Abstract
Gliomas are the most common and severe malignant tumors of the brain. The diagnosis and grading of gliomas are typically based on MRI images and pathology images. To improve the diagnosis accuracy and efficiency, we intend to design a framework for computer-aided diagnosis combining the two modalities. Without loss of generality, we first take an individual network for each modality to get the features and fuse them to predict the subtype of gliomas. For MRI images, we directly take a 3D-CNN to extract features, supervised by a cross-entropy loss function. There are too many normal regions in abnormal whole slide pathology images (WSI), which affect the training of pathology features. We call these normal regions as noise regions and propose two ideas to reduce them. Firstly, we introduce a nucleus segmentation model trained on some public datasets. The regions that has a small number of nuclei are excluded in the subsequent training of tumor classification. Secondly, we take a noise-rank module to further suppress the noise regions. After the noise reduction, we train a gliomas classification model based on the rest regions and obtain the features of pathology images. Finally, we fuse the features of the two modalities by a linear weighted module. We evaluate the proposed framework on CPM-RadPath2020 and achieve the first rank on the validation set.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Gliomas are the most common primary intracranial tumors, accounting for 40% to 50% of all cranial tumors. Although there are many systems for gliomas grading, the most common one is the World Health Organization (WHO) grading system, which classifies gliomas into grade 1 (least malignant and best prognosis) to grade 4 (most malignant and worst prognosis). According to the pathological malignancy of the tumor cells, brain gliomas are also classified into low-grade gliomas (WHO grade 1 to 2) and high-grade gliomas (WHO grade 3 to 4).
Magnetic resonance imaging (MRI) is the common examination method for glioma. In MRI images, different grades of gliomas show different manifestations: low-grade gliomas tend to show T1 low signal, T2 high-signal. They are mainly located in the white matter of the brain and usually have a clear boundary with the surrounding tissue. However, high-grade gliomas usually show heterogeneous signal enhancement, and the boundary between tumor and surrounding brain tissue is fuzzy. As mentioned in previous works [4, 5], MRI is mainly used to identify low-grade gliomas and high-grade gliomas, while it alone cannot accurately identify astrocytoma (grade II or III), oligodendroglioma (grade II or III), and glioblastoma (grade IV).
The current standard diagnosis and grading of brain tumors are done by pathologists according to Hematoxylin and Eosin (H&E) staining tissue sections fixed on glass slides under an optical microscope after resection or biopsy. Different subtypes of stained cells often have unique characteristics that pathologists use to classify glioma subtypes. However, some gliomas with a mixture of features from different subtypes are difficult for pathologists to distinguish accurately. In addition, the entire process is time-consuming and is prone to sampling errors. The previous studies [6,7,8, 10] have shown that CNN can be helpful for improving the accuracy and efficiency of pathologists. The whole slide pathological image is too large for end-to-end training, hence, typical solutions are patch-based, in which they assign the whole image label directly to the randomly sampled patches. This process introduces quite a lot of noisy samples inevitably and affects the network training.
In this paper, we intend to reduce the noise samples in the training stage. Main contributions of this paper can be highlighted as follows:
-
(1)
A nucleus segmentation model is adopted to filter patches with a small number of nuclei.
-
(2)
We stack the nucleus segmentation mask with the original image as the input to increase the attention on the nucleus.
-
(3)
The noise-rank module in study [9] is used to further suppress noise samples. In addition to CNN features, we also introduce the LBP feature as complementary.
2 Related Work
The goal of CPM-RadPath is to assess automated brain tumor classification algorithms developed on both radiology and histopathology images. Pei et al. [11] proposed to use the tumor segmentation results for glioma classification, and experimental results demonstrated the effectiveness. However, the pathological images were not employed in their work. Weng et al. [12] used the pre-trained VGG16 models to extract pathological features and a pyradiomics module [13] to extract radiological features, which were not updated by the CPM-RadPath data. Yang et al. [14] introduced a dual path residual convolutional neural network model and trained it by MRI and pathology images simultaneously. This work did not impose any constraints in the patch sampling process from the whole slide images, which could introduce noisy patches during training inevitably. [15] won the first-place in CPM-RadPath 2019, which set several constraints to prevent sampling in the background. Unfortunately, not all the patches sampled in the foreground contained diseased cells. These noisy patches have no discriminative features and will affect the training. In order to remove the noisy patches as much as possible, and dig more informative patches, we propose a delicate filtering strategy and a noise-rank module in this work.
3 Dataset and Method
In this section, we describe the dataset and our solution including preprocessing and networks.
3.1 Dataset
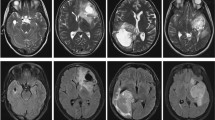
The training dataset of CPM-RadPath-2020 [1] consists of 221 paired radiology scans and histopathology images. The pathology data are in.tiff format, and there are four types of MRI data: Flair, T1, T1Ce, and T2. The corresponding pathology and radiology examples are shown in Fig. 1.
The patients' age information was also provided in the dataset. The age distribution of different subtypes was shown in Fig. 2. Low-grade astrocytoma and glioblastoma have different age distributions. Low-grade astrocytoma is predominantly in the lower age groups, while glioblastoma is predominantly in the higher age groups.
CPM-RadPath2020 aims to distinguish between three subtypes of brain tumors, namely astrocytoma (Grade II or III), oligodendroglioma (Grade II or III) and glioblastoma (Grade IV). The number of each subtype in the training data is shown in Table 1.
Except for the CPM-RadPath2020 dataset, two public datasets are also utilized: the Multimodal Brain Tumor Segmentation Challenge 2019 (BraTS-2019) [2] and MoNuSAC [3]. The BraTS-2019 dataset is used to train a tumor segmentation model of MRI images and MoNuSAC dataset is used to train a nucleus segmentation model of pathology images.
3.2 Preprocessing
Pathology.
Due to limited computational resources, it is not feasible to process the whole image directly, so we performed patch extraction on the whole slide image. In each WSI, only a few cells were stained, most of the regions were white background, we need to find an efficient way to extract valid information from WSI. In this paper, the WSI was first down-sampled to generate a thumbnail, and then these thumbnails were binarized to distinguish between foreground and background. We only sampled patches for training from the foreground. In addition, some of the data also had staining anomalies, we refer to the work [15] to constrain the extracted patch.
The above operations can filter out most of the background patches, but there are still some patches with less information or abnormal staining (e.g. Fig. 3c or d). Therefore, the nucleus segmentation model was added to suppress the noise patches by counting the proportion of nuclei in the patch. Figure 3 lists four different types of patch data.
The size of WSI in the training set has a wide range, which means the number of the extracted patches also differs from each WSI. To balance the training patches from different WSIs, we set a maximum number of extracted patches from one WSI as 2000. The extracted patch was assigned the same label as the corresponding WSI, as most studies [5] do. This process is unavoidable to bring in some noisy patches, which may affect the model training. We provide some solutions detailed in the next section.
Due to differences in the staining process of the slices, WSIs have a big variance in color. The general practice is color normalization [17]. For the sake of simplicity, this paper adopted the same strategy as study [15] to directly convert RGB images into gray images.
The whole pipeline of the preprocessing step is shown in Fig. 4.
The pre-processing failed for one WSI (CPM19_TCIA06_184_1.tiff), depicted in Fig. 5. We could not sample any valid patches from it. Hence, this WSI was discarded during the training.
Radiology.
For each patient, there are four types of MRI images, with a size of 240 * 240 * 155. We first used BraTS2019 dataset to train a segmentation model of the tumors and then extracted the tumor regions according to the segmentation mask. The extracted regions were resized to a fixed size of 160 * 160 * 160. The extracted regions from different MRIs of the same patient were concatenated forming a 4D data, with a size of 4 * 160 * 160 * 160. Z-score normalization was performed for a fast convergence.
3.3 Classification Based on MRI Images and Noise Reduced Pathology Images
The pipeline of the proposed framework is shown in the Fig. 6, including a pathological classification network, a radiological classification network and a features fusion module. Next, we will detail each module.
Pathological Classification Network.
The nucleus segmentation Model was trained on the MoNuSAC dataset. After the training, we used it to filter the patches whose total area of nuclei was less than 7% of the patch size. The rest patches were transformed into gray images and resized to 256 * 256. To force the classification model to focus on the nuclei, we stitched the original gray image with the nucleus segmentation mask. The final input size of the classification model was 2 * 256 * 256. We adopted a unified Densenet network structure [16] for feature extraction, and set the numbers of dense blocks in different stages as 4, 8, 12, 24.
Directly assigning the whole image label to the patches inevitably introduced noisy samples, so we took a noise-rank [9] module to filter out those samples with high noise probability. In the noise-rank module, we first used K-means to cluster in the feature space for each subtype. Besides the CNN features, we also utilized LBP features [18] for the clustering, which is visually discriminative and a complementary to CNN features. The center of each cluster was regarded as a prototype, with an original label of its corresponding subtype. We can also obtain a predicted label by KNN for each prototype. Based on the original and predicted label of these prototypes, the posterior probability that indicates the label noise for all the samples was estimated. We then ranked all the samples according to the probability and dropped the top 20% samples in the training of classification model. The detail of noise-rank could be found in the study [9]. The noise-rank module and the classification model were trained alternatively.
The loss function was cross entropy. To avoid overfitting, the same augmentation methods as the study [19] were used, including Random-Brightness, Random-Contrast, Random-Saturation, Random-Hue, Flip and Rotation.
Radiological Classification Network.
We extracted regions of interest (ROI) by a lesion segmentation model pre-trained on BraTS2019 [2] as the input to the classification network. The backbone of the network was a 3D-Densenet [16]. The numbers of dense blocks in different stages were 4, 8, 12. The loss function was cross entropy. Dropout and augmentation were also used to avoid over-fitting.
Features Fusion.
After the training of the above two classification models, we extracted the features from the two models. To keep the same feature length of different pathological images, we only selected a fixed number of patches to represent the whole pathological image. Then we predicted a weight for each modality to fuse the two features. The fused features were sent to a multi-layer perception (MLP) for glioma classification.
4 Experiments
4.1 Implementation Details
The tumor segmentation model of radiology images was trained on TensorFlow [20] and all the other models were based on MXNet [21]. Parameters were optimized by SGD [22], and the weight decay and momentum were set to 1e−4 and 0.95 respectively. All of our models were trained for 200 epochs on a TeslaV100 GPU. The learning rate was initially set to 0.001 and was divided by 10 at 50% and 75% of the total training epochs. The Noise-Rank module was updated every 20 epochs during the training.
4.2 Results and Discussion
In this section, we will present our experimental results on CPM-RadPath-2020. Since the number of training data is small, we perform 3-forder cross validation on the training data for all the experiments.
Figure 2 shows that low-grade astrocytoma and glioblastoma have different age distributions. We tried to combine the age information with the CNN features after global average pooling. However, the results did not show any improvement brought by the age information. So we discarded the age information in the subsequent experiments.
We first show the classification results based on only pathology images in Table 2. The baseline model was trained on original gray image patches directly with a lot of noisy samples. When we filtered out some noisy patches with a small number of nuclei and concatenated the nuclei segmentation mask with the original gray image patches, all the indexes got a significant increase. And noise-rank module brought a further increase. These results demonstrated the effectiveness of the proposed noise reduction algorithm.
The results of radiology images were quite lower than the pathology images, as shown in Table 3. We figure out that the reason is that MRI images have poor discriminative capacity between astrocytoma and oligodendroglioma. When fusing the features of the two modalities by the proposed linear weighted module, the F1-micro was increased from 91.7% to 93.2 on the cross-validation dataset. The results indicated that MRI images could provide some complementary features to pathology images, despite the big performance difference.
Table 4 presents the results on the online validation set. Since the online validation sets are the same as the last year, we compared our results with the first-place methods [15] in CPM-RadPath-2019. We achieved higher performances on both pathology only and radiology only settings. The results further demonstrated the effectiveness of the proposed noise reduction in pathology images. The ROI extraction step in MRI images could make the model focus on tumor regions, so it brought improvements. We evaluated our solution on the validation set of CPM-RadPath-2020. The final F1-micro reached 0.971 and ranked 1st among 61 teams.
5 Conclusion
In this paper, we proposed a framework combining MRI Images and pathology Images to identify different subtypes of glioma. To reduce the noisy samples in pathology images, we leverage a nucleus segmentation model and a noise-rank module. With the help of noise reduction, we obtain a more precise classification model. Fusing the two modalities on feature space provides a more complete representation. Our results ranked in the first place on the validation set, which demonstrates the effectiveness of the proposed framework.
References
Computational Precision Medicine Radiology-Pathology challenge on Brain Tumor Classification (2020). https://www.med.upenn.edu/cbica/cpm2020.html
Bakas, S., et al.: Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge (2018)
Kumar, N., et al.: A multi-organ nucleus segmentation challenge. IEEE Trans. Med. Imaging 39, 5 (2017)
Choi, K.S., et al.: Prediction of IDH genotype in gliomas with dynamic susceptibility contrast perfusion MR imaging using an explainable recurrent neural network. NEURO-ONCOLOGY 21, 9 (2019)
Decuyper, M., et al.: Automated MRI based pipeline for glioma segmentation and prediction of grade, IDH mutation and 1p19q co-deletion. MIDL 2020 (2020)
Li, Y., et al.: Classification of breast cancer histology images using multi-size and discriminative patches based on deep learning. IEEE Access 7, 21400–21408 (2019)
Das, K., et al.: Multiple instance learning of deep convolutional neural networks for breast histopathology whole slide classification. In: 15th International Symposium on Biomedical Imaging (2018)
Hashimoto, N., et al: Multi-scale domain-adversarial Multiple-instance CNN for cancer subtype classification with unannotated histopathological images. In: CVPR 2020 (2020)
Sharma, K., Donmez, P., Luo, E., Liu, Y., Yalniz, I.Z.: NoiseRank: unsupervised label noise reduction with dependence models. In: Vedaldi, A., Bischof, H., Brox, T., Frahm, J.-M. (eds.) ECCV 2020. LNCS, vol. 12372, pp. 737–753. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-58583-9_44
Kurc, T., et al.: Segmentation and classification in digital pathology for glioma research: challenges and deep learning approaches. Front. Neurosci. 14, 1–15 (2020). https://doi.org/10.3389/fnins.2020.00027
Pei, L., et al.: Brain tumor classification using 3D convolutional neural network. In: MICCAI 2019 Workshop (2019)
Weng, Y.-T., et al.: Automatic classification of brain tumor types with the mri scans and histopathology images. In: MICCAI 2019 Workshop (2019)
Van Griethuysen, J.J.M., et al.: Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77, 21 (2017)
Yang, Y., et al.: Brain tumor classification with tumor segmentations and a dual path residual convolutional neural network from MRI and pathology images. In: MICCAI 2019 Workshop (2019)
Ma, X., et al.: Brain tumor classification with multimodal MR and pathology images. In: MICCAI 2019 Workshop (2019)
Huang, G., et al.: Densely connected convolutional networks. Computer Era. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708 (2017)
Tellez, D., et al.: Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med. Image Anal. 58, 101544 (2019)
Chenni, W., et al.: Patch Clustering for Representation of Histopathology Images. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-23937-4_4
Liu, Y., et al.: Detecting Cancer Metastases on Gigapixel Pathology Images. In: MICCAI Tutorial (2017)
Abadi, M., et al.: TensorFlow: a system for large-scale machine learning. Oper. Syst. Des. Implementation 2016 (2016)
Chen, T., et al.: MXNet: a flexible and efficient machine learning library for heterogeneous distributed systems. arXiv preprint arXiv:1512.01274 (2015)
Niu, F., et al.: HOGWILD!: a lock-free approach to parallelizing stochastic gradient descent. In: Advances in Neural Information Processing Systems, vol. 24 (2011)
Acknowledgements
This work was supported by the National Natural Science Foundation of China under Grant 61472393.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Yin, B., Cheng, H., Wang, F., Wang, Z. (2021). Brain Tumor Classification Based on MRI Images and Noise Reduced Pathology Images. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2020. Lecture Notes in Computer Science(), vol 12659. Springer, Cham. https://doi.org/10.1007/978-3-030-72087-2_41
Download citation
DOI: https://doi.org/10.1007/978-3-030-72087-2_41
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-72086-5
Online ISBN: 978-3-030-72087-2
eBook Packages: Computer ScienceComputer Science (R0)