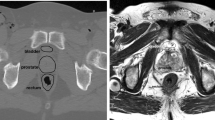
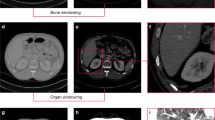
Abstract
Medical image segmentation is a common image analysis task that can impact diagnostic accuracy. Image quality varies depending on application, acquisition parameters and patient preparation. This requires developers’ data preparation efforts prior training. Training on an explicit data nature imposes limitations on generalization. This requires further data pre/post processing efforts for each dataset of different nature. Hence, it’s essential to embed a core component in the segmentation algorithm to automatically handle data processing tasks. In this work, we developed a fully-automated system that transforms computed tomography (CT) images into a standard format prior to segmentation learning and then inverse-transform segments to the original image format to preserve co-registration. Furthermore, we have developed a multi neural network segmentation module that incorporates three convolutional task-specific neural networks that utilize global (two networks) and local (one network) information, while making the segmentation based on agreement of contextual information using probability heatmaps. For baseline non-standard training and testing on a diverse liver dataset, there was no learning convergence and testing results performed relatively poorly. In contrast, after standardized training on the same dataset, all networks converged and yielded dice of +90%. Using the same standardized training, standard and non-standard testing were compared after applying single variable variation. Standardization consistently improved performance by 2.5%, 34.7% and 1.5% for field-of-view, slice-thickness and anatomy, respectively. We also found that global-based segmentation outperformed local-based, and network cooperation significantly outperformed individual networks with dice 95.20%. This offers a segmentation system with better generalization capabilities to serve variable clinical environments.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Lötjönen, J., Wolz, R., Koikkalainen, J., Thurfjell, L., Waldemar, G., Soininen, H.: Fast and robust multi-atlas segmentation of brain magnetic resonance images. Neuroimage 49(3), 2352–2365 (2010)
Wang, H., Suh, J., Das, S., Pluta, J., Craige, C., Yushkevich, P.: Multi-atlas segmentation with joint label fusion. IEEE Trans. Pattern Anal. Mach. Intell. 35(3), 611–623 (2012)
Schmidhuber, J.: Deep learning in neural networks: an overview. Neural Netw. 61, 85–117 (2015)
LeCun, Y., Bottou, L., Bengio, Y., Haffner, P.: Gradient-based learning applied to document recognition. Proc. IEEE 86(11), 2278–2324 (1998)
Krizhevsky, A., Sutskever, I., Hinton, G.: Imagenet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, pp. 1097–1105 (2012)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015)
Zhou, X., Takayama, R., Wang, S., Hara, T., Fujita, H.: Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method. Med. Phys. 44(10), 5221–5233 (2017)
Vania, M., Mureja, D., Lee, D.: Automatic spine segmentation from CT images using convolutional neural network via redundant generation of class labels. J. Comput. Design Eng. 6(2), 224–232 (2019)
Cordier, N., Delingette, H., Ayache, N.: A patch-based approach for the segmentation of pathologies: application to glioma labelling. IEEE Trans. Med. Imaging 35(4), 11 (2015)
Sun, J., Chen, W., Peng, S., Liu, B.: DRRNet: Dense Residual Refine Networks for automatic brain tumor segmentation. J. Med. Syst. 43(7), 221 (2019)
Wolz, R., Chu, C., Misawa, K., Fujiwara, M., Mori, K., Rueckert, D.: Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE Trans. Med. Imaging 32(9), 1723–1730 (2013)
Tang, M., Zhang, Z., Cobzas, D., Jagersand, M.: Segmentation-by-detection: a cascade network for volumetric medical image segmentation. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 1356–1359 (2018)
Xu, X., Zhou, F., Liu, B., Fu, D., Bai, X.: Efficient multiple organ localization in CT image using 3D region proposal network. IEEE Trans. Med. Imaging 38(8), 1885–1898 (2019)
Ma, L., Guo, R., Zhang, G., Tade, F., Schuster, D., Nieh, P., Fei, B.: Automatic segmentation of the prostate on CT images using deep learning and multi-atlas fusion. In: Medical Imaging 2017: Image Processing, International Society for Optics and Photonics, vol. 10133, pp. 1013320 (2017)
3-D Brain Tumor Segmentation Using Deep Learning. https://www.mathworks.com/help/images/segment-3d-brain-tumor-using-deep-learning.html (Matlab Documentation). Accessed Feb 2020
Budak, Ü., Guo, Y., Tanyildizi, E., Şengür, A.: Cascaded deep convolutional encoder-decoder neural networks for efficient liver tumor segmentation. Med. Hypotheses 134, 109431 (2020)
Bagade, S.S., Shandilya, V.K.: Use of histogram equalization in image processing for image enhancement. Int. J. Softw. Eng. Res. Practices 1(2), 6–10 (2011)
Roth, H., Shen, C., Oda, H., Oda, M., Hayashi, Y., Misawa, K., Mori, K.: Deep learning and its application to medical image segmentation. Med. Imaging Technol. 36(6), 63–71 (2018)
Salman, O.S., Klein, R.: Anatomical region identification in medical x-ray computed tomography (CT) scans: development and comparison of alternative data-analysis and vision-based methods. Neural Comput. Appl. (NCAA). Springer (2020)
Yan, Z., Zhan, Y., Peng, Z., Liao, S., Shinagawa, Y., Zhang, S., Metaxas, D.N., Zhou, X.: Multi-instance deep learning: discover discriminative local anatomies for bodypart recognition. IEEE Trans. Med. Imaging 35(5), 1332–1343 (2016)
Roth, H., Lee, C., Shin, H., Seff, A., Kim, L., Yao, J., Lu, L., Summers, R.: Anatomy-specific classification of medical images using deep convolutional nets. In: 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), pp. 101–104 (2015)
Cho, J., Lee, K., Shin, E., Choy, G., Do, S.: Medical Image Deep Learning with Hospital PACS Dataset. arXiv preprint arXiv:1511.06348 (2015)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 424–432 (2016)
Sudre, C., Li, W., Vercauteren, T., Ourselin, S., Cardoso, M.: Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, pp. 240–248. Springer, Cham (2017)
Kingma, D., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980. (2014)
Liver Tumours. http://medicaldecathlon.com/ (Medical Segmentation Decathalon). Accessed Feb 2020
MSD Challenge. http://medicaldecathlon.com/results.html (Medical Segmentation Decathlon). (2018). Accessed Feb 2020
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Salman, O.S., Klein, R. (2021). Developing an Automatic Cooperating Neural Networks and Image Standardization Approach for Segmentation of X-Ray Computed Tomography Images. In: Arai, K., Kapoor, S., Bhatia, R. (eds) Proceedings of the Future Technologies Conference (FTC) 2020, Volume 1. FTC 2020. Advances in Intelligent Systems and Computing, vol 1288. Springer, Cham. https://doi.org/10.1007/978-3-030-63128-4_29
Download citation
DOI: https://doi.org/10.1007/978-3-030-63128-4_29
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-63127-7
Online ISBN: 978-3-030-63128-4
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)