS, Dunn R, Jackson AA, Poston L, Hanson MA (2006) Folate supplementation during pregnancy improves offspring cardiovascular dysfunction induced by protein restriction. Hypertension 47:982–987
Urakami Y, Okuda M, Saito H, Inui K (2000) Hormonal regulation of organic cation transporter OCT2 expression in rat kidney. FEBS Lett 473:173–176
van Abeelen AF, de Rooij SR, Osmond C, Painter RC, Veenendaal MV, Bossuyt PM, Elias SG, Grobbee DE, van der Schouw YT, Barker DJ, Roseboom TJ (2011) The sex-specific effects of famine on the association between placental size and later hypertension. Placenta 32:694–698
van Nas A, Guhathakurta D, Wang SS, Yehya N, Horvath S, Zhang B, Ingram-Drake L, Chaudhuri G, Schadt EE, Drake TA, Arnold AP, Lusis AJ (2009) Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology 150:1235–1249
Vucetic Z, Kimmel J, Totoki K, Hollenbeck E, Reyes TM (2010) Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology 151:4756–4764
Waddell J, McCarthy MM (2012) Sexual differentiation of the brain and ADHD: what is a sex difference in prevalence telling us? Curr Top Behav Neurosci 9:341–360
Wagschal A, Feil R (2006) Genomic imprinting in the placenta. Cytogenet Genome Res 113:90–98
Wallace JM, Horgan GW, Bhattacharya S (2012) Placental weight and efficiency in relation to maternal body mass index and the risk of pregnancy complications in women delivering singleton babies. Placenta 33:611–618
Wang Y, Pringle KG, Sykes SD, Marques FZ, Morris BJ, Zakar T, Lumbers ER (2011) Fetal sex affects expression of renin-angiotensin system components in term human decidua. Endocrinology 153:462–468
Waterland RA, Travisano M, Tahiliani KG (2007) Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. Faseb J 21:3380–3385
Watson ED, Cross JC (2005) Development of structures and transport functions in the mouse placenta. Physiology (Bethesda) 20:180–193
Wauthier V, Sugathan A, Meyer RD, Dombkowski AA, Waxman DJ (2010) Intrinsic sex differences in the early growth hormone responsiveness of sex-specific genes in mouse liver. Mol Endocrinol 24:667–678
Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D (2005) Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37:853–862
Xu J, Burgoyne PS, Arnold AP (2002) Sex differences in sex chromosome gene expression in mouse brain. Hum Mol Genet 11:1409–1419
Xu J, Deng X, Disteche CM (2008) Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS One 3:e2553
Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ (2006) Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res 16:995–1004
Acknowledgments
These studies were supported by the Foundation Cœur et Artères (FCA No 05-T4), the Institut Benjamin Delessert, the Agence Nationale pour la Recherche (ANR 06-PNRA-022-01) and Contrat Cadre d’Aide au Projet d’Innovation Stratégique Industrielle “IT-Diab” OSEO-ISI (18/12/2008).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Gabory, A. et al. (2014). Male and Female Placentas Have Divergent Transcriptomic and Epigenomic Responses to Maternal Diets: Not Just Hormones. In: Seckl, J., Christen, Y. (eds) Hormones, Intrauterine Health and Programming. Research and Perspectives in Endocrine Interactions, vol 12. Springer, Cham. https://doi.org/10.1007/978-3-319-02591-9_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-02591-9_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-02590-2
Online ISBN: 978-3-319-02591-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)