Abstract
Semantic segmentation has been popularly addressed using fully convolutional networks (FCNs) with impressive results if the training set is diverse and large enough. However, FCNs often fail to achieve satisfactory results due to a limited number of manually labelled samples in medical imaging. In this paper, we propose a conjugate fully convolutional network (CFCN) to address this challenging problem. CFCN is a novel framework where pairwise samples are input and synergistically segmented in the network for capturing a rich context representation. To avoid overfitting introduced by appearance and shape changes in a small number of training samples, a fusion module is designed to provide proxy supervision for the network training process. Quantitative evaluation shows that the proposed method has a significant performance improvement on pathological liver segmentation.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Semantic segmentation, a computer vision task, aims at predicting semantic labels for each pixel of an image. Benefiting from the recent development of deep learning, it has achieved great success in natural image segmentation [4, 8]. While the 2D/3D medical image segmentation can leverage similar technologies (e.g., U-Net [13] and V-Net [9]) to achieve a sound result, there are a few challenges that deserve broad attention. Firstly, semantic segmentation is a task to assign a consistent semantic label to a category of objects, rather than to each single pixel. Actually, all the objects, not only the ones to be segmented, in medical images/volumes usually lie in a low-dimensional manifold and modeling the intrinsic relations of them is of great significance for segmentation. For example, in liver segmentation, the relative positions of the surrounding organs are very important for locating the liver and helpful to the liver segmentation. Secondly, individual differences such as smoothness and pixel intensity of target objects in different images need a large number of training samples to be discriminative. With limited training samples, how to trade-off between exactly modeling the manifold of target objects and robustly representing the individual difference is a bottleneck to a fully convolutional network (FCN) and its variants, e.g., U-Net [13] and V-Net [9].
There are some remarkable works for modeling the manifold or prior knowledge of the target objects. One group leveraged the intrinsic relations among the same category of pixels to improve the performance of FCNs. For example, a dense conditional random field (CRF) was attached to the FCN as a post-processing step to preserve the boundary of the object in [3]. Similarly, Ke et al. [6] proposed an adaptive affinity field to encode spatial structural information and geometric regularities through the label relations in the training process. Another group of methods improved the segmentation performance of FCNs by explicitly or implicitly modeling high-order prior knowledge in-between different objects in medical images/volumes, such as shape, topological structure, etc. Chen et al. [2] took gland objects and contours as auxiliary information under a multi-task learning framework to boost the gland segmentation from histology images. In [12], a non-linear shape model was pre-learned by convolutional autoencoders (CAE) to model the shape manifold space and then incorporated in an FCN. Ravishankar et al. [11] proposed a novel framework based on deep learning to jointly learn the foreground, background and shape to improve segmentation accuracy. In [1], a topology-aware loss was proposed to train the FCN for coding geometric and topological priors of containment and detachment on histology gland segmentation.
Considering semantic segmentation is a structured prediction task, a target object lying in different images/volumes should have consistent labels. This implies that objects in different slices of volumes or different patches of images usually have intrinsic relations among context, shape and location. To model these intrinsic relations, we propose a conjugate fully convolutional network (CFCN) to pairwisely segment the medical objects. Generally speaking, our CFCN has two conjugate sub-networks, each for segmenting one single sample of a paired input. To capture the intrinsic relations of pairwise input and encode the manifold of target objects, the two sub-networks share the same weights in the encoder backbone, which implies that the low-level features captured by CFCN should be sufficient to represent the target object and robust for distinguishing backgrounds. In the remaining layers, the two conjugate sub-networks have independent weights, which enables CFCN to learn discriminative features for representing each of the two inputs. As medical image segmentation is a location-aware task where the relative position of the anatomical structure is very important for locating the target object, we design a fusion sub-network to provide proxy supervision for modeling intra-class inconsistency and prior shape knowledge. Different from the mini-batch training manner of FCNs, in the CFCN, features of the pairwise input interact and guide each other in the fusion module, which offers a new data augmentation method that one sample could be paired with (multi-shot by) different samples.
We demonstrate the efficiency of our approach on the typical problem of pathological liver segmentation. Compared with the traditional FCNs, the main contributions of this paper are three-fold: Firstly, a new conjugate network framework is proposed to pairwisely mine the semantic information of samples and implicitly model the correlation between the two input samples. Secondly, a novel fusion module is designed to provide proxy supervision for eliminating intra-class inconsistency and modeling shape prior of target objects to improve segmentation accuracy. Thirdly, pairwise input can augment the magnitude of training samples and provide a new solution for medical image segmentation with limited training samples.
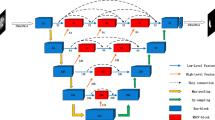
The overview of the proposed conjugate fully convolutional network. The architecture consists of three parts: encoder module, decoder module, and fusion module. The network takes two different inputs and outputs the corresponding probability maps for the segmentation throughout two sub-networks of the decoder module. Besides, an additional output of the fusion module is employed to fit the proxy supervision.
2 Method
In this section, we present the details of the proposed pairwise segmentation network CFCN. The CFCN model consists of three parts: an encoder module, a decoder module and a fusion module. The encoder module takes a pair of samples as inputs and jointly captures their representations. The decoder module consists of two conjugate sub-networks which take the low-level and high-level features from the encoder module as their input and learn the discriminative features for segmenting each input under the supervision of their own groundtruth maps. The fusion module uses the summation of low-level features from the encoder module, and the summation of high-level features from the decoder module, as its input to capture the location-aware representation under the proxy supervision. Our CFCN for medical image segmentation is illustrated in Fig. 1.
Before detailed description, we summarize the notations used in this paper. The training data is composed of an image sequence \(\mathbf X = \{\mathbf x^1, \mathbf x^2, \cdots , \mathbf x^N \}\) from 3D medical volumes and a corresponding mask sequence \(\mathbf Y = \{\mathbf y^1, \mathbf y^2, \cdots , \mathbf y^N \}\), with \(\mathbf x^i \in \mathbb R^{h\times w}\) and \(\mathbf y^i \in \{0,1\}^{h\times w}, i=1,2,\cdots , N\), where h, w denote the height and width of the medical image, respectively.
2.1 Encoder Module
The encoder module has two parallel fully convolutional sub-networks with shared weights and each of them consists of a group of cascading residual blocks [5] for gradually extracting more abstract features. After then, each of the two sub-networks is followed by an Atrous Spatial Pyramid Pooling (ASPP) module to capture the multi-scale semantic information as [4]. In order to reduce the number of parameters and improve computation efficiency, the encoder module in this paper is constructed based on ResNet-18 which has four residual blocks with 18 layers in total.
Different from the traditional medical image segmentation networks, our encoder module takes pairwise samples as inputs and try to make the siamese sub-networks guide each other during the training process. As aforementioned, the medical image/volume globally lies in a low-dimensional manifold and pairwise input usually has intrinsic relations in context, shape, location, etc. Using this strategy, the encoder module can learn rich semantic information representing the target object while robustly distinguishing it from the background. In the fusion module we further exploit the intrinsic relations of paired inputs. The outputs of each sub-networks are high-level and low-level features from ASPP and the first residual blocks of ResNet-18, respectively, which serve as the inputs of the decoder module and the fusion module.
2.2 Decoder Module
The decoder module consists of two conjugate convolutional sub-networks (Sub-network A and Sub-network B) for segmenting each sample of the paired inputs. Each decoder sub-network uses the same architecture as [4], where the high-level features from the encoder’s ASPP module are filtered by a convolutional layer and up-sampled to the same size of the low-level features, and then concatenated with the low-level features from ResNet-18. The resulting feature maps are fed into a convolutional layer with \(3 \times 3\) convolutions and bilinearly up-sampled to the same size as the mask maps. Note that, if removed one of the two conjugate sub-networks, the other one attached with the encoder module can be separately regarded as DeepLabv3+ [4] with a ResNet-18 backbone, which is a state-of-the-art architecture in semantic segmentation. Please refer to [4] for more details.
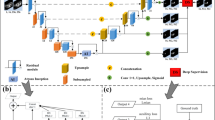
Two typical challenges faced in the medical image segmentation. (a) Intra-class inconsistency: a pair of slices are from two different volumes, where the livers differ greatly in appearance. And the local contexts are very different on the below pathological liver. Synergistically predicting the intersection and difference of the two masks can eliminate intra-class inconsistency, as shown in the central subgraph. (b) Shape modeling in an end-to-end manner. To address this, a pair of slices are sampled from one volume with a small interval along the axial direction, and the difference of the two masks can encode the shape prior of the liver, as the central subgraph shows. Note that the lesions are masked for better visualization. (Color figure online)
2.3 Fusion Module
The fusion module is designed by considering the following motivations: the first one is intra-class inconsistency where the target objects in different images/volumes share the same semantic label but different appearances. For example, in liver segmentation, individual anatomy difference and imaging device difference can both result in intra-class inconsistency as shown in Fig. 2(a). Especially with limited training data, the network is sensitive to intra-class inconsistency and prone to overfitting. The second motivation is shape learning in an end-to-end manner. As shown in Fig. 2(b), if the two inputs are sampled from the same volume with a small interval along the axial direction, then the difference of the mask (where region masked in red denotes only one of the two inputs’ pixels belonging to the target objects) can encode the shape prior of the target object. To this end, we design a fusion module to learn location-aware features through a proxy supervision in the training process. Suppose \(\mathbf x^i\) and \(\mathbf x^j\) are a pair of inputs, \(\mathbf y^i\) and \(\mathbf y^j\) are their corresponding groundtruth maps, then the proposed proxy is simply implemented by \(\mathbf y^{ij}=\mathbf y^i+\mathbf y^j\), where the mask \(\mathbf y^{ij} \in \{0, 1, 2\}^{h\times w}\) and 0, 1, 2 imply the two inputs’ pixels, at the index, are both belonging to the background, one for the target object and both belonging to the target object, respectively. In other words, we can implement the proxy supervision by an additional 3-category segmentation task, where the labels constitute three maps, including the intersection of background, the difference and intersection of the target object (corresponding to the label 0, 1, 2), respectively.
As aforementioned, the proxy supervision is abstract prior knowledge which requires the fusion module to exploit semantic information of the low-level and high-level features of the paired inputs. To this end, we element-wisely add the low-level and high-level features, and take the overall features as the input of the fusion module. Furthermore, inspired by [14], we adopt a channel attention block to refine the low-level features. With this design, the fusion module can adaptively learn global context information with a negligible computation cost. In pursuit of further fusion, we sum the features from the channel attention block and the high-level features. Then, the features are fed into a \(3 \times 3\) convolutional layer and bilinearly up-sampled to the same size as output corresponding to the proxy supervision maps.
In this paper, we employ multi-class Dice loss to learn the proxy supervision:
where \(\mathbf p^{ij}\) and \(\mathbf y^{ij}\) are the CFCN’s output probability maps and groundtruth maps corresponding to \(\mathbf x^{ij}\), respectively; s, t are the height and width indices of the two maps; and \(\ell \) is the category index of the proxy supervision. With the proxy supervision, the network is prone to predicting the target objects on different inputs as the same category, and the difference mask map can encode the contour information.
The overall loss of the proposed CFCN model can then be presented:
where \(\mathbf y^i\) and \(\mathbf y^j\) are the mask maps of \(\mathbf x^i\) and \(\mathbf x^j\); \(\mathbf p^i\) and \(\mathbf p^j\) are their corresponding predicted probability maps output by the Sub-network A and Sub-network B of the decoder module, respectively; \(\mathcal L_1\) and \(\mathcal L_2\) refer to the pixel-level segmentation losses for \(\mathbf x^i\) and \(\mathbf x^j\), respectively. In this paper, the Dice loss is adopted for \(\mathcal L_1\) and \(\mathcal L_2\). Note that \(\lambda \) is a user-preset weight used to balance the contribution of the paired inputs’ loss and the proxy loss.
3 Experiments and Discussions
We then substantiate the robustness and generalization capability of our proposed framework on the 2D segmentation of abnormal liver. Since the presence of any pathology or abnormality may seriously distort the scanned texture, accurate pathological liver segmentation remains a challenge to deep FCNs, especially with small or moderate amount of training data.
Dataset: We evaluate our method on the public benchmark dataset of the Liver Tumor Segmentation Challenge DatasetFootnote 1 (LiTS), which consists of 201 contrast-enhanced abdominal CT volumes acquired from multiple clinical sites. Since the challenge organizers only provide a subset of 131 volumes with manually labelled liver masks, we perform all our experiments on this subset.
Implementation: All experiments are implemented with the PyTorch framework [10]. We use the Adam Optimizer [7] with batch size of 20, weight decay of \(5e^{-4}\), and learning rate of \(1e^{-4}\) in training. As for \(\lambda \), we empirically set it as 0.2. For each of paired inputs, we adopt a 2.5D input with five adjacent slices.
We use a fixed proportion 20% (26 volumes) of the 131 volumes as the test set and the remaining volumes as the training set. In order to verify that our model can achieve a reasonable segmentation accuracy with a limited number of training samples, additional experiments are designed with 5% and 1% of the training subset (corresponding to 7 volumes and 1 volume, respectively), and the training samples are augmented by the aforementioned multi-shot ways, i.e. paired inputs are sampled from two random volumes with each slice paired twice, and sampled from one volume at intervals of 5, 9, and 13 slices. We use the average Dice score of all volumes to evaluate the performance of the proposed method, and compare the results with U-Net [13] and DeepLabv3+ [4] with a ResNet-18 backbone due to their sound performance and close relation with our CFCN model, as mentioned in Sect. 2.2.
The results are listed in Table 1. As shown, our CFCN achieves a Dice score of 96.43% with \(80\%\) training data, which outperforms U-Net [13] and DeepLabv3+ [4] with Dice scores of 95.88% and 95.93%, respectively. Especially, with \(5\%\) training data, the performance of CFCN is 94.45%, outperforming \(1.8\%\) compared with the two state-of-the-art networks. As shown in Fig. 3, CFCN is superior to the two comparison methods in delineating the boundary and maintaining intra-class consistency of the liver segmentation. Unsurprisingly, the CFCN model focuses on modeling the manifold of target objects and eliminating the effect of intra-class inconsistence, which is of great significance for network training with a small training set. Furthermore, we also try to train deep models with a extremely limited training set of one volume, and the performance of CFCN is 85.26%, outperforming \(4.1\%\) compared with U-Net [13] and DeepLabv3+ [4].
To investigate the effect of each component of our CFCN model, we perform an ablation study on the training set with \(5\%\) volumes. We respectively study the ablation for the fusion module, pairwise input from different volumes as Fig. 2(a) shows, and pairwise input from the same volume as Fig. 2(b) shows. For simplicity, we call the three ablation schemes as Siamese DeepLabv3+, heterogeneous inputs and homogeneous inputs, respectively. Note that the latter two are with the fusion module. As Table 2 shows, these schemes can gradually and effectively improve the segmentation performance with a limited training set.
4 Conclusion
In this paper, we proposed the CFCN model to pairwisely segment pathological livers on CT, which employs location correlation to eliminate intra-class inconsistence and shape priors to model the manifold of target objects in an end-to-end training manner. The experimental result demonstrates that CFCN can significantly improve segmentation accuracy with a limited number of training data. The model can be naturally extended to other medical segmentation applications and we will further exploit relevant prior knowledge to incorporate into deep learning models.
References
BenTaieb, A., Hamarneh, G.: Topology aware fully convolutional networks for histology gland segmentation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 460–468. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_53
Chen, H., Qi, X., Yu, L., Heng, P.A.: DCAN: deep contour-aware networks for accurate gland segmentation. In: CVPR, pp. 2487–2496 (2016)
Chen, L.C., Papandreou, G., Kokkinos, I., et al.: DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE TPAMI 40(4), 834–848 (2018)
Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) ECCV 2018. LNCS, vol. 11211, pp. 833–851. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01234-2_49
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: CVPR, pp. 770–778 (2016)
Ke, T.-W., Hwang, J.-J., Liu, Z., Yu, S.X.: Adaptive affinity fields for semantic segmentation. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) ECCV 2018. LNCS, vol. 11205, pp. 605–621. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01246-5_36
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: CVPR, pp. 3431–3440 (2015)
Milletari, F., Navab, N., Ahmadi, S.A.: V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: Fourth International Conference on 3D Vision, pp. 565–571 (2016)
Paszke, A., Gross, S., Chintala, S., et al.: Automatic differentiation in PyTorch. In: NIPS Workshop Autodiff, pp. 1–4 (2017)
Ravishankar, H., Thiruvenkadam, S., Venkataramani, R., Vaidya, V.: Joint deep learning of foreground, background and shape for robust contextual segmentation. In: Niethammer, M., et al. (eds.) IPMI 2017. LNCS, vol. 10265, pp. 622–632. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-59050-9_49
Ravishankar, H., Venkataramani, R., Thiruvenkadam, S., Sudhakar, P., Vaidya, V.: Learning and incorporating shape models for semantic segmentation. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10433, pp. 203–211. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66182-7_24
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Yu, C., Wang, J., Peng, C., et al.: Learning a discriminative feature network for semantic segmentation. In: CVPR, pp. 1857–1866 (2018)
Acknowledgments
This work was supported by the China NSFC (11690011, 61661166011, 61721002, 81830053, U1811461) and the Key Area Research and Development Program of Guangdong Province, China (2018B010111001).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Wang, R., Cao, S., Ma, K., Meng, D., Zheng, Y. (2019). Pairwise Semantic Segmentation via Conjugate Fully Convolutional Network. In: Shen, D., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science(), vol 11769. Springer, Cham. https://doi.org/10.1007/978-3-030-32226-7_18
Download citation
DOI: https://doi.org/10.1007/978-3-030-32226-7_18
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32225-0
Online ISBN: 978-3-030-32226-7
eBook Packages: Computer ScienceComputer Science (R0)