Abstract
Internet of Things for medical devices is revolutionizing healthcare industry by providing platforms for data collection via cloud gateways and analytic. In this paper, we propose a process for developing a proof of concept solution for sleep detection by observing a set of ambulatory physiological parameters in a completely non-invasive manner. Observing and detecting the state of sleep and also its quality, in an objective way, has been a challenging problem that impacts many medical fields. With the solution presented here, we propose to collect physiological signals from wearable devices, which in our case consist of a smart wristband equipped with sensors and a protocol for communication with a mobile device. With machine learning based algorithms, that we developed, we are able to detect sleep from wakefulness in up to 93% of cases. The results from our study are promising with a potential for novel insights and effective methods to manage sleep disturbances and improve sleep quality.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Sleep monitoring
- Mobile health
- Data collection
- Actigraphy
- Quantified-self
- Wearables analytics
- Classification
1 Introduction
Sleep disorders are affecting a significant percentage of adult population, where insomnia, sleep apnea, and somnolence [18] are being the most prevalent. The lack of sleep has been associated with irritability and a negative mood, lack of energy and problems with memory and retaining information, and even weight gain and health issues. It has been recently shown that lack of sleep, besides resulting in lowered cognitive and physical performance is even linked to diabetes and cardiovascular diseases [16]. In addition, many chronic conditions such as diabetes, sleep apnea, and cardiovascular diseases actually result in an increased risk of an attack during sleep. Therefore new methods for detecting, monitoring, and managing sleep disorders are of essential importance today.
Physiological changes during sleep and sleep disorders have been extensively studied via polysomnography methods. Such methods involve collecting signals like the electroencephalography (EEG) for observing brain activity, electrocardiography (ECG) for heart rhythm, electronystagmography (ENG), electrooculography (EOG) for eye movement, and electromyography (EMG) for muscle activity or skeletal muscle activation. However, these methods are costly and require hospitalization of the patient for the duration of the procedure. Thus, increased research efforts are oriented towards a non-invasive monitoring of human physiological parameters as well as activity parameters. A wide range of wearable sensors are being developed for real-time non-invasive health monitoring, which opens up possibilities to follow sleep patterns and analyze the dependence between sleep and measured physiological variables.
Physiological variables of healthy individuals are controlled optimally during wakefulness to respond to the body’s functioning. The body temperature, blood pressure, and levels of oxygen, carbon dioxide, and glucose in the blood remain quite constant and optimally regulated during wakefulness. During sleep, however, physiological demands are reduced and, for example, temperature and blood pressure drop. In general, many of the physiological functions such as brain wave activity, breathing, and heart rate are quite variable when a person is awake or during rapid eye movement (REM) sleep, but are extremely regular when a person is in non-rapid eye movement (NREM) sleep.
In this paper, we propose a method for monitoring sleep by use of physiological signals collected solely from a bracelet-like and sensor-equipped wristband which is comfortable, discreet, and non-invasive and therefore perfectly suitable for ambulatory settings. We collect physiological signals that correspond to electrodermal activity, heart rate variability, body temperature, and body movements. Our goal is to propose a system that objectively measures and estimates the periods of sleep and wakefulness, respectively, and we aim to achieve that by a machine learning classifier which classifies based on physiological signals solely. However, for training the classifier, we needed a labeled training set. For this purpose, we developed a mobile application that provides an intuitive user interface through which a user can register his/her sleep and wake hours while wearing the sensor-equipped wristband. The data provided by the user in this way are used to construct a training dataset which is further used to train a support vector machine based classifier. We show that by optimizing parameters of the classifier, we are able to correctly classify sleep and wakefulness in up to 93% of the cases. We present two applications, one implemented as a part of the mobile application and another on a cloud back-end application. We show that both types of classifiers ultimately achieve similar performance.
This paper is organized as follows: in Sect. 2 we discuss the related works, in Sect. 3 we present the problem formulation and our contributions, in Sect. 4 we present our data collection process, tools, and methods, the wearable sensor and the type of signals that we are able to collect with it, and the architecture of our system. We also explain our machine learning methods and classification results in this section. In Sect. 5 we present our mobile application and the user interface and we conclude in Sect. 6.
2 Related Work
Recent works related to ambulatory monitoring of sleep regularity focus on the type of sensors and sensing signals that can be obtained and the type of data analysis and the knowledge that can be extracted from the collected data. Furthermore, various studies focus on different types of sleep disorders among which the most frequent are insomnia or lack of sleep, somnolence or sleepiness, and sleep apnea [18].
In order to properly diagnose various sleep disorders, data regarding physiological signals are collected in hospitals via controlled laboratory experiments and this method is known as polysomnography (PSG). The polysomnography is the “gold standard” for sleep assessment [4], is very expensive, and can be intrusive to sleep itself, making the search for alternatives essential to the field. Study participants come to a sleep laboratory where multiple channels of data are collected. PSG provides general sleep measures, such as total sleep time (TST = sleep duration) and sleep efficiency (SE = TST/time in bed), light sleep (nREM1 + nREM2), and deep sleep (SWS + REM). The phases nREM1 and nREM2 are known as non-rapid eye movement and are providing measures of specific sleep stages referred to as light sleep, while REM, known as rapid eye movement together with SWS, known as slow-wave sleep, is often referred to as deep sleep. Polysomnography recordings include electroencephalography (EEG), electrooculogram (EOG), and electromyogram (EMG) of patient data among other signals [1].
In the last couple of years, the method known as actigraphy appeared together with the democratization and increasing usage of the non-invasive devices in many fields including health domain. With real-time information from the patient, collected in natural environment, this rich source of information allows users to change their lifestyle, predict and prevent hazardous events, optimize physical activities and sleep patterns [9].
Actigraphy is a non-invasive method, which can be used to infer sleep/wake patterns based on periods of activity versus inactivity, by analyzing raw data through specific algorithms [3]. The detection of movement is calculated by the accelerometer of the bracelet or by combining the accelerometer on the wrist with the accelerometer in a mobile phone. Actigraphy, in the domain of sleep medicine, has been a research method for studying a person’s sleep-wake cycle, as demonstrated by the recent studies [11, 15].
There are three main actigraphy approaches [1] for inferring sleep quality: (1) objective approach based on measure metrics such as total sleep time (TST) and sleep efficiency (SE), (2) subjective approach based on self-reports via surveys and diaries, and (3) machine learning based, the approach that often combines the two previous methods, whose goal is to improve the objectivity and reliability of the observations from data collection via machine learning algorithms.
Sensor-equipped devices are being increasingly used to monitor patients with various chronic diseases, such as cardiovascular and neurological. Different sensors are combined and incorporated into wrist-wearable devices to be used in a plethora of applications providing feedback to users and also to health professionals. Currently, we observe a trend towards strengthening the customization or personalization of feedback to the users. In the domain of personalized health-care services [13] the evolution of sensor-equipped smartphones offers tremendous opportunities for monitoring. A variety of dedicated mobile applications offer new opportunities in psychological research [6] and many other areas. QuantifyMe platform [17] is one of the recent attempts that are starting to emerge in the quantified-self field. The goal is to allow to the non-experts to try to conduct self-experiments by use of appropriate methodology, in an automated way, using their smartphones. This technology is supposed to allow the users to find their personal optimal behavioral variables (e.g., bed time or physical activity) or to achieve their goals (e.g., productivity or happiness) based on evidence-based experimentation.
In a recent study [10] the authors outline platform aimed at processing physiological data from several wearable sensors and devices, namely Microsoft Band 2, Empatica E4, eHealth Sensor Platform, and BITalino (http://bitalino.com/). In this study critical comparison of the quality of HR and GSR signals is given. Most of the devices used in this study are wristbands, which are low-cost and accessible, thus creating opportunity for real life applications.
Several recent studies have compared the two most prominent approaches: polysomnography and actigraphy. In [15] the authors compared results for sleep monitoring obtained using PSG and the results from commercially available devices such as Fitbit Flex and Actiwatch among others. Different aspects were compared in this study: (1) regarding sleep efficiency (SE), it was shown that the device had low correlation with PSG, (2) regarding total sleep (TST), for all devices, a strong correlation with the PSG method was shown, (3) regarding light sleep time: significant difference with respect to PSG was reported, and (4) regarding deep sleep time measurements, the results were correlated with the PSG measurement. This study reveals strengths and limitations between actigraphy and PSG.
In [11] the authors evaluate the applicability of data obtained from a wearable activity tracker, Fitbit Charge HR, to medical research. This study was performed by comparing the wearable activity tracker, Fitbit Charge HR, with an actigraphy device, Actiwatch 2 (Royal Philips), for sleep evaluation and circadian rest-activity rhythm measurement. The findings of this study showed that the Fitbit Charge HR is a valid, reliable, and alternative device for use for sleep evaluations and circadian rest-activity rhythm measurements compared with actigraphy in healthy young adults. However, the sensitivity of the Fitbit Charge HR for accurately identifying activity was lower than actigraphy.
In [16] the link between electrodermal activity, skin temperature, and sleep was explored. They presented various feature sets for sleep/wake identification using electroencephalography (EEG) and actigraphy, while [7] elaborates more in detail about the EDA signal (phasic and tonic) patterns during sleep, and incorporates this information into a relatively simple method for identifying sleep and wake epochs using EDA data only. The results of this study indicate that electrodermal activity is not only a robust parameter for describing sleep, but also a potentially suitable method for ambulatory sleep monitoring. Automatic sleep classification based on support vector machine method and using EEG signal was presented in [2].
The heart rate signal for sleep monitoring was explored in [11] and [1], the reported results are encouraging but require complementarity with other signals. In [12] electrocardiogram (ECG) and heart rate variability (HRV) signals were used to classify sleep in infants and the correct classification reported was between 85% and 87%. In [5] different algorithms are used to predict sleep quality, using the heart rate and temperature signals. Our work differs compared to these works in the sense that it uses more comprehensive set of different physiological signals and their derived features.
The user interface and its different aspects and their impact on user together with practical considerations for designing have been discussed in [14].
3 Problem Formulation
Sleep regularity and disorders have usually been monitored and analyzed using polysomnography methods. Polysomnography is a comprehensive recording of the biophysiological changes that occur during sleep. A polysomnogram will typically record a minimum of 12 channels requiring a minimum of 22 wire attachments to the patient.Footnote 1 It is obvious that such procedures must be performed by a trained medical expert and require hospitalization of the patient.
Our objective in this work is to study and show to what extent it is possible to accurately detect the regularity of the sleep patterns in ambulatory conditions. For this purpose we use a wearable device that is equipped with multiple sensors and is usually worn on the wrist. The device is comfortable, easy to use, and poses absolutely no harm or risk to the user. Our contributions are the following:
-
To the best of our knowledge we are the first to use at the same time, combined electrodermal activity (EDA), heart rate variability (HRV), accelerometer data, and blood volume pulse (BVP) together with body temperature in order to classify sleep and wakefulness. Previous works for sleep classification have concentrated on one or the combination of some of the aforementioned signals.
-
We provide a study about various features and their relevance to the sleep classification problem.
-
We have developed and presented a mobile application that is connected with the sensor-equipped bracelet to collect the physiological signals, provide data visualization and information about the regularity and duration of sleep patterns.
-
We have proposed a support vector machine (SVM) based model to classify sleep and wakefulness, tested several kernels, and optimized the parameters such that the classification F1 score is up to 93%.
4 Methodology
Several wrist-wearable monitoring devices have recently appeared on the market among which the most prominent are Fitbit, Apple, Xiaomi, Garmin, and Fossil. However, the most comprehensive acquisition and access to raw data is currently provided by Empatica E4 sensorFootnote 2 and thus we decided to use it for the purpose of this study.
4.1 Wearable Sensor and Architecture
The Empatica E4 bracelet, the device that has been used for this project, offers the acquisition in real time of physiological data and provides full access to data for research purposes. The Empatica company has made available the Empatica Connect platform,Footnote 3 which allows to visualize the graphs corresponding to different signals. The bracelet works in two modes: (a) recording mode: the wristband records the data in the internal memory, and it can record up to 60 h. (b) Streaming mode: the bracelet connects via Bluetooth to the application.
The Empatica E4 bracelet is equipped with the following physiological sensors:
-
EDA Sensor (or GSR Sensor): measures the fluctuating changes of certain electrical properties of the skin. It is measured in microsiemens.
-
Infrared Thermopile: measures the temperature of the skin and gives the data measured in Celsius degrees.
-
3-axis Accelerometer: measures motion based activity, contains the data of the 3-axis (x, y, and z) accelerometer sensor. It measures continuous gravitational force (g) applied to each of the three spacial dimensions (x, y, and z). The scale is limited to ±2g (by default) and can be extended to ±8g with custom firmware. The accelerometer in our application is configured to measure acceleration in the range [−2g, 2g]. The unit is expressed in g which is 9.81 m/s.
-
PPG Sensor (Photoplethysmography Sensor): measures the blood volume pulse (BVP) from which two signals can be derived: (1) heart rate (HR) and the interbeat interval (IBI). The blood volume pulse is measured in nanoWatts, heart rate variability HRV is measured in beats per minute (bpm), and interbeat interval is measured in time between two consecutive beats.
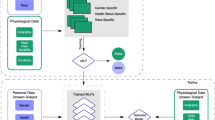
The architecture of our system is represented in Fig. 1. Figure 1 represents our system architecture. It consists of three main parts: (Subsystem 1) Machine Learning (top part, green-dotted rectangle) which aims at building the best performant model by iterating the different steps of the pipeline which are prepare data, extract features, evaluate and tune model, and finally build the best performant one. The final pipeline step consists of deploying the model on the repository S3 on AWS Cloud after being serialized. (Subsystem 2) Android mobile application (left part, black-dotted rectangle) whose aim is to interact with the Cloud Backend and make a Bluetooth connection with Empatica E4 wristband, gather values received from all the sensors embedded in it and sending them to the Backend. It also fetches the results from the Backend and visualizes them in a graphical and statistical way. (Subsystem 3) Backend and Cloud (right part, brown-dotted rectangle) composed by the three Cloud model PaaS (Platform as a Service) (S3 Bucket repository where we store some data and code), DBaaS (Database as a Service) (DynamoDB databases), and FaaS (Function as a Service) (Lambda Function where different Python scripts are triggered to execute many tasks like the prediction algorithm). The objective is to gather raw data received by the mobile application and sample them with a 600-s window. The prediction process is automated and launched after each recorded patient session and the results are stored in the dataset.
4.2 Data Collection
In order to collect data, each participating subject wore the Empatica E4 wristband for several days and for several long uninterrupted periods during day and night. Three subjects, two men and one woman, participated in this study. Each day they recorded the exact time of falling asleep and was recorded waking up. On some occasions, when they woke at night, such as going to the bathroom or drinking water, such periods were recorded as a wakeness hours. The subjects wore the bracelet a few hours before going to bed and a few hours after waking up, which favors a balanced dataset.
Empatica E4 provides six different measures: accelerometer data (ACC), blood volume pulse (BVP), electrodermal activity (EDA), heart rate (HR) (which is) automatically derived from BVP, interbeat interval (IBI) also calculated from BVP, and temperature (TEMP). These measures however do not have the same frequency, some are sampled at 1 Hz frequency like, for example, the heart rate (HR) and some like the blood volume pulse (BVP) at 64 Hz. For this study, we decided to downsample the signals at the same frequency of 1 Hz. Using the collected data, we created a training set containing all the features labeled by each participant. The labeling is manually done using the mobile application, with the two values 0 and 1, respectively, corresponding to “sleep” and “awake,” respectively. We have 21 days of data collected, and the global dataset contains recordings of 1,058,374 s in total.
We collected data from three subjects as three is the maximum participants’ number that we can use to implement and test an experimental model. In the future we aim to make this application openly accessible for use by multiple subjects with the consent from ethics committee.
4.3 Features Extraction
In order to better characterize the data collection, we summarized several statistics from the collected set of signals. We focus on measures such as the average values and the standard deviation in order to find out which are good features candidate for the classification algorithm. In the discussion below we summarized the measures related to a session whose total time is 15 h, 42 min and 16 s. The sleep and wakefulness period duration was 5 h and 18 min and 10 h and 22 min.
We have looked more in detail to discover how each signal is affected during sleep and wakefulness. In Fig. 2 we have represented the EDA signal for the three subjects during periods of sleep and wakefulness. Here we observe a significant drop when the sleep period starts and significantly increases when the wakefulness period starts. This observation is consistent with the EDA analysis in [8]. We observe differences between subjects, though. The drop in EDA amplitude at sleep period is more pronounced for subject 1 and the increase in EDA amplitude is more pronounced for subjects 2 and 3.
In Figs. 3, 4, and 5 we show the heart rate variability (HRV), the blood volume pulse (BVP), and the interbeat interval (IBI). As we mentioned previously, BVP is the directly measured from the PPG sensor and HRV and IBI are derived from BVP. As for the BVP signal from Fig. 4, we observe that the variance is significantly higher during wake periods than during sleep. This observation is consistent for the three subjects. Thus, the standard BVP deviation appears to be a good significant difference between sleep and wakefulness periods. Regarding HRV signal from Fig. 3, we notice that both the variance and the average values increase during wakefulness periods (compared to sleep), and this is also consistent for the three subjects. This is in line with the observations in [12] where the ECG signal is measured to derive the HRV for sleep and wakefulness classification and the average of HRV is considered as a feature for classification. As for the IBI signal, Fig. 5, we observe that its amplitude increases during sleep periods as the breathing slows down. This is consistent for the three subjects, as we notice that during the period of sleep the signal amplitude of the signal that represents the time between two beats gets longer. We see however that this feature does not improve the classification accuracy and shows that it can be omitted for the final classification without disturbing the performance. We discuss these details in Sect. 4.4.
A summary about the values of average and standard deviation measured for the same period of EDA, HRV, and BVP together with the measurements from the accelerometer (ACC) and body temperature (TEMP) is illustrated in Fig. 6.
The following conclusions can be drawn regarding the different values of the mean and standard deviations for the observed signals:
For the HRV signal, when comparing the differences between sleep and wakefulness periods, the difference between the two average values is greater than the difference between standard deviations. Although the difference is not that high, we tend to choose the HRV mean as a feature. For the ACC signal, the difference between the two mean values is greater than that of the standard deviation; thus, we tend to choose the mean of ACC as a feature. For the EDA signal the difference in mean values between sleep and wakefulness is larger than that of the standard deviation; thus, we tend to choose the EDA mean as a feature. For the BVP signal the difference between the standard deviations between sleep and wakefulness is larger than that of the mean; thus, we tend to choose the BVP standard deviation as a feature.
We further built our model and tested the performance for separate features, doing optimization (tuning) and cross-validation for the training set. Some signals have shown a tendency to deteriorate the performance of the model, and that is why we did not choose them in the final model, TEMP and IBI. Finally we extracted the following features: average values for the heart rate (HR), the electrodermal activity (EDA), the accelerometer (ACC), and the standard deviation for the BVP signal.
4.4 Experimental Results
Our objective is to classify the collected physiological data into two classes, namely sleep and wakefulness, and for this purpose we have decided to use support vector machine (SVM) algorithms. SVM method has been already successfully used for classifying physiological data in other settings such as for classifying sleep and wake using ECG signal [12]. The SVM success is mainly due to the classifier’s ability to create an optimal, highly complex decision boundary for the training data. SVMs use kernel functions to map the input space to a higher dimensional feature space. Optimization techniques are then applied to find the separating hyperplane that maximizes the margin between two classes in the feature space. This creates an arbitrarily complex decision boundary ideal for non-linearly separable data. We have thus explored both linearly and non-linearly separable approaches to investigate which one is the best for separating into the two classes sleep and wake.
Out of all the collected data, we created a training and a testing set by selecting 1764 records for the training set and 441 records for the testing set. The sleep and wakefulness periods were registered manually via mobile application dataset and serve as labels for the training set. Those labels were removed from the testing set. The features such as the mean values and standard deviation were calculated for a period of 1 s.
Next we tested combinations of selected features and performed fine-tuning of the SVM parameters to optimize its performance. We used scikit library to achieve tuning of the hyper-parameters of the classifiers. The library GridSearchCV exhaustively generates candidates from a grid of parameter values: when “fitting” it on a dataset all the possible combinations of parameter values are evaluated and the best combination is retained. Thus, using GridSearchCV we optimized the following parameters:
-
Estimator object: in our case we estimated the best choice for the SVM kernel by testing the three options “linear,” “rbf,” and “sigmoid.”
-
Cross-validation parameter cv determines the cross-validation splitting strategy and the number of folds in stratified k-fold.
-
c: penalty parameter used for the error term. Common to all SVM kernels, this parameter trades off misclassification of training examples against simplicity of the decision surface. A low c makes the decision surface smooth, while a high c aims at classifying all training examples correctly.
-
gamma: kernel coefficient for “rbf” and “sigmoid.” Gamma defines how much influence a single training example has. The larger gamma is, the closer other examples must be to be affected.
In our case the best value for cv of ten was estimated; thus, a linear, radial basis function (RBF) kernel and sigmoid kernels are used, and the optimal model parameters are determined using tenfold cross-validation of the training set.
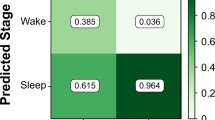
We further tested and evaluated each signal with its mean and standard deviation to estimate their contribution value when used as inputs for the classification. We have observed that blood volume pulse, heart rate variability, and accelerometer data give fairly good classification results even when used as single feature. We decided to use F1 score to evaluate the classification methods, as F1 is usually more useful than accuracy in cases when there is an uneven class distribution, such as ours is. For example, we observe that the standard deviation of BVP, std(BV P) with linear kernel gives F1 score of 0.82, the mean of accelerometer data mean(ACC) with sigmoid kernel results in 0.93 F1 score, and the mean(HRV ) results in 0.84 when used with sigmoid and rbf kernels. We then combined pairs of these features and fed them as inputs to the classifier for each kernel type. The best result was obtained by combining mean(ACC) and std(BV P) with linear kernel, which resulted in 0.92 F1 score.
Finally we combined and tested sets of three and four features. The best results with three features were obtained with mean(EDA), mean(ACC), and std(BV P) with linear kernel and that result is 0.92 F1 score. With four features, we have obtained 0.93 F1 score and the best results were obtained with mean(HRV ), mean(EDA), mean(ACC), and std(BV P) with the “linear” kernel. As the HRV signal cannot be obtained directly via the mobile application the final combination of features that includes mean(HRV) can only be used through the cloud back-end application. Therefore when only mobile application is available for the purpose of training the classificator we can use mean(EDA), mean(ACC), and std(BV P) without much deterioration in performance. For the cloud back-end application, in addition to these three we use mean(HRV ). These results are summarized in Table 1. In both cases, with three and four features, it is obvious that the best results are obtained using the linear kernel. This result holds for the two classifications that are with and without the HRV.
5 Mobile Application
Our mobile application communicates directly and collects the data from the Empatica E4 device and provides real-time information to the user about the regularity of his sleep pattern via an intuitive user interface. This application, developed for Android, allows connection via Bluetooth with the Empatica E4 bracelet. Using the application we are able to collect real-time data from the various sensors embedded in the bracelet E4, and these data are subsequently sent to the Amazon Web Services server where a database is stored, in order to make a machine learning processing at the server side. The application displays the EDA, BVP, and HRV signals for the user in the three different tabs of Fig. 7. This feature allows the user to observe the graphical representation of his vital signals and at the same time to make sure that the application works and that the signal values are successfully retrieved in real time. In order to save the battery and improve the application performance, a switch button “start/stop graph” is provided to start or stop the graphical representation in the background, when the display is not essentially needed. This allows to save the battery in critical periods. The results regarding the sleep patterns are displayed in a clock-like pie chart that represents the entire 24-h day data collection, Fig. 7. Using a spinner button, the user can select a date to view the results. The results are projected on two clock-like pie charts, the first in light green color is for sleep, and the second in pink color is for wakefulness. The periods that were not recorded and labeled are not visible.
In addition, the user has the possibility to manage application modes through other controls: (1) track sleep allows to start or stop sleep monitoring and control the data sending to the AWS server, (2) streaming: the “streaming” button allows to turn off the bracelet and stops the Bluetooth connection via the application. It also allows to reset the Empatica SDK to be ready to create a new Bluetooth connection. (3) start/stop graph: starts or stops the graphs display in the EDA, BVP, and HRV tabs, which saves battery and smartphone resources.
6 Conclusions and Future Work
In this work we present a system that integrates a wearable sensor-equipped bracelet, a mobile application, and a cloud-based back-end application that allows non-invasive monitoring of sleep patterns. We further analyzed a set of physiological signals and their features to explore which features among them indicate significant differences between sleep and wakefulness. Our study is fairly comprehensive with regard to the variety of physiological signals that we collect in ambulatory settings without disturbing the patient. In our analysis various signals such as electrodermal activity, heart rate variability, blood volume pulse, and patients’ movements registered via an accelerometer are taken into consideration. We show that by carefully estimating a rich set of parameters for support vector machine based classification algorithm we can achieve up to 93% of correct classification when classifying sleep and wake periods for the analyzed subjects. Our study shows that such a system can be of great use for medical health practitioners who are interested to follow sleep patterns and regularity for patients whose condition is at risk either during sleep or due to lack of sleep.
In the future we aim to investigate more in details the different types and phases of sleep such as nREM1, nREM2, slow-wave, and rapid eye movement, REM. For this purpose, we plan to compare a completely non-invasive methods such as the method proposed here, with hospital grade quality sleep evaluation methods. Integrating user feedback about the mobile application is also planned as a part of our future work.
References
Alfeo, A.L., Barsocchi, P., Cimino, M.G., et al.: Sleep behavior assessment via smartwatch and stigmergic receptive fields. Person. Ubiquit. Comput. 22(2), 227–243 (2018)
Alickovic, E., et al.: Ensemble SVM method for automatic sleep stage classification. IEEE Trans. Instrum. Meas. 67(6), 1258–1265 (2018). https://doi.org/10.1109/TIM.2018.2799059
Anderson, W.M., Boehlecke, B., Claman, D., et al.: Clinical guidelines for the use of unattended portable monitors in the diagnosis of obstructive sleep apnea in adult patients. J. Clin. Sleep Med. 3(07), 737–747 (2007)
Blackwell, T., Redline, S., Ancoli-Israel, S., et al.: Comparison of sleep parameters from actigraphy and polysomnography in older women: the SOF study. Sleep 31(2), 283–291 (2008)
De Arriba-Pérez, F., Caeiro-Rodríguez, M., et Santos-Gago, J.M.: How do you sleep? Using off the shelf wrist wearables to estimate sleep quality, sleepiness level, chronotype and sleep regularity indicators. J. Amb. Intell. Human. Comput. 19, 1–21 (2017)
Harari, G.M., Lane, N.D., Wang, R., et al.: Using smartphones to collect behavioral data in psychological science: opportunities, practical considerations, and challenges. Perspect. Psychol. Sci. 11(6), 838–854 (2016)
Herlan, A., et al.: Electrodermal activity patterns in sleep stages and their utility for sleep versus wake classification. J. Sleep Res. 28 e12694 (2018)
Hwang, S.H., et al.: Sleep period time estimation based on electrodermal activity. IEEE J. Biomed. Health Inform. 21, 115–122 (2017)
Kamišalić, A., Fister, I., Turkanović, M., et al.: Sensors and functionalities of non-invasive wrist-wearable devices: a review. Sensors 18(6), 1714 (2018)
Kutt, K., et al.: Towards the development of sensor platform for processing physiological data from wearable sensors. In: International Conference on Artificial Intelligence and Soft Computing. Springer, Cham (2018)
Lee, H.-A., Lee, H.-J., Moon, J.-H., et al.: Comparison of wearable activity tracker with actigraphy for sleep evaluation and circadian rest-activity rhythm measurement in healthy young adults. Psych. Investig. 14(2), 179–185 (2017)
Lewicke, A., Sazonov, E., Corwin, M.J., et al.: Sleep versus wake classification from heart rate variability using computational intelligence: consideration of rejection in classification models. IEEE Trans. Biomed. Eng. 55(1), 108–118 (2008)
Li, H., Zhang, Q., et Lu, K.: Integrating mobile sensing and social network for personalized health-care application. In : Proceedings of the 30th Annual ACM Symposium on Applied Computing, pp. 527–534. ACM, New York (2015)
Liu, X., Ai, W., Li, H., et al.: Deriving user preferences of mobile apps from their management activities. ACM Trans. Inform. Syst. (TOIS) 35(4), 39 (2017)
Mantua, J., Gravel, N., et Spencer, R.: Reliability of sleep measures from four personal health monitoring devices compared to research-based actigraphy and polysomnography. Sensors 16(5), 646 (2016)
Sano, A.: Measuring college students’ sleep, stress, mental health and wellbeing with wearable sensors and mobile phones. 2016. Thèse de doctorat. Massachusetts Institute of Technology
Taylor, S., Sano, A., Ferguson, C., et al.: QuantifyMe: an open-source automated single-case experimental design platform. Sensors 18(4), 1097 (2018)
Zorick, F.J.: Patterns of sleepiness in various disorders of excessive daytime somnolence. Sleep: J. Sleep Res. Sleep Med. 5, S165–S174 (1982)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Assaf, M., Rizzotti-Kaddouri, A., Punceva, M. (2020). Sleep Detection Using Physiological Signals from a Wearable Device. In: Inácio, P., Duarte, A., Fazendeiro, P., Pombo, N. (eds) 5th EAI International Conference on IoT Technologies for HealthCare. HealthyIoT 2018. EAI/Springer Innovations in Communication and Computing. Springer, Cham. https://doi.org/10.1007/978-3-030-30335-8_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-30335-8_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-30334-1
Online ISBN: 978-3-030-30335-8
eBook Packages: EngineeringEngineering (R0)