Abstract
Ultrasound simulation based on ray tracing enables the synthesis of highly realistic images. It can provide an interactive environment for training sonographers as an educational tool. However, due to high computational demand, there is a trade-off between image quality and interactivity, potentially leading to sub-optimal results at interactive rates. In this work we introduce a deep learning approach based on adversarial training that mitigates this trade-off by improving the quality of simulated images with constant computation time. An image-to-image translation framework is utilized to translate low quality images into high quality versions. To incorporate anatomical information potentially lost in low quality images, we additionally provide segmentation maps to image translation. Furthermore, we propose to leverage information from acoustic attenuation maps to better preserve acoustic shadows and directional artifacts, an invaluable feature for ultrasound image interpretation. The proposed method yields an improvement of 7.2% in Fréchet Inception Distance and 8.9% in patch-based Kullback-Leibler divergence.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Ultrasound (US) is a low-cost, real-time, and portable diagnostic imaging technique without ionizing radiation, hence widely used in gynecology and obstetrics. Since its interpretation can be nontrivial due to ultrasound-specific artifacts such as acoustic shadows and tissue-specific speckle texture, sonographer training is crucial. For an education tool, ray tracing can be used for US simulation [3, 14], where US wavefront is represented with rays on the GPU to simulate interaction with tissue layers, whereas speckle patterns are simulated with a convolutional model of tissue speckle noise. With stochastic Monte-Carlo sampling of rays [11], this can produce realistic looking images. However, interactive computational constraints often necessitate a compromise in image quality, e.g. with limited number of rays or by disabling or reducing essential simulation features.
Deep learning has achieved great success in various computer vision and graphics tasks. In particular, generative adversarial networks (GANs) [5] have been demonstrated as a powerful tool for image synthesis and translation [8, 23]. GANs have been widely adapted for various medical image synthesis tasks, such as image inpainting [2] and cross modality translation in both supervised [1, 13] and unsupervised [20, 22] settings. In US image synthesis, a two-stage stack GAN was introduced in [17] for simulating intravascular US imagery conditioned on tissue echogenicity map. In [7], freehand US images are generated conditioned on calibrated physical coordinates. Recently in [18], feasibility of improving the realism of ray-traced US images has been demonstrated using cycleGAN [23].
In this work we propose a deep learning based approach for improving the quality of simulated US images that are obtained using a ray tracing algorithm, such that computationally simpler (low quality) images can be used to generate higher quality images mimicking a computationally sophisticated simulation that may not be feasible at interactive frame rates. Access to a simulation framework together with comprehensive anatomical models allows us to obtain realistic paired images of differing quality aligned with anatomical models. Therefore, we tackle this problem in an image-to-image translation setting with paired low and high quality images. Our framework leverages conditional GANs [12] to recover image features that are missing in the low quality images. Since low quality images may have missing anatomical structures, which introduces ambiguities in the image translation process, we propose to additionally leverage information that is readily available from the underlying simulation algorithm. For this purpose, we use 2D segmentation map slices at given transducer locations, to provide any anatomical information missing from low quality images. Since major acoustic effects such as shadows are integral along wave path and hence global in nature, they would require large network receptive fields to model. Thus, we further propose to incorporate integral attenuation maps as additional input to the network. Such segmentation and attenuation maps can be easily obtained as by-products of ray-based simulation frameworks [3, 11, 14].
2 Materials and Methods
Data Generation. Simulated B-mode US images are generated using a Monte-Carlo ray tracing framework on a custom geometric fetal model for obstetric training [11]. US wave interactions are simulated using a surface ray tracing model to find the ray segments between tissue boundaries. Tissue properties such as acoustic impedance, attenuation and speed-of-sound are assigned to each tissue type from literature and based on sonographers’ visual inspection. Along each extracted ray segment, a ray-marching algorithm is applied on the GPU to emulate US scatterer texture by convolving a locally changing point-spread-function with an underlying tissue scatterer representation generated randomly using Gaussian distributions per tissue type [10]. Simulated RF data is post-processed with envelope detection, time-gain compensation, log compression and scan-conversion into Cartesian coordinates, yielding a gray-scale B-mode image.
US Images. For each regularly-sampled key frame of a simulated US fetal exam, paired low and high quality images are generated using two simulation passes: low quality images using one primary ray per US scanline and one elevational layer; and high quality images using 32 primary rays per scanline and three elevational layers [11]. Other simulation parameters are kept identical for both simulation passes, cf Table 1. Example B-mode images are shown in Fig. 1(a–b).
Image Mask. A fixed binary image mask demarcating the imaging region after scan-conversion for the convex probe is also provided as input to the network, in order to constrain the meaningful image translation region and help to save generator capacity.
Segmentation Maps. As additional input for our method, segmentation maps as the cross-section of input triangulated anatomical surfaces are also output by the simulation, corresponding to each low-/high-quality image, cf Fig. 1(c).
Attenuation Maps. A characteristic feature in real US images is the presence of directional artifacts, which is also valuable for the interpretation of images, for instance in diagnosis of pathology. It is therefore important to accurately simulate such artifacts for training purposes. Besides reflection and refraction effects, a major source of directional US artifacts is attenuation, which is caused by a reduction in acoustic intensity along the wave travel path due to local tissue effects such as absorption, scattering, and mode conversion. Since such artifacts are not only a function of local tissue properties but an integral function along the viewing direction, we propose to directly provide this integrated information to the translation network, hypothesized to improve the quality of translation.
Acoustic intensity arriving at a depth z can be modeled as \(I(z)=I_0 e^{-\mu z}\), where \(\mu \) is the attenuation constant at a given imaging frequency and \(I_0\) is the initial intensity. Given that the waves travel through different tissue layers with varying attenuation constants \(\mu (z)\), the total intensity arriving at a point z can be approximated by
To approximate such attenuation effect, we create attenuation integral maps \(a=e^{-\sum _{i=0}^z \mu [i]}\), accumulated for each image point along the respective ultrasound propagation path. For better dynamic range and to avoid outliers, these maps are normalized by the 98 %ile of image intensities and then scan-converted into the same Cartesian coordinate frame as the simulated B-mode images. Figure 1(d) shows sample integral attenuation maps.
Image Translation Network. Our image-to-image translation framework is based on the pix2pix network proposed in [8]. Simulated low and high quality US images are considered as source and target domain, respectively, where a translation network G learns a mapping from the source to the target domain. Specifically, G maps the low quality US image x, the binary mask m, the segmentation map s, and the attenuation integral map a to the high quality US image y, i.e.: \(G: \{x,m,s,a\}\rightarrow \{y\}\). The discriminator D is trained to distinguish between real and fake high quality images conditioned on the corresponding inputs to the generator. The objective function of the conditional GAN consists of a weighted sum between a GAN loss \(L_\mathrm {GAN}\) and a data fidelity term \(L_\mathrm {F}\), i.e.,
where \(\tilde{x} = (x,m,s,a)\). Before computing the losses, the output is element-wise multiplied with the binary mask to restrict the loss to the relevant output regions.
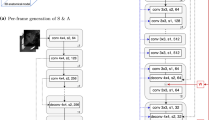
Similarly to [8], we use a deterministic G parametrized using a 8-layer Unet with skip connections and D using a 4-layer convolutional network, i.e. a patchGAN discriminator. Instance normalization is applied before nonlinear activation. The full field-of-view B-mode images from the simulation are of size \(1000\times 1386\) pixels. Applying pix2pix directly at such high resolution may lead to unsatisfactory results, as reported in [19]. We therefore use randomly cropped patches of a smaller size. A patch size of \(512\times 512\) pixels is found empirically to provide sufficient anatomical context, without degradation in image quality. Figure 2 shows an overview of our network architecture.
3 Experiments and Results
Implementation Details and Network Training. We use the Adam optimizer [9] with a learning rate of 0.0002 and exponential decay rates \(\beta _1=0.5\) and \(\beta _2=0.999\). Since GANs in general underfit [21] and the Nash equilibrium is often not reached in practice, we early stop training at 50k iterations, by when FID of a randomly-sampled training subset saturates. We use a batch size of 16 and set \(\lambda =100\). Our dataset consists of 6669 4-tuples (x, y, s, a) and a constant binary mask m covering the beam shape for all samples. We use randomly-selected 6000 images for training and the rest for evaluation. To quantitatively evaluate our models, from each test image we randomly crop four patches of size \(512\times 512\), yielding an evaluation set of 2676 image patches that are not seen during training. Note that our original dataset consists of images that are temporally far apart, thus the test images cannot be temporally consecutive and thus inherently similar to any training images.
Comparative Evaluation. To demonstrate the effectiveness of the proposed additional inputs from the image formation process, we conduct an ablation study by considering different combinations of network inputs. We refer the pix2pix network with low quality image and binary mask in the input channel as our baseline L2H\(_\mathrm {M}\). We compare this baseline with the following variants: 1) L2H\(_\mathrm {MS}\): L2H\(_\mathrm {M}\) with segmentation map s as additional input; 2) L2H\(_\mathrm {MSA}\): L2H\(_\mathrm {MS}\) with attenuation integral map a as additional input.
Qualitative Results. Figure 3 shows a visual comparison of the three model variants on four examples. The baseline L2H\(_\mathrm {M}\) fails to preserve anatomical structures due to missing structural information in the input images. Resulting ambiguities in the network prediction cause artifacts such as blur in regions that feature fine details such as bones. Providing segmentation maps as additional input (L2H\(_\mathrm {MS}\)) greatly reduces such artifacts as shown in Fig. 3(c). However, L2H\(_\mathrm {MS}\) still struggles in modeling complex non-local features such as directional occlusion artifacts, note the lack of acoustic shadows in Fig. 3(c). In contrast, our final model L2H\(_\mathrm {MSA}\) is able to accurately synthesize these features and produces translations significantly closer to the target, as demonstrated in Fig. 3(d). In particular, our proposed model with segmentation and attenuation integral maps is able to recover both missing anatomical structures and directional artefacts.
Quantitative Results. The effectiveness of the proposed model is further evaluated using the following quantitative metrics:
-
1)
PSNR: Peak signal-to-noise ratio between two images A and B is defined by \(\text {PSNR} = 10\log _{10}(\frac{255}{\text {MSE}})\) with mean squared error MSE between A and B.
-
2)
SSIM: Structural similarity index quantifies the visual changes in structural information as \(\text {SSIM}(A, B)=\frac{(2\mu _A\mu _B+c_1)(2\sigma _{AB}+c_2)}{(\mu _A^2+\mu _B^2+c_1)(\sigma _A^2+\sigma _B^2+c_2)}\) with regularization constants \(c_1\) and \(c_2\), local means \(\mu _A\) and \(\mu _B\), local standard deviations \(\sigma _A\) and \(\sigma _B\), and cross covariance \(\sigma _{AB}\). We use the default parameters of the MATLAB implementation to compute the metric.
-
3)
pKL: Speckle appearance, relevant for tissue characterization in US images [15], affects image histogram statistics. Hence, discrepancy in histogram statistics can quantify differences in tissue-specific speckle patterns. Kullback-Leibler divergence compares normalized histograms \(h_A\) and \(h_B\) of two images A and B as: \( \text {KL} (h_A || h_{B}) = \sum _{l=1..d} h_A[l] \log \left( \frac{h_A[l]}{h_{B}[l]}\right) \). We set the number of histogram bins d to 50. To emphasize structural differences, we calculate KL divergence locally within \(32\times 32\) sized non-overlapping patches and report the metric mean, called patch KL (pKL) herein.
-
4)
FID: Fréchet Inception Distance compares the distributions of generated samples and real samples by computing the distance between two multivariate Gaussians fitted to hidden activations of Inception network v3. This is a widely used metric to evaluate GAN performance, capturing both perceptual image quality and mode diversity. For this purpose, center crops of test images are sub-divided into four pieces of \(299\times 299\), to match Inception v3 input size.
Table 2 summarizes quantitative results for all models and all metrics, with the additional comparison to the discrepancy between low quality and high quality images as reference. A preliminary baseline experiment without GAN loss resulted in very blurry images with an FID score of 184.71. The results in Table 2 demonstrate that L2H\(_\mathrm {MSA}\) achieves the best translation performance in terms of all proposed metrics. The effectiveness of providing informative inputs to the network is well demonstrated in the gradual improvement in PSNR, SSIM and pKL, showing higher fidelity in anatomical structures and directional shadow artifacts. The metric pKL gives further indication of closer speckle appearance achieved by L2H\(_\mathrm {MSA}\). Based on Wilcoxon signed-rank tests, improvements of L2H\(_\mathrm {MSA}\) over L2H\(_\mathrm {MS}\) and those two over the baseline L2H\(_\mathrm {M}\) are statistically significant (\(\text {p}<10^{-5}\)) for all evaluation metrics. Moreover, FID score indicates higher statistical similarity between the target and generated images using the proposed final model, with an improvement of \(7.2\%\) compared to L2H\(_\mathrm {M}\).
Full Field-of-View Images. Above image translation has been demonstrated on patches. For the entire field-of-view (FoV) US images, patch fusion from image translation of non-overlapping patches would cause artifacts at image seams. Averaging overlapping patches, on the other hand, would blur the essential US texture. Although seamless tiling of US images is possible using graphical models [4], this requires prohibitively long computation time. Herein, we instead directly apply our trained generator on full FoV low-quality images, since the generator is fully convolutional and thus can operate on images of arbitrary size. Figure 4 shows two examples of translated images by L2H\(_\mathrm {MS}\) and L2H\(_\mathrm {MSA}\), demonstrating direct inference on full FoV images. While anatomical structures are well preserved and the effect of attenuation integral map is apparent, speckle texture appearance is seen to degrade slightly especially in the top image regions, where the ultrasound texture looking particularly different due to focusing difference and near-field effects.
4 Discussion and Conclusions
We have proposed a patch-based generative adversarial network for improving the quality of simulated US images, via image translation of computationally low-cost images to high quality simulation outputs. Providing segmentation and attenuation integral maps to the translation framework greatly improves preservation of anatomical structures and synthesis of important acoustic shadows. Continuous simulation parameters, such as transmit focus and depth-dependent lateral resolution, are implicitly captured by our framework thanks to training on image patches. For discrete simulation parameters such as imaging mode and transducer frequency that can take a handful of different values in typical clinical imaging, it is feasible to train a separate GAN for each such setting.
Image rendering time highly depends on chosen simulation parameters and 3D mesh model complexity. For instance, high framerates are reported for a simpler model in [16]. Rendering high and low quality images herein takes 75 ms and 40 ms, respectively. Our network inference time with a non-optimized code is 12.6 ms on average for full FoV images on a GTX 2080 Ti using TensorRT. This timing improvement is rather a lower-bound, since network inference can be further accelerated, e.g. with FPGAs [6]. Furthermore, since a pass through the network runs in constant time, potential time gain can be arbitrarily high depending on the desired complexity of the target simulation. With our proposed framework a trade-off between image quality and computational speed is obviated, thus enabling interactive framerates even with sophisticated anatomical scenes and computationally-taxing simulation settings. Although the convolutional network can process arbitrary sized image, translating full FoV images without any artifacts is still a challenge.
References
Armanious, K., et al.: MedGAN: medical image translation using gans. Comput. Med. Imaging Graph. 79, 101684 (2020)
Armanious, K., Mecky, Y., Gatidis, S., Yang, B.: Adversarial inpainting of medical image modalities. In: ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 3267–3271. IEEE (2019)
Burger, B., Bettinghausen, S., Radle, M., Hesser, J.: Real-time GPU-based ultrasound simulation using deformable mesh models. IEEE Trans. Med. Imaging 32(3), 609–618 (2013)
Flach, B., Makhinya, M., Goksel, O.: PURE: panoramic ultrasound reconstruction by seamless stitching of volumes. In: Tsaftaris, S.A., Gooya, A., Frangi, A.F., Prince, J.L. (eds.) SASHIMI 2016. LNCS, vol. 9968, pp. 75–84. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46630-9_8
Goodfellow, I., et al.: Generative adversarial nets. In: Advances in Neural Information Processing Systems, pp. 2672–2680 (2014)
Guo, K., Zeng, S., Yu, J., Wang, Y., Yang, H.: A survey of FPGA-based neural network accelerator. arXiv preprint arXiv:1712.08934 (2017)
Hu, Y., et al.: Freehand ultrasound image simulation with spatially-conditioned generative adversarial networks. In: Cardoso, M.J., et al. (eds.) CMMI/SWITCH/RAMBO -2017. LNCS, vol. 10555, pp. 105–115. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67564-0_11
Isola, P., Zhu, J.Y., Zhou, T., Efros, A.A.: Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1125–1134 (2017)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Mattausch, O., Goksel, O.: Scatterer reconstruction and parametrization of homogeneous tissue for ultrasound image simulation. In: 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 6350–6353. IEEE (2015)
Mattausch, O., Makhinya, M., Goksel, O.: Realistic ultrasound simulation of complex surface models using interactive Monte-Carlo path tracing. Comput. Graph. Forum 37, 202–213 (2018)
Mirza, M., Osindero, S.: Conditional generative adversarial nets. arXiv preprint arXiv:1411.1784 (2014)
Nie, D., et al.: Medical image synthesis with deep convolutional adversarial networks. IEEE Trans. Biomed. Eng. 65(12), 2720–2730 (2018)
Salehi, M., Ahmadi, S.-A., Prevost, R., Navab, N., Wein, W.: Patient-specific 3D ultrasound simulation based on convolutional ray-tracing and appearance optimization. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9350, pp. 510–518. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24571-3_61
Shankar, P.M., Reid, J.M., Ortega, H., Piccoli, C.W., Goldberg, B.B.: Use of non-rayleigh statistics for the identification of tumors in ultrasonic B-scans of the breast. IEEE Trans. Med. Imaging 12(4), 687–692 (1993)
Starkov, R., Zhang, L., Bajka, M., Tanner, C., Goksel, O.: Ultrasound simulation with deformable and patient-specific scatterer maps. Int. J. Comput. Assist. Radiol. Surg. 14(9), 1589–1599 (2019). https://doi.org/10.1007/s11548-019-02054-5
Tom, F., Sheet, D.: Simulating patho-realistic ultrasound images using deep generative networks with adversarial learning. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 1174–1177. IEEE (2018)
Vitale, S., Orlando, J.I., Iarussi, E., Larrabide, I.: Improving realism in patient-specific abdominal ultrasound simulation using CycleGANs. Int. J. Comput. Assist. Radiol. Surg. 15(2), 183–192 (2019). https://doi.org/10.1007/s11548-019-02046-5
Wang, T.C., Liu, M.Y., Zhu, J.Y., Tao, A., Kautz, J., Catanzaro, B.: High-resolution image synthesis and semantic manipulation with conditional gans. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 8798–8807 (2018)
Wolterink, J.M., Dinkla, A.M., Savenije, M.H.F., Seevinck, P.R., van den Berg, C.A.T., Išgum, I.: Deep MR to CT synthesis using unpaired data. In: Tsaftaris, S.A., Gooya, A., Frangi, A.F., Prince, J.L. (eds.) SASHIMI 2017. LNCS, vol. 10557, pp. 14–23. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-68127-6_2
Wu, Y., Burda, Y., Salakhutdinov, R., Grosse, R.: On the quantitative analysis of decoder-based generative models. arXiv preprint arXiv:1611.04273 (2016)
Zhang, Z., Yang, L., Zheng, Y.: Translating and segmenting multimodal medical volumes with cycle-and shape-consistency generative adversarial network. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 9242–9251 (2018)
Zhu, J.Y., Park, T., Isola, P., Efros, A.A.: Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 2223–2232 (2017)
Acknowledgments
Funding was provided by the Swiss Innovation Agency Innosuisse.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhang, L., Portenier, T., Paulus, C., Goksel, O. (2020). Deep Image Translation for Enhancing Simulated Ultrasound Images. In: Hu, Y., et al. Medical Ultrasound, and Preterm, Perinatal and Paediatric Image Analysis. ASMUS PIPPI 2020 2020. Lecture Notes in Computer Science(), vol 12437. Springer, Cham. https://doi.org/10.1007/978-3-030-60334-2_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-60334-2_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-60333-5
Online ISBN: 978-3-030-60334-2
eBook Packages: Computer ScienceComputer Science (R0)