Abstract
In response to increased global industrialization and environmental pollution by toxic xenobiotics, foreign substances transgressing an ecosystem or a living organism, the need for new cost-effective decontamination and remediation methods is greater than ever. This chapter focuses on the role and potential application of fungal enzymes and their respective mechanisms in the bioremediation of various xenobiotics. Fungal bioremediatory enzymes exhibit uniquely broad substrate ranges. Among the enzymes examined herein are laccases (LAC), lignin peroxidases (LiP), manganese peroxidases (MnP), versatile peroxidases (VP), cytochrome P450 monooxygenases (P450), and unspecific peroxygenases (UPO). Each of these enzymes has been demonstrated to transform and remediate toxic xenobiotics to less hazardous or bioactive forms. However, there are nuanced considerations for each class of enzyme. Laccases and peroxidases each have their advantages and limitations; laccases can use molecular oxygen, while peroxidases require a supply of H2O2. On the other hand, peroxidases can function in a wider pH range and carry a high redox potential. P450s represent an incredibly promising and diverse superfamily of enzymes which remain to fully understood as is the case with UPOs. By isolating these enzymes and immobilizing them, the potential adverse effects of introducing a novel species into an already polluted and stressed environment are mitigated. Furthermore, while fungi are quite robust they are limited by environmental toxicity – a limitation that is overcome by employing isolated fungal enzymes. Enzymatic efficiency and longevity are, however, dependent on many variables such as pH, temperature, and environmental availability of various minerals and co-substrates. As a result, further investigation into fungal enzyme bioremediation and enzyme optimization is warranted to realize the tremendous biotechnical potential of these enzymes in the field of xenobiotic bioremediation. Genetic engineering for enzyme optimization and new enzyme immobilization approaches to generate stable and efficient enzyme-based detoxification systems are exciting prospects in development.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
19.1 Introduction
Xenobiotics are natural or synthetic organic compounds foreign to an organism that are potentially toxic and entail negative ecological or physiological consequences, in the form of pollution and disease, respectively (Olicon-Hernandez et al. 2017). Industry and agriculture are two major sources of environmentally prevalent xenobiotics which include fertilizers, insecticides, pesticides, dyes, plastics, and hydrocarbon derivatives (Sharma et al. 2018). Chemically, the vast majority of these compounds are aromatic with one or more frequently substituted phenyl functional groups. Many of these xenobiotic compounds are of major health and ecological concern as they are frequently carcinogenic and teratogenic, thereby disrupting development and reproductive capabilities in humans, fish, fish-eating birds, and other animals. A serious environmental and health issue is the accumulation of persistent, toxic chemical pollutants requiring new cost-effective and efficient ways to tackle the growing threat of environmental toxicity in the modernized world.
The majority of xenobiotic compounds can be decomposed or modified by microbes (Cameron et al. 2000). Bioremediation utilizes the metabolic potential of biological organisms to degrade or transform hazardous compounds in the environment into less toxic or nontoxic forms (Watanabe 2001; Yadav et al. 2019a, b). In particular, the use of fungi, referred to as mycoremediation, has attained widespread attention. Although bacteria are also versatile decomposers of xenobiotic pollutants, certain fungal strains are able to tolerate higher levels of pollutants. In some cases, such as the degradation of polyaromatic hydrocarbons (PAHs), bacteria can metabolize them and utilize them as carbon and energy sources, but they cannot mineralize them completely the way fungi can (Mougin et al. 2009). Fungi are capable of metabolizing a wider range of pollutants than their prokaryotic counterparts due to both extra- and intracellular degradation mechanisms and the powerful, nonspecific nature of the enzymes involved in both processes (Christian et al. 2005a). Additionally, the enzymes in question are tolerant of an active under diverse conditions including broad pH ranges, making fungi and their enzymes desirable for bioremediation (Verma and Madamwar 2002) (Rastegari et al. 2019; Yadav et al. 2017, 2018).
Filamentous fungi, particularly from the group called white rot fungi and mainly from Basidiomycetes, demonstrate a striking ability for oxidative decomposition of lignin, a recalcitrant component of wood composed of polyphenols (Mougin et al. 2009). These enzymes were evolved by such fungi to assist in the breakdown and detoxification of potentially hazardous by-products resulting from the decomposition of wood and other organic matter (Morel et al. 2013). The presence of these enzymes in many species of white rot fungi has been confirmed using comparative genomics to identify the so-called xenome , which consists of genes involved in xenobiotic detoxification. In the last two decades, these enzymes have been extensively adapted for bioremediation of pollutants in a low-cost, eco-friendly manner.
Bioremediation involves environments toxic to the survival of biological systems and despite fungal robustness and innate degradation infrastructure, there is a limit to the organisms’ tolerance of xenobiotic-induced environmental toxicity. Frequently, the polluted areas are too nutrient-poor to support microbial growth. Moreover, the fungi capable of detoxifying a pollutant may be sensitive to other pollutants in the environment (Mougin et al. 2009). To circumvent these problems, one option is to utilize isolated fungal enzymes or enzyme mixtures. The use of fungal enzymes rather than complete fungal populations is also a more eco-friendly approach in contrast to using live microbes as it allows for the mitigation of any adverse environmental impact resulting from the introduction of novel species (Sharma et al. 2018; Kour et al. 2019; Rana et al. 2019a, b). This is especially desirable as release of genetically modified fungi or other organisms is contingent on both acceptance by regulatory bodies like EPA and the general public (Ang et al. 2005).
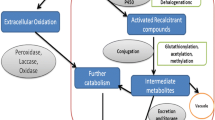
The most prominent groups of enzymes utilized in xenobiotic bioremediation transformations are oxidoreductases : peroxidases, laccases, and oxygenases (Sharma et al. 2018). Oxidoreductases can detoxify compounds by catalyzing oxidative coupling reactions using oxidizing agents to support the reactions. Laccases (LACs) and CYP monooxygenases (P450s) use molecular oxygen as the electron acceptor, while peroxidases use hydrogen peroxide to oxidize the substrates; both reactions result in the formation of water as a by-product. Peroxidases are heme-containing proteins, and the major types of peroxidases involved in detoxification processes in fungi are manganese peroxidase (MnP), lignin peroxidase (LiP), and versatile peroxidases (VP) (Doddapaneni et al. 2005). Xenobiotic detoxification enzymes are produced by a great diversity of fungi, but many are often produced by a large physiological group called the white rot fungi. These fungi actively degrade lignin – a large, complex, aromatic-containing networked polymer, creating a bleached appearance in the host, hence the name white rot fungi (Pointing 2001). Given the complexity of lignin and the structural motifs common to both lignin and various xenobiotics, these enzymes are well suited for xenobiotic detoxification (Fig. 19.1). A few genera of fungi are used extensively in bioremediation processes; these include Trametes, Pleurotus, Phanerochaete spp., etc.
The relationship and commonalities between organically produced lignin and varied xenobiotic chemical structures. (a) The structure of lignin according to Laurichesse and Averous (Laurichesse and Avérous 2014) and the monomeric alcohols which polymerize to form the cross-linked complex. (b) Common xenobiotic classes and associated structures as lignin model compounds targeted by lignin-degrading enzymes. Alkylated phenolic structures are common to both EDCs and lignin, as is the geminal phenyl motif of BPA. Nitrogen is not commonly present in lignin structures, nor are fused polycyclic aromatics nor polychlorinated phenols making lignin-degrading enzymatic degradation of these compounds less obvious
In this chapter, we will review the utility of fungal enzymes in bioremediation processes. In particular, we focus on the detoxification of organic xenobiotics by oxidoreductase enzymes including the extracellular peroxidases and laccases as well as the intracellular CYP450.
19.2 Sources of Xenobiotic Pollutants
Synthetic dyes released from paper, textile, plastic, cosmetic, food, and drug industries can be toxic and carcinogenic (Asgher et al. 2008; Levin et al. 2005). Polycyclic aromatic hydrocarbons (PAHs) like anthracene, pyrene, benzopyrene, and naphthalene are toxic, potentially carcinogenic xenobiotics that are produced from fuel combustion, gas plants, and industrial applications. Endocrine disrupting chemicals (EDCs) include alkylphenols such as nonylphenol and octylphenol as well as biphenyls such as bisphenol A (BPA), stilbene, and genistein estrogens. These are pharmaceutically active compounds that disrupt endocrine homeostasis in animals and are of grave concern to human health (Asgher et al. 2008). Other sources of xenobiotics include bleach plant effluents from paper and pulp industry, which contain toxic polychlorinated phenols (PCPs) and other organic compounds used for bleaching, and include dyes like azo dye and crystal violet (D’Souza et al. 2006). Chlorinated aromatic compounds like dichloro- and trichlorophenol and derivatives such as DDT, chlordane, and lindane are used in pesticides.
19.3 Metabolism of Xenobiotic Pollutants by Fungi
Bioremediation through fungi may be achieved by direct metabolism, whereby a fungus may completely degrade a xenobiotic compound to innocuous end products like carbon dioxide and other simple inorganic compounds (Mougin et al. 2009). This is particularly likely under nutrient-limiting conditions when the substrate compound could serve the carbon and/or energy needs of the organism. While this metabolic pathway is preferred as the toxic compound is eliminated or nearly so, the more common method of detoxification is co-metabolism. In this process, a cosubstrate is used to transform a xenobiotic compound without utilizing the compound for growth or energy needs. Generally, this only results in minor changes in the structure of the pollutant. For instance, substrate-free radicals may be generated which subsequently cross-link the substrates with themselves or with environmental structures such as soil components, thereby forming adducts with decreased bioavailability and toxicity (Bollag 1992). Alternatively, these modified compounds could be further metabolized by other organisms leading to further detoxification. A third detoxification pathway involves conjugation or oligomerization of a pollutant. In this process, the resulting products of enzymatic action have larger and more complex structures than the parent compounds, but the structural modification or aggregation reduces the bioavailability of the compound, thereby negating its biological potency. In conjugation, typically, the compound may be methylated, acetylated, alkylated, or conjugated with sugars and amino acids and subsequently excreted or sequestered into storage structures. In oligomerization, a xenobiotic is coupled with a second molecule of the same or different xenobiotic following oxidation , aggregating into structures that become less bioactive.
19.4 Fungal Enzymes in Xenobiotic Remediation
The most commonly studied enzymes involved in mycoremediation all catalyze oxidoreduction reactions in the transformation of xenobiotics (Sharma et al. 2018). Among these enzymes are some active extracellularly and other which are intracellularly involved in the transformation and subsequent detoxification of xenobiotics. Within these oxidoreduction-catalyzing enzymes, some such as laccases (LACs) and CYP monooxygenases (P450s) use molecular oxygen as an eventual electron acceptor, while peroxidases use hydrogen peroxide as electron acceptors in their respective reaction pathways. One specific class of enzyme common to many xenobiotic degradation mechanisms are the heme peroxidases of which the most extensively involved and studied are manganese peroxidase (MnP), lignin peroxidase (LiP), and versatile peroxidases (VP) (Doddapaneni et al. 2005). The relatively lower substrate specificity of the extracellular enzymes in oxidative degradation allows them to target a wider variety of xenobiotic compounds and is as such of great interest in bioremediation processes (Harms et al. 2011). As with many secreted proteins, these peroxidases and laccases are glycosylated, at times heavily; for instance, certain laccases show 16% sugar content (Salony et al. 2006). Many fungal metabolites bearing diverse functional groups can serve as redox mediators for the oxidative function of these enzymes and can enhance their activity (Asgher et al. 2008). These include veratryl alcohol, N-hydroxyacetanilide, and acetosyringone to name a few. Mediators are particularly important when the substrates are too large to be accommodated into the active site of the enzyme (Mougin et al. 2009). The toxic xenobiotics or the products formed following their metabolism by extracellular enzymes can be further detoxified by intracellular enzymes including CYPs that are ubiquitously present in all organisms (Morel et al. 2013). These enzymes can catalyze various reactions including dealkylation, hydroxylation, and sulfoxidation and frequently function by inserting molecular oxygen into various substrates.
19.4.1 Laccase
LACs, originally identified as ligninolytic enzymes, are present in ascomycetes, basidiomycetes, and deuteromycetes and are commonly found in gene families (Harms et al. 2011). Laccases are secreted blue multicopper oxidases that catalyze single-electron oxidation of phenolic and other aromatic substrates to create free radicals with the concomitant reduction of molecular oxygen to water. The radical products cross-link by self-coupling or cross-coupling to form less toxic polymers; during this process they may undergo further decomposition reactions such as decarboxylation, dechlorination, and demethoxylation. Following LAC-dependent oxidation events, metabolites commonly exhibit oxidative coupling or radical polymerization resulting in compounds of greater molecular masses than the parent compound (Junghanns et al. 2005) which potentially may further negate the biological effects of such foreign compounds (Tsutsumi et al. 2001). The use of abundantly available oxygen as an oxidizing agent with the formation of water as a harmless by-product makes laccases a desirable bioremediation enzyme. Compared to peroxidases, different laccases can tolerate a wider pH range (2–10) and have broader substrate specificities (Xu 1996). The broad substrate range of laccases encompasses various xenobiotic compounds contaminating soil and water, i.e., PAHs, organophosphorus insecticides, and toxic dyes which include aromatic compounds like phenols, trichlorophenols, aminophenols (anilines), phenylenediamine derivatives, and benzenethiols (Amitai et al. 1998; Kues 2015; Sharma et al. 2018; Xu 1996; Yadav et al. 2016).
The potential of LACs in xenobiotic remediation has long since been a point of interest and investigation. Laccases from Trametes versicolor and Pleurotus ostreatus have been used in the detoxification of PCBs (polychlorinated biphenyls) which were used as insulators in electrical equipment until they were banned in 1979 for their high toxicity (Keum and Li 2004). Laccases facilitate substrate oligomerization and dechlorination, leading to detoxification of these compounds. LACs catalyze the initial oxidation of polycyclic aromatic hydrocarbons (PAHs) , beginning their extracellular degradation (Pozdnyakova et al. 2018b). A LAC isolated from Coriolopsis gallica oxidizes the PAHs carbazole, N-ethylcarbazole, and dibenzothiophene in concert with proper mediators such as 1-hydroxybenzotriazole and 2.2′-azino-bis-(3-ethylbenzothiazoline)-6-sulfonic acid (Viswanath et al. 2014). When free laccase was applied to PAH-contaminated soil, 15 PAHs including anthracene and benzopyrene were degraded, demonstrating the potential of direct enzyme application for bioremediation (Wu et al. 2008). Covalently immobilized laccases from Trametes versicolor were shown to efficiently degrade the PAHs, anthracene, naphthalene, and phenanthrene (Bautista et al. 2015). Multiple studies revealed the ability of laccases from Pleurotus ostreatus, Trametes versicolor, and other strains to effectively degrade BPA and other endocrine disruptors like alkylphenols. The detoxification of these compounds by laccases may be brought about by oligomerization or mineralization to carbon dioxide, and this action was enhanced by supplementation with mediators (Macellaro and Pezzella 2014; Margot et al. 2013; Zhang et al. 2015). Laccases from Pycnoporus sanguineus, Myceliophthora thermophila, Trametes trogii, and Ceriporiopsis subvermispora effectively metabolized the toxic dyes like bromophenol blue, methyl violet, and malachite green and other dyes used in the textile industry (Antosova et al. 2018; Chmelova and Ondrejovic 2016; Mougin et al. 2009). Synthetic laccases are also used in the paper and pulp industries to bleach paper and clothes (Sharma et al. 2018). The addition of mediator compounds can dramatically enhance the detoxification of substrates as witnessed in the transformation of halogenated pesticides by laccases from the fungus, Coriolopsis gallica (Torres-Duarte et al. 2009); in this study, acetosyringone and syringaldehyde proved to be the most effective mediators. Finally, laccases from Trametes versicolor have been used to oxidize the pharmaceutical drugs, diclofenac and mefenamic acid in municipal wastewater (Margot et al. 2013). Beyond naturally occurring LACs, some researchers have explored the possibility of engineering LACs with the intention of improving transformational efficiency and demonstrating transient expression by means of directed evolution (Camarero et al. 2012; Gu et al. 2014; Mate and Alcalde 2015; Theerachat et al. 2012; Wong et al. 2013b).
19.4.2 Peroxidase
The ligninolytic peroxidases of interest in bioremediation carry high redox potentials (>1.4 V) and catalyze the oxidative breakdown of lignin and other compounds with aromatic ring structures using hydrogen peroxide as a cosubstrate and with the help of certain mediator compounds like veratryl alcohol (Piontek et al. 2001; Sharma et al. 2018). These peroxidases are glycosylated secreted proteins carrying an iron protoporphyrin (heme) ring at the catalytic center. Peroxidases can oxidize and generate phenolic radicals which may aggregate and precipitate. There are several groups of secreted peroxidases in fungi:
19.4.2.1 Lignin Peroxidase
LiPs are heme-containing enzymes which can metabolize various aromatic compounds many of which are generally refractory to breakdown, with pH optima in the acidic range (2–5) (Shrivastava et al. 2005). Since the 1983 discovery of P. chrysosporium, LiP in extracellular media, isozyme forms among various basidiomycetes have been isolated ranging in weight from 38 kDa to 43 kDa (Falade et al. 2017). LiPs have been shown to completely oxidize both methylated lignin model and non-methylated lignin model compounds as well as PAHs (Kadri et al. 2017). Through powerful, nonspecific catalytic transformative activity, the aptly named LiP is capable of direct transformation of up to 90% of lignin structural components (Falade et al. 2017).
LiPs are commonly produced by Phanerochaete and Trametes spp. As demonstrated by LiP isolated from P. chrysosporium which transforms the PAHs benzo[a]pyrene, anthracene, 1-methylanthracene, 2-methylanthracene, 9-methylanthracene, fluoranthene, acenaphthene, and dibenzothiophene, these enzymes possess broad substrate ranges (Pozdnyakova 2012). LiPs also demonstrate expanded substrate ranges in the presence of veratryl alcohol which increases oxidation of weak or terminal LiP substrates. With the advantageous kinetics conferred by mediators like veratryl alcohol, LiP transforms most aromatic compounds with an ionization potential less than 8 eV. LiP also has the unique ability to cleave esters in non-phenolic aromatics, thereby further demonstrating the significance of LiPs to bioremediation efforts of diverse, aromatic-containing xenobiotics (Pozdnyakova 2012).
19.4.2.2 Manganese Peroxidase
MnPs are also heme-containing secreted enzymes which function under relatively less acidic conditions (pH 4–7) than those of LiP (Asgher et al. 2008). As a heme peroxidase, MnPs share much of their characteristics and mechanism with LiPs (Deshmukh et al. 2016). MnP appears not to occur in the same large gene families characteristic of other ligninolytic enzymes such as LAC (Torres-Farrada et al. 2017) and is only found in Basidiomycota (Harms et al. 2011). MnP is capable of aromatic ring cleavage within monoamino-dinitrotoluene and chlorophenol derivatives (Harms et al. 2011). The oxidative potential of MnP relies on the oxidation of Mn2+ to Mn3+ by the enzyme followed by the indirect, single-electron oxidation of subsequent substrates as Mn3+ is reduced, thereby reverting to Mn2+.
Due to the indirect mechanism, the substrate range is broad, and the extent of transformation of resulting metabolites is near complete. MnP oxidizes phenols, aromatic amines, and dye compounds as well as mineralizes CO2 from various quinones – common products of PAH radical polymerization – by ring fission transformations. MnPs isolated from Anthracophyllum discolor clearly demonstrate this PAH-substrate promiscuity as they are able to oxidize pyrene, anthracene, fluoranthene, and phenanthrene as well as various derivatives of these compounds (Pozdnyakova 2012). A MnP from the white rot fungus, Trametes, displayed a strong ability to degrade azo and indigo dyes as well as PAHs (Zhang et al. 2016). Another MnP from Peniophora incarnata not only displayed the potential to break down the PAH anthracene, but this ability was transferable by heterologous expression in yeast, signifying a bioremediatory potential. MnP from Ganoderma lucidum used as cross-linked enzyme aggregates efficiently degraded the endocrine disrupting nonylphenol and triclosan (Bilal et al. 2017a).
MnP is of great interest in bioremediation efforts as it has been shown to be stable under adverse conditions; however, a complicating factor of its use in remediation effort and applications is the mechanistic need for a suitable chelator (Bogan et al. 1996). Such chelators are commonly organic acids such as oxalic or malonic acid derivatives (Kadri et al. 2017). These compounds complex with Mn3+, enabling the oxidation of substrate lignin model compounds. In addition to a mechanistic dependence on chelators, MnP activity significantly increases in the presence of redox mediators such as Tween 80. With the added effect of Tween 80, MnP has been shown to transform compounds with ionization potentials of 8.2 eV. Despite the powerful oxidative potential and resistance to adverse conditions of MnP, the application of these enzymes in biotechnical pursuits is complicated by not only chelator requirements and redox mediator reliance for elevated efficiency but also by LAC-dependent initiation of Mn3+ complexing (Schlosser and Hofer 2002). No Mn3+ complexing was observed in in vitro mixtures of semi-purified MnP, Mn2+, and oxalate or malonate when H2O2 sources were excluded. However, the addition of LAC stimulated Mn3+ complexing and ultimate MnP-stimulated substrate oxidation. In response to PAH-polluted media, MnP secretion by Fomes is very high, reaching concentrations of approximately 1299 U/L after 21 days of xenobiotic exposure (Godoy et al. 2016).
19.4.2.3 Versatile Peroxidase
Versatile peroxidases (VP) are a hybrid between LiPs and MnPs as they contain a heme group and oxidize Mn2+to Mn3+, inducing indirect oxidations, as well as oxidize phenolic and non-phenolic substrates; VPs conjoin mechanisms and substrate ranges between these two enzymes (Kues 2015; Pozdnyakova 2012). VPs have thus far only been identified in Basidiomycetes (Harms et al. 2011). In the initial characterization of the first identified VP – specifically, PS1 isolated from Pleurotus eryngii – it was shown to possess both the Mn oxidation domain of MnPs and the aromatic substrate oxidation center (AS) of LiPs (Camarero et al. 1999). Furthermore, PS1 and subsequently characterized VPs have been shown to actually retain LiP- or MnP-like enzymatic activity in conditions that would inactivate LiPs or MnPs, respectively. Little is understood about the role of VPs in the mediation of xenobiotic transformation; however, it is known that VP production is induced by the presence of PAH pollutants (Pozdnyakova et al. 2018b). In addition to PAHs, VPs can also degrade polyhalogenated aromatic pesticides containing diverse functional groups as demonstrated by VP isolated from Bjerkandera adusta which successfully transformed dichlorophen (an antimicrobial polycyclic), bromoxynil (a nitrile herbicide), and pentachlorophenol (PCP) – a potent pesticide (Davila-Vazquez et al. 2005). Given the hybridization of LiP and MnP substrate ranges, they have the potential to be a major component of the subsequent remediation of many chemically diverse xenobiotics. Due to their broad substrate range and various transformation mechanisms, VPs warrant further characterization and investigation regarding their potential application in bioremediation and biotechnical efforts. Examples of fungal laccases and peroxidases employed for xenobiotic detoxification are presented in Table 19.1.
19.4.3 Cytochrome P450 Monooxygenase
Many fungi also possess intracellular transformation enzymatic machinery capable of further degradation of xenobiotics (Syed and Yadav 2012). They belong to the larger group of oxygenases that are the principal intracellular enzymes involved in the aerobic degradation of aromatic xenobiotics using oxygen (Sharma et al. 2018). These are also heme-containing enzymes that add one or more oxygens to destabilize aromatic rings to break down and even solubilize the compound. Oxygenases could catalyze the addition of one oxygen molecule (monooxygenase) or two molecules (dioxygenase) to the substrate. Many halogenated herbicides, fungicides, and pesticides are detoxified using oxygenases, and the best-studied enzyme belongs to the cytochrome P450 enzyme family, which utilized NADPH as a cofactor to catalyze redox reactions (Doddapaneni et al. 2005).
P450s are very distinct from other xenobiotic-degrading fungal enzymes as it is active intracellularly (Deshmukh et al. 2016). Furthermore, CYP450 are found in massive gene families with multiple subfamilies as discovered in the genome of P. chrysosporium with at least 149 isozymes (Doddapaneni et al. 2005; Olicon-Hernandez et al. 2017; Syed et al. 2011). Such gene families have been identified in ascomycetes, basidiomycetes, mucoromycetes, and chytridiomycetes (Harms et al. 2011). As monooxygenases, P450 incorporates a single atom from molecular oxygen into the substrate while reducing the remaining oxygen to water. Compared to CYPs involved in primary metabolism, CYPs mediating detoxification processes are generally less specific (Cresnar and Petric 2011). P450 monooxygenases are a major component in the degradation of various xenobiotics as they are quite nonspecific and demonstrate very powerful enzymatic activity in the transformations of such diverse chemical motifs. However, as these are intracellularly active proteins, their application in fungal enzyme isolation and immobilization is further complicated.
Within each class of P450s, multiple subfamilies exist sharing general function and some sequence identity. Many class-II subfamilies are involved in biosynthetic pathways and homeostatic processes. The subfamily CYP51 has been indicated in maintaining cell wall integrity while CYP61 (a fungi-specific subfamily) is responsible for spore outer wall formation. Various Class II P450s transform PAHs, alkyl phenols (APs), alkanes, and polychlorinated dibenzo-p-dioxins (PCDDs) (Harms et al. 2011; Kues 2015). CYPs from the basidiomycete Phanerochaete chrysosporium have been shown to catabolize PAHs including anthracene and the endocrine disrupting alkylphenols (Hirosue et al. 2011; Syed et al. 2011). Among Class II P450 subfamilies, CYP57 has been shown to detoxify pisatin, a heterocyclic, aromatic-containing fungus growth inhibitor produced in pea plants upon microbial attack. Furthermore, CYP53 has been shown to degrade and detoxify benzoate derivatives within multiple species from both the Ascomycota and Basidiomycota phyla. Additionally, CYP504 is known to degrade phenylacetate derivatives. The significance of Class II P450 activity is further emphasized in observed biodegradation by non-ligninolytic fungi such as Scopulariopsis brevicaulis which was shown to completely transform anthracene (a tricyclic PAH), producing the same major metabolite – 9,10-anthraquinone – as ligninolytic fungi such as the basidiomycete Fomes , despite the absence of extracellular enzymes (LAC, LiP, and MnP) demonstrating the versatility and potency of Class II P450-dependent intracellular transformation pathways (Godoy et al. 2016). CYPs have been tapped to detoxify a variety of pharmaceutical compounds, including antibiotics, anti-inflammatories, and β-blockers (Olicon-Hernandez et al. 2017). Naproxen is an environmentally prevalent pharmaceutical pollutant and has even invaded drinking water systems due to overuse in treating human and animal diseases. CYPs were shown to detoxify this drug through demethylation and hydroxylation (Aracagok et al. 2017). A fundamental limitation in utilizing CYP enzymes in cell-free systems is the fact that CYP functions in intracellular networks that also involve other enzymes (Haroune et al. 2017). Shotgun proteomics of proteins in response to the PAH, anthracene in Penicillium oxalicum, revealed regulation of hundreds of proteins, and intracellular metabolism of such xenobiotics typically follows two distinct phases with the CYPs working in conjunction with other enzymes like epoxide hydrolases and transferases (Lucero Camacho-Morales et al. 2018).
19.4.4 Unspecific Peroxygenase
UPOs are secreted hybrid enzymes which combine the functionalities of heme peroxidases and P450 monooxygenases (Karich et al. 2017). Among this class of peroxygenases, two common structural motifs have been observed: short UPOs are approximately 29 kDa, while long UPOs are approximately 44 kDa. Short UPOs are found across all fungal phyla, but long UPOs are exclusive to ascomycetes and basidiomycetes. Both short and long UPOs have immense substrate ranges, acting in association with hydrogen peroxide as a cofactor, a marketed improvement in simplicity from the nuanced requirements of MnP. Furthermore, 41 of EPA-listed priority pollutants have been shown to be transformed by UPOs, and more than 300 other aromatic, poly- and heterocyclic, and aliphatic substrates have been identified thus far. Despite the recent findings regarding xenobiotic substrates, the physiological role of UPOs remains to be identified (Olicon-Hernandez et al. 2017); however, Karich et al. theorized that UPOs and P450s may work in harmony with UPOs crudely transforming xenobiotics extracellularly to reduce negative biological effects, while P450s “fine-tune” the resulting metabolites so that they may be further transformed and rendered inert within the cell or even consumed as carbon sources due to the incredibly diverse functionality of these enzymes. Thus far, present understanding explains that UPOs’ substrate transformations are limited by steric hindrance, bioavailability and potential substrate solubility, or strong inactivation of an aromatic ring by electron withdrawing groups (Karich et al. 2017). Despite these regulatory factors in UPOs’ substrate specificity, UPO isolated from Agrocybe aegerita demonstrated significant transformation of naphthalene, phenol, anisole, toluene, ethylbenzene, acenaphthylene, acenaphthene, fluorene, phenanthrene, anthracene, pyrene, benzo[a]anthracene, 1,2-diphenylhydrazine, benzidine, and 2,4-dimethylphenol as well as various phthalate esters, nitroarenes, and polychlorinated benzenes (Karich et al. 2017). Among these known substrates exist several previously only thought to be transformed by P450s or MnPs and LiPs, again demonstrating the remarkable potential of these enzymes. Such versatile enzymes in extracellular degradation processes carry great implications for biotechnical development of xenobiotic mitigation strategies.
19.5 Mechanisms of Xenobiotic Detoxification by Fungal Enzymes
Fungal extracellular xenobiotic degradation occurs in two conserved steps (Rao et al. 2010). Firstly, the hydrolytic system targets macromolecules for degradation by hydrolase activity. The hydrolyzed substrates are then further transformed by the ligninolytic system comprised of nonspecific oxidative enzymes. The enzymatic mechanisms we focus on in this section are the oxidoreductases whose mechanisms are well researched (Fig. 19.2).
Simplified mechanism of fungal enzymes used in bioremediation. (a) Laccase enzymatic cycle. Mediators include veratry alcohol (VA), Tween 80, and other small organic molecules. (b) Lignin peroxidase enzymatic cycle. Both oxidized states of LiP are able to oxidize substrates and mediators. Mediator molecules are used when the substrates cannot access enzyme active site. (c) Manganese peroxidase enzymatic cycle. MnP preferentially oxidizes Mn2+ in the first redox reaction but can also oxidize other mediators. Mn2+ oxidation is highly specific for the second redox reaction which restores the base state. (d) CytP450 enzymatic cycle. The reduction of heme-bound molecular oxygen is either directly catalyzed by the reductase coenzyme or indirectly via the use of cytochrome B5 as a redox mediator. (Adapted from Kues (2015) and Guengerich (2001b))
19.5.1 Laccases
Laccases are multicopper oxidases with low substrate specificity. Within basidiomycetes these families range from 5 to 17 isozymes (Yang et al. 2017). Within LAC isozymes, enzymatic copper involvement is conserved with four copper atoms at +2 oxidation states in the resting enzyme. The four copper atoms are characterized as T1, T2, and T3 – which is binuclear. The T1 copper facilitates substrate oxidation, while T2 and T3 copper atoms store the resulting electrons which then convert diatomic oxygen to water. They catalyze coupled redox reaction between a substrate and molecular oxygen resulting in the formation of a radical cation and water, respectively. The type I copper is found in a wide cavity of the enzyme surface, which allows it to bind to many different types of substrates (Su et al. 2018).
The significance of LACs in the remediation of xenobiotics is only amplified by laccase-mediator systems (LMSs) which act as electron transfer chains and thereby further broaden the substrate range as well as increase kinetic favorability of transformation (Wong 2013a). LMSs share the common mechanism of facilitating the indirect oxidation of a substrate following the primary oxidation event of the mediator by LAC. The enzymatic cycle of laccases can be simplified as a four-electron abstraction from the substrate by the oxidized enzyme followed by the reduction of molecular oxygen to water to regenerate the active enzyme (Fig. 19.2a). The enzyme primarily exists in the resting oxidized state (RO) with each of the copper atoms oxidized. The catalytic reaction is initiated by the donation to the type I copper of four electrons from suitable substrates (reductants). Three electrons are transferred through a conserved His-Cys-His sequence of amino acids to the trinuclear cluster (TNC) consisting of the type II copper and the two type III copper atoms, resulting in the fully reduced enzyme. The electrons are used to reduce molecular oxygen which binds to the TNC, forming a peroxide intermediate (PI). The reduction then proceeds by donation of a hydrogen by a nearby glutamic acid residue resulting in the cleavage of the oxygen-oxygen bond and the native intermediate (NI) state of the enzyme. This state can directly proceed to the fully reduced state in the presence of large amounts of suitable substrates, producing two water molecules or proceed to the fully oxidized state with loss of a single water molecule in the absence of significant levels of reductants (Jones and Solomon 2015).
Laccases tend to accommodate substrates with relatively low redox potentials such as phenolic compounds which are oxidized to phenoxyl radicals which may undergo coupling reactions or isomerization to form quinones. Laccases are generally unable to directly oxidize compounds with high redox potentials or steric hindrance. Instead, such compounds are oxidized via the laccase-mediator system which involves oxidation of micromolecular organics which can then oxidize target compounds through redox reactions leading to cleavage of bonds (Su et al. 2018).
19.5.2 Peroxidases
Of the various peroxidases discussed herein, only the general mechanism of lignin peroxidase and manganese-dependent peroxidases has been elucidated. The shared mechanism of these two types of peroxidases involves the oxidation of the enzyme by two electron abstractions by the H2O2 cosubstrate followed by two one-electron transfer steps where the oxidized enzyme abstracts an electron from the substrate (Mougin et al. 2009).
19.5.2.1 Lignin Peroxidases
In LiPs, Fe(III) in the heme ring is coordinated with four heme tetrapyrrole nitrogens and to a histidine residue. LiP has been shown to fold into a globular profile measuring about 50 × 40 × 40 ¯; this form is subdivided into proximal and distal domains relative to the heme group. Two small molecular channels allow for heme accessibility despite its fixed position within the protein’s tertiary structure. LiPs are capable of cleaving Cα-Cβ bonds as well as bonds between aryl Cα (Kadri et al. 2017). LiP is further differentiated from other peroxidases by the optimal pH of approximately 3.0 (Falade et al. 2017). Lignin peroxidases are activated by a two-electron oxidation of the native enzyme by the cosubstrate H2O2 (Mougin et al. 2009). This results in compound I which is reduced by the substrate as part of a one-electron redox reaction to compound II. Compound II then abstracts a second electron from the substrate resulting in regeneration of the native enzyme. The activated forms of the enzyme have a high redox potential allowing it to oxidize compounds such as lignin that other peroxidases are unable to transform. LiPs act through three different mechanisms based on the availability of its heme cofactor toward the substrate. First, LiPs act directly on certain phenolic and non-phenolic compounds which can access the heme group. This can lead to breakage of carbon-carbon bonds in substrates leading to conformational changes (Fig. 19.2b). The second lignin peroxidase mechanism is indirect, acting through a redox mediator to oxidize compounds that cannot access the heme group (Fig. 19.2b). Lignin peroxidase oxidizes mediators such as veratryl alcohol (VA) to a cation radical (VA+) which then oxidizes compounds through a redox reaction. The third mechanism occurs through further reactions of VA. The cation radical oxidizes organic acids into anion radicals which act as reductant. These radicals can also reduce molecular oxygen to a dioxygen anion which acts as a reductant. Through the reduction of ferrous ions, this dioxygen anion can also reduce hydrogen peroxide to a hydroxyl radical which can oxidize compounds through the non-enzymatic Fenton’s reaction (Christian et al. 2005b).
19.5.2.2 Manganese Peroxidase
Manganese peroxidases, also known as manganese-dependent peroxidases, are oxidized to a two-electron-deficient state (compound I) and restore its basal oxidation state by two single-electron abstracting steps (Christian et al. 2005b). The first step which reduces the compound I to a single-electron-deficient state (compound II) is not very specific and can use either Mn2+ or small compounds such as VA as reductants. The second step which restores the basal oxidation state of the enzyme is highly specific to Mn2+ (Fig. 19.2c). Both of the activated enzyme states oxidize Mn2+ to Mn3+ which acts as a nonspecific small, diffusible redox mediator. Mn3+ are chelated to carboxylic acids such as oxalic acid and are thus able to cause one-electron abstraction oxidation of various compounds (Fig. 19.2c). They can also react with carboxylic acids to produce radicals such as VA+ and superoxide. Manganese peroxidases are also able to act indirectly as reductors. This is done through oxidation of hydroquinone to semiquinone radicals which reduce highly oxidized compounds. This generates a quinone which is then reduced back to hydroquinone by quinone reductase enzyme.
19.5.3 Cytochrome P450
Cytochrome P450 enzymes are part of a superfamily with remarkable variation. Due to this variation within the CYP superfamily, the reactions associated with P450 enzymes are classified based on reactionary motifs and various protein involvements into ten distinct classes, three of which have been observed in fungi: class II, class VIII, and class IX (Cresnar and Petric 2011). Most commonly, CYP450 enzymes act as monooxygenases that incorporate an atom of oxygen into substrates (van den Brink et al. 1998).
P450 enzymes therefore serve as versatile catalysts for region-specific and stereospecific oxidation resulting in hydroxylation, heteroatom oxygenation, dealkylation, and epoxidation of C=C bonds (Deshmukh et al. 2016; Olicon-Hernandez et al. 2017). However, the basis of all these reactions can be summarized as being
with R-H as the substrate of cytochrome P450. Among these transformations, NADH or NADPH frequently acts as an electron donor to the second oxygen as it is reduced to water (Kues 2015). The reaction does not usually produce a phenol but instead an oxide which isomerizes to the more favorable phenolic conformation (Christian et al. 2005b). The phenolic form is activated toward further enzyme-catalyzed reactions, resulting in trans-dihydrodiols (Tongpim and Pickard 1999).
Much like the peroxidases, P450 enzymes are heme-based, with their catalytic active site containing a heme-bound iron atom (Guengerich 2001a). The substrate first binds to the heme iron Fe3+ which is reduced by an accessory enzyme, NADPH reductase (Fig. 19.2d). This reduced iron Fe2+ then binds with molecular oxygen. The accessory enzyme then reduces molecular oxygen which is then protonated. This leads to cleavage of the O-O bond, causing the protonated oxygen to be released as H2O. The resulting FeO3+complex radicalizes the substrate by proton (or electron abstraction). The radical then accepts the hydroxyl group (or oxygen atom in case of electron abstraction) before being released, simultaneously returning the iron to its base state. There exists a shunt pathway in which a peroxide is used as cosubstrate instead of molecular oxygen, skipping the need for the reductase coenzyme (Fig. 19.2d).
The monooxygenation mechanism of P450 is not limited to carbons but can also apply to heteroatoms such as nitrogen, sulfur, phosphorus, and iodine. In these cases, the oxygen transfer is more complicated, with the oxygen being added to the substrate after two successive electron transfer steps.
19.6 Strategies and Considerations in Xenobiotic Bioremediation
Either fungi or enzymes isolated from fungi could be used for the detoxification of xenobiotic compounds. Some fungal phyla possess more versatile degradation systems relative to bacterial strains (Pozdnyakova et al. 2018a). The fundamental limitations in using isolated fungal enzymes include productivity as well as stability and retention of activity (Sharma et al. 2018). Laccases, for example, have an acidic pH optimum that may not be available in effluent soils and waters, presenting a major limitation to their use. Furthermore, cost of application, intolerance of enzymes to high levels of cosubstrate hydrogen peroxide, and issues with enzyme reusability have hampered the application of these enzymes in the field (Nicell and Wright 1997). Once applied to sites for remediation, the enzymes are also under threat of denaturation and destruction by physical and chemical forces and by the action of microbes and their enzymes. Besides, excessive exposure of heme-containing proteins to oxidative species can lead to inactivation and subsequent degradation of the protein (Valderrama et al. 2002). Indeed, these limitations are so critical that they have delayed or limited large-scale application of these enzymes in bioremediation processes (Ayala et al. 2008).
One approach to the identification of potential fungal strains with significant remediation potential and biotechnical application is to isolate populations from xenobiotic-polluted environments (Godoy et al. 2016). Doing so results, in part, in fungi with innate tolerance to the environmentally present pollutants. The pollutant-resistant detoxifying fungi often produce biodegradative enzymes that are too limited in amount to isolate and utilize. To scale up production of these enzymes, genetic engineering could be used to overexpress fungal enzymes in fungi, plants, or other organisms that could colonize the polluted substratum. Alternatively, enzymes could be mass-extracted from such organisms by heterologous expression and utilized for bioremediation. This is a cost-effective strategy that not only allows purification of large amounts of enzyme with stability and activity, the purification process of recombinant proteins is also simpler as they can be isolated using cleavable tags (Alcalde et al. 2006). Furthermore, the stability, activity, and other features of an enzyme can also be enhanced using genetic and enzyme engineering approaches. By introducing mutations using strategies like DNA shuffling, error-prone PCR, and site-directed mutagenesis (Dua et al. 2002), enzyme engineering could be accomplished, where a change in protein sequence from mutations results in a possible change in enzyme structure or regulation with improved traits including increased stability and activity; increased xenobiotic substrate specificity or wider substrate range; tolerance to a wider range of pH, temperature, and stress conditions; and decreased susceptibility to proteases in the natural environment (Rayu et al. 2012). One such mutated laccase protein from Pleurotus ostreatus identified in a screen gained greater enzymatic activity and higher stability at acidic pH, widening its pH tolerance in detoxifying toxic industrial dyes (Miele et al. 2010). Similarly, directed evolution of a laccase resulted in 3.5-fold increase in acetonitrile catabolism while tolerating relatively high concentrations of it (Alcalde et al. 2005).
As enzymes are sensitive to physical and chemical environmental changes, various strategies have been evolved to fortify and preserve their structure and function while exposed to adverse conditions. Enzyme immobilization in carbohydrate-based matrices has been shown to be effective in prolonging the half-life and enhancing the stability and catalytic function of the remediative enzymes. Enzymes can be immobilized to solid support systems for more effective use in bioremediation. Enzyme immobilization can be done by covalent linkage or adsorption of enzymes onto solid surfaces like glass, affinity tag-bearing beads or nylon membranes, suspension in polymeric gels like chitosan, gelatin and encapsulation in solid matrices such as sodium alginate and could also accompany enzyme crosslinking using glutaraldehyde (Diano et al. 2007; Koyani and Vazquez-Duhalt 2016; Sirisha et al. 2016). Immobilization has been shown to improve the stability, catalytic function, and longevity of enzymatic function of the fungal enzymes and additionally protects the enzymes from proteolytic degradation (Asgher et al. 2007; Cheng et al. 2007; Jing and Kjellerup 2018). This is brought about in part by the increased resistance to physical, chemical, and biological denaturing agents afforded by the immobilization. Additionally, immobilized enzymes can be recovered and reused, thus economizing the process. Laccases immobilized on glass beads from Trametes retained 90% of their activity and showed greater resistance to proteases (Bilal et al. 2017b; Dodor et al. 2004). Similar benefits have been observed for other fungal enzymes like MnPs (Bilal and Asgher 2016). Enzymatic nanoreactors with dendritic copolymers bearing glycosidic groups anchoring laccases not only showed improved enzymatic activity but also increased thermostability (Gitsov et al. 2008). Similarly, vault nanoparticles with hollow cores covalently anchoring multiple MnP enzymes showed enhanced stability while displaying a threefold increase in phenol degradation (Wang et al. 2015).
The addition of cofactors, cosubstrates, and mediators is important to drive the action of the enzymes in bioremediation scenarios; this is particularly important for recalcitrant pollutants. Additionally, the presence of mediator compounds can enhance enzyme activity. The fungal metabolites, veratryl alcohol and 2-chloro-1,4-dimethoxybenzene, can stimulate LiP enzyme activity, and this is particularly useful when treating recalcitrant substrates (Asgher et al. 2008), while acetohydroxamic acid serves as an excellent mediator for laccases (Minussi et al. 2007). In some cases, the addition of glucose as a carbon source to fungal cultures also enhanced the detoxification process (Asgher et al. 2008).
Marine fungi are adapted to much harsher conditions than terrestrial fungi. Their enzymes are able to withstand high salinity and concentration of phenolic compounds. Certain marine fungi have shown bioremediation potential for water-soluble crude oil fractions between 0.01 and 0.25 mg/mL (Deshmukh et al. 2016). Other extremophilic fungi such as Pestalotiopsis palmarum are able to survive in high concentration of extra-heavy crude oil and salt while producing oxidative exoenzymes (laccases and lignin peroxidases) which degrade the maltene and asphaltene fractions of oil for use as a carbon and energy source (Naranjo-Briceno et al. 2013). The isolation of enzymes from marine organisms can select for traits such as heat and cold tolerance and salt and pressure tolerance (Lima and Porto 2016).
Recently, non-protein enzyme mimics or next-generation artificial enzymes in nanoparticles or nanocomposites have been used to replace enzymes in bioremediation processes (Gao and Yan 2016). Desired for their low cost and higher stability, these structures display enzyme-like properties under physiological conditions. Nanozymes, although lacking an active site, bind substrates specifically and catalyze their transformation. The Fe3O4-based magnetic nanoparticles mimic peroxidases and can degrade toxic dye compounds like methylene blue (Wu et al. 2015). Carbon-based nanomaterials made of graphene oxide (GO) also display peroxidase-like activity (Ma et al. 2017). Similarly, a guanosine monophosphate (GMP) coordinated copper nanocomposite mimicked laccases in being able to degrade phenolic compounds including hydroquinone and naphthol (Liang et al. 2017).
Although it is desirable to identify and isolate detoxification enzymes in polluted environments, a basic handicap is the ability to culture these microbial species. Only a handful of microbes are culturable, and these may not include the microbes that make the enzyme of interest. This problem is circumvented by recent developments in metagenomics which can identify potential detoxification enzyme-coding genes. These genes could be expressed in the heterologous system to mass-produce the enzymes of interest for bioremediation. Clues to the functionalities of these genes could be revealed by metatranscriptomic and metaproteomic approaches, which could also reveal biochemical pathways and synthetic pathways for xenobiotic-transforming enzyme production.
In silico approaches can be employed to understand the evolution of xenobiotic detoxification by phylogenetic analyses. More recently, bioinformatic analysis in the form of molecular docking tools has been developed to predict pollutant substrates of the detoxification enzymes. This approach has been particularly useful for laccases which have broad substrate specificity; in one study, laccase enzyme structures were screened against a database of toxic compounds to identify putative substrates (Suresh et al. 2008). The study found that 30% of the studied compounds that were recognized as environmental pollutants could be potentially metabolized by fungal laccases. Similar approaches could be employed for other enzymes with known structures to test if they could be used to detoxify pollutants of interest.
19.7 Conclusions and Future Perspectives
Increased global industrialization has presented many challenges including the production of ecotoxic industrial wastes that also present health threats as xenobiotic compounds. Fungi have specialized enzymes that are highly efficient in detoxifying these pollutants. Harnessing the power of these enzymes is proving to be an effective strategy for the bioremediation of the xenobiotic compounds. In particular, oxidoreductase enzymes including laccase, peroxidase, and oxygenases like CYP from various fungal species are employed to detoxify a wide range of pollutants. In addition to bioremediation by detoxification, enzymes can also be employed for the detection and quantification of xenobiotics in the environment. White rot fungi (WRF) are a versatile group of microbes that are capable of oxidative detoxification of a variety of chemical pollutants and xenobiotic compounds that are environmentally harmful. These organisms and their enzymes have the ability to not only tolerate the xenobiotics but in many cases also metabolize or help sequester them. The limitation in biodegradation of xenobiotic compounds is frequently the initial stages of degradation. The oxidoreductase enzymes are all extracellular and specialized for less specific activity in initial stages of xenobiotic metabolism and, as such, as ideal candidates for detoxification of these compounds.
Development of heterologous expression systems and industrial scale expression is still a limitation. Limitations of enzyme quantity and stability are being overcome by the adoption of genetic engineering technology and enzyme immobilization approaches. Fungal enzymes can also be modified for enhanced catalytic activity in addition to thermostability to maximize the efficiency of the detoxification process. Creation of enzymes with increased redox potentials, especially in those such as laccases which have a lower redox potential, could widen the substrate range. The utility of laccases and CYPs may be limited by the availability of molecular oxygen. Development of peroxidases to target those substrates could be one possible approach. Present bioengineering tactics are focused on developing superior enzymes for the bioremediation of known pollutants, but such approaches may be extended to target novel pollutants, not known to be biodegradable. Alternatives to fungal enzymes in the form of nanozymes are also being explored.
An inherent limitation in the use of single enzymes in bioremediation systems is the fact that detoxification processes are often multi-step pathways, requiring the action of multiple enzymes in a sequence. The ability to use fungal enzymes in synthetic pathways for biotransformation of toxic pollutants into economically valuable compounds would be a great future direction. Since fungal-fungal and fungal-bacterial consortia have been employed for remediation of xenobiotics like PAHs, a similar strategy with enzymes could follow suit.
Structural studies of enzymes as well as directed evolution can facilitate changes in structure that could substantially enhance activity and xenobiotic metabolism. Better structural understanding of laccases as well as other enzymes could lead to minimization of the use of redox mediators, especially in in situ bioremediation scenarios in environmental settings, where the massive release of these eco-unfriendly compounds may be undesirable. The solving of crystal structures of fungal detoxification enzymes enables molecular docking analyses to predict the ability to metabolize various xenobiotic substrates. Similarly, the application of enzymes also requires a fine understanding of soil structure and soil-water interactions as these can significantly affect enzyme activity. Recent advances in this area are likely to benefit soil modeling studies and assessment of enzyme function for future bioremediation projects. Adopting a bioprospecting-like approach to find new strains of fungi with superior ligninolytic enzymes in environments like rainforests could be a promising way forward in continued efforts to combat environmental pollutants of industrial origin in cost-effective and environmentally sustainable tactics.
References
Alcalde M, Bulter T, Zumarraga M, Garcia-Arellano H, Mencia M, Plou FJ, Ballesteros A (2005) Screening mutant libraries of fungal laccases in the presence of organic solvents. J Biomol Screen 10:624–631. https://doi.org/10.1177/1087057105277058
Alcalde M, Ferrer M, Plou FJ, Ballesteros A (2006) Environmental biocatalysis: from remediation with enzymes to novel green processes. Trends Biotechnol 24:281–287. https://doi.org/10.1016/j.tibtech.2006.04.002
Amitai G, Adani R, Sod-Moriah G, Rabinovitz I, Vincze A, Leader H, Chefetz B, Leibovitz-Persky L, Friesem D, Hadar Y (1998) Oxidative biodegradation of phosphorothiolates by fungal laccase. FEBS Lett 438:195–200. https://doi.org/10.1016/S0014-5793(98)01300-3
Ang EL, Zhao H, Obbard JP (2005) Recent advances in the bioremediation of persistent organic pollutants via biomolecular engineering. Enzym Microb Technol 37:487–496. https://doi.org/10.1016/j.enzmictec.2004.07.024
Antosova Z, Herkommerova K, Pichova I, Sychrova H (2018) Efficient secretion of three fungal laccases from Saccharomyces cerevisiae and their potential for decolorization of textile industry effluent-a comparative study. Biotechnol Prog 34:69–80. https://doi.org/10.1002/btpr.2559
Aracagok YD, Goker H, Cihangir N (2017) Biodegradation of micropollutant naproxen with a selected fungal strain and identification of metabolites. Z Naturforsch C 72:173–179. https://doi.org/10.1515/znc-2016-0162
Asgher M, Asad MJ, Bhatti HN, Legge RL (2007) Hyperactivation and thermostabilization of Phanerochaete chrysosporium lignin peroxidase by immobilization in xerogels. World J Microbiol Biotechnol 23:525–531. https://doi.org/10.1007/s11274-006-9255-9
Asgher M, Bhatti HN, Ashraf M, Legge RL (2008) Recent developments in biodegradation of industrial pollutants by white rot fungi and their enzyme system. Biodegradation 19:771–783. https://doi.org/10.1007/s10532-008-9185-3
Ayala M, Pickard MA, Vazquez-Duhalt R (2008) Fungal enzymes for environmental purposes, a molecular biology challenge. J Mol Microbiol Biotechnol 15:172–180. https://doi.org/10.1159/000121328
Balcazar-Lopez E, Mendez-Lorenzo LH, Batista-Garcia RA, Esquivel-Naranjo U, Ayala M, Kumar VV, Savary O, Cabana H, Herrera-Estrella A, Folch-Mallol JL (2016) Xenobiotic compounds degradation by heterologous expression of a Trametes Sanguineus Laccase in Trichoderma atroviride. PLoS One 11:e0147997. https://doi.org/10.1371/journal.pone.0147997
Bautista LF, Morales G, Sanz R (2015) Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by laccase from Trametes versicolor covalently immobilized on amino-functionalized SBA-15. Chemosphere 136:273–280. https://doi.org/10.1016/j.chemosphere.2015.05.071
Bilal M, Asgher M (2016) Enhanced catalytic potentiality of Ganoderma lucidum IBL-05 manganese peroxidase immobilized on sol-gel matrix. J Mol Catal B Enzym 128:82–93. https://doi.org/10.1016/j.molcatb.2016.03.013
Bilal M, Asgher M, Iqbal HM, Hu H, Zhang X (2017a) Bio-based degradation of emerging endocrine-disrupting and dye-based pollutants using cross-linked enzyme aggregates. Environ Sci Pollut Res Int 24:7035–7041. https://doi.org/10.1007/s11356-017-8369-y
Bilal M, Asgher M, Parra-Saldivar R, Hu H, Wang W, Zhang X, Iqbal HMN (2017b) Immobilized ligninolytic enzymes: an innovative and environmental responsive technology to tackle dye-based industrial pollutants – a review. Sci Total Environ 576:646–659. https://doi.org/10.1016/j.scitotenv.2016.10.137
Bogan BW, Schoenike B, Lamar RT, Cullen D (1996) Manganese peroxidase mRNA and enzyme activity levels during bioremediation of polycyclic aromatic hydrocarbon-contaminated soil with Phanerochaete chrysosporium. Appl Environ Microbiol 62:2381–2386. https://aem.asm.org/content/62/7/2381
Bollag JM (1992) Decontaminating soil with enzymes. Environ Sci Technol 26:1876–1881. https://doi.org/10.1021/es00034a002
Camarero S, Sarkar S, Ruiz-Duenas FJ, Martinez MJ, Martinez AT (1999) Description of a versatile peroxidase involved in the natural degradation of lignin that has both manganese peroxidase and lignin peroxidase substrate interaction sites. J Biol Chem 274:10324–10330. https://doi.org/10.1074/jbc.274.15.10324
Camarero S, Pardo I, Canas AI, Molina P, Record E, Martinez AT, Martinez MJ, Alcalde M (2012) Engineering platforms for directed evolution of Laccase from Pycnoporus cinnabarinus. Appl Environ Microbiol 78:1370–1384. https://doi.org/10.1128/aem.07530-11
Cameron MD, Timofeevski S, Aust SD (2000) Enzymology of Phanerochaete chrysosporium with respect to the degradation of recalcitrant compounds and xenobiotics. Appl Microbiol Biotechnol 54:751–758. https://doi.org/10.1007/s002530000459
Chen A, Zeng G, Chen G, Fan J, Zou Z, Li H, Hu X, Long F (2011) Simultaneous cadmium removal and 2,4-dichlorophenol degradation from aqueous solutions by Phanerochaete chrysosporium. Appl Microbiol Biotechnol 91:811–821. https://doi.org/10.1007/s00253-011-3313-4
Cheng X-B, Jia R, Li P-S, Zhu Q, Tu S-Q, Tang W-Z (2007) Studies on the properties and Co-immobilization of Manganese Peroxidase. Chin J Biotechnol 23:90–96. https://doi.org/10.1016/S1872-2075(07)60006-5
Chmelova D, Ondrejovic M (2016) Purification and characterization of extracellular laccase produced by Ceriporiopsis subvermispora and decolorization of triphenylmethane dyes. J Basic Microbiol 56:1173–1182. https://doi.org/10.1002/jobm.201600152
Christian V, Shrivastava R, Shukla D, Modi H, Vyas BRM (2005a) Mediator role of veratryl alcohol in the lignin peroxidase-catalyzed oxidative decolorization of Remazol Brilliant Blue R. Enzym Microb Technol 36:327–332. https://doi.org/10.1016/j.enzmictec.2004.09.006
Christian V, Shrivastava R, Shukla D, Modi HA, Vyas BR (2005b) Degradation of xenobiotic compounds by lignin-degrading white-rot fungi: enzymology and mechanisms involved. Indian J Exp Biol 43:301–312. http://nopr.niscair.res.in/bitstream/123456789/23105/1/IJEB%2043%284%29%20301-312.pdf
Cresnar B, Petric S (2011) Cytochrome P450 enzymes in the fungal kingdom. Biochim Biophys Acta 1814:29–35. https://doi.org/10.1016/j.bbapap.2010.06.020
Davila-Vazquez G, Tinoco R, Pickard MA, Vazquez-Duhalt R (2005) Transformation of halogenated pesticides by versatile peroxidase from Bjerkandera adusta. Enzym Microb Technol 36:223–231. https://doi.org/10.1016/j.enzmictec.2004.07.015
Deshmukh R, Khardenavis AA, Purohit HJ (2016) Diverse metabolic capacities of fungi for bioremediation. Indian J Microbiol 56:247–264. https://doi.org/10.1007/s12088-016-0584-6
Diano N, Grano V, Fraconte L, Caputo P, Ricupito A, Attanasio A, Bianco M, Bencivenga U, Rossi S, Manco I, Mita L, Del Pozzo G, Mita DG (2007) Non-isothermal bioreactors in enzymatic remediation of waters polluted by endocrine disruptors: BPA as a model of pollutant. Appl Catal B Environ 69:252–261. https://doi.org/10.1016/j.apcatb.2006.07.004
Doddapaneni H, Chakraborty R, Yadav JS (2005) Genome-wide structural and evolutionary analysis of the P450 monooxygenase genes (P450ome) in the white rot fungus Phanerochaete chrysosporium: evidence for gene duplications and extensive gene clustering. BMC Genomics 6:92. https://doi.org/10.1186/1471-2164-6-92
Dodor DE, Hwang H-M, Ekunwe SIN (2004) Oxidation of anthracene and benzo[a]pyrene by immobilized laccase from Trametes versicolor. Enzym Microb Technol 35:210–217. https://doi.org/10.1016/j.enzmictec.2004.04.007
D’Souza DT, Tiwari R, Sah AK, Raghukumar C (2006) Enhanced production of laccase by a marine fungus during treatment of colored effluents and synthetic dyes. Enzym Microb Technol 38:504–511. https://doi.org/10.1016/j.enzmictec.2005.07.005
Dua M, Singh A, Sethunathan N, Johri A (2002) Biotechnology and bioremediation: successes and limitations. Appl Microbiol Biotechnol 59:143–152. https://doi.org/10.1007/s00253-002-1024-6
Falade AO, Nwodo UU, Iweriebor BC, Green E, Mabinya LV, Okoh AI (2017) Lignin peroxidase functionalities and prospective applications. Microbiol Open 6. https://doi.org/10.1002/mbo3.394
Gao L, Yan X (2016) Nanozymes: an emerging field bridging nanotechnology and biology. Sci China Life Sci 59:400–402. https://doi.org/10.1007/s11427-016-5044-3
Gitsov I, Hamzik J, Ryan J, Simonyan A, Nakas JP, Omori S, Krastanov A, Cohen T, Tanenbaum SW (2008) Enzymatic nanoreactors for environmentally benign biotransformations. 1. Formation and catalytic activity of supramolecular complexes of laccase and linear−dendritic block copolymers. Biomacromolecules 9:804–811. https://doi.org/10.1021/bm701081m
Godoy P, Reina R, Calderon A, Wittich RM, Garcia-Romera I, Aranda E (2016) Exploring the potential of fungi isolated from PAH-polluted soil as a source of xenobiotics-degrading fungi. Environ Sci Pollut Res Int 23:20985–20996. https://doi.org/10.1007/s11356-016-7257-1
Gu C, Zheng F, Long L, Wang J, Ding S (2014) Engineering the expression and characterization of two novel laccase isoenzymes from Coprinus comatus in Pichia pastoris by fusing an additional ten amino acids tag at N-terminus. PLoS One 9:e93912. https://doi.org/10.1371/journal.pone.0093912
Guengerich FP (2001a) Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol 14:611–650. https://doi.org/10.1021/tx0002583
Guengerich FP (2001b) Uncommon P450-catalyzed reactions. Curr Drug Metab 2:93–115. https://doi.org/10.2174/1389200013338694
Han Y, Shi L, Meng J, Yu H, Zhang X (2014) Azo dye biodecolorization enhanced by Echinodontium taxodii cultured with lignin. PLoS One 9:e109786. https://doi.org/10.1371/journal.pone.0109786
Harms H, Schlosser D, Wick LY (2011) Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat Rev Microbiol 9:177. https://doi.org/10.1038/nrmicro2519
Haroune L, Saibi S, Cabana H, Bellenger JP (2017) Intracellular enzymes contribution to the biocatalytic removal of pharmaceuticals by trametes hirsuta. Environ Sci Technol 51:897–904. https://doi.org/10.1021/acs.est.6b04409
Hirosue S, Tazaki M, Hiratsuka N, Yanai S, Kabumoto H, Shinkyo R, Arisawa A, Sakaki T, Tsunekawa H, Johdo O, Ichinose H, Wariishi H (2011) Insight into functional diversity of cytochrome P450 in the white-rot basidiomycete Phanerochaete chrysosporium: involvement of versatile monooxygenase. Biochem Biophys Res Commun 407:118–123. https://doi.org/10.1016/j.bbrc.2011.02.121
Jing R, Kjellerup BV (2018) Biogeochemical cycling of metals impacting by microbial mobilization and immobilization. J Environ Sci 66:146–154. https://doi.org/10.1016/j.jes.2017.04.035
Jones SM, Solomon EI (2015) Electron transfer and reaction mechanism of laccases. Cell Mol Life Sci 72:869–883. https://doi.org/10.1007/s00018-014-1826-6
Junghanns C, Moeder M, Krauss G, Martin C, Schlosser D (2005) Degradation of the xenoestrogen nonylphenol by aquatic fungi and their laccases. Microbiology 151:45–57. https://doi.org/10.1099/mic.0.27431-0
Kadri T, Rouissi T, Kaur Brar S, Cledon M, Sarma S, Verma M (2017) Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by fungal enzymes: a review. J Environ Sci 51:52–74. https://doi.org/10.1016/j.jes.2016.08.023
Karich A, Ullrich R, Scheibner K, Hofrichter M (2017) Fungal unspecific peroxygenases oxidize the majority of organic EPA priority pollutants. Front Microbiol 8:1463. https://doi.org/10.3389/fmicb.2017.01463
Keum YS, Li QX (2004) Fungal laccase-catalyzed degradation of hydroxy polychlorinated biphenyls. Chemosphere 56:23–30. https://doi.org/10.1016/j.chemosphere.2004.02.028
Kour D, Rana KL, Yadav N, Yadav AN, Singh J, Rastegari AA, Saxena AK (2019) Agriculturally and industrially important fungi: current developments and potential biotechnological applications. In: Yadav AN, Singh S, Mishra S, Gupta A (eds) Recent advancement in white biotechnology through fungi, Volume 2: perspective for value-added products and environments. Springer International Publishing, Cham, pp 1–64. https://doi.org/10.1007/978-3-030-14846-1_1
Koyani RD, Vazquez-Duhalt R (2016) Laccase encapsulation in chitosan nanoparticles enhances the protein stability against microbial degradation. Environ Sci Pollut Res Int 23:18850–18857. https://doi.org/10.1007/s11356-016-7072-8
Kues U (2015) Fungal enzymes for environmental management. Curr Opin Biotechnol 33:268–278. https://doi.org/10.1016/j.copbio.2015.03.006
Laurichesse S, Avérous L (2014) Chemical modification of lignins: towards biobased polymers. Prog Polym Sci 39:1266–1290. https://doi.org/10.1016/j.progpolymsci.2013.11.004
Lee AH, Kang CM, Lee YM, Lee H, Yun CW, Kim GH, Kim JJ (2016) Heterologous expression of a new manganese-dependent peroxidase gene from Peniophora incarnata KUC8836 and its ability to remove anthracene in Saccharomyces cerevisiae. J Biosci Bioeng 122:716–721. https://doi.org/10.1016/j.jbiosc.2016.06.006
Levin L, Forchiassin F, Viale A (2005) Ligninolytic enzyme production and dye decolorization by Trametes trogii: application of the Plackett–Burman experimental design to evaluate nutritional requirements. Process Biochem 40:1381–1387. https://doi.org/10.1016/j.procbio.2004.06.005
Liang H, Lin F, Zhang Z, Liu B, Jiang S, Yuan Q, Liu J (2017) Multicopper laccase mimicking nanozymes with nucleotides as ligands. ACS Appl Mater Interfaces 9:1352–1360. https://doi.org/10.1021/acsami.6b15124
Lima RN, Porto AL (2016) Recent advances in marine enzymes for biotechnological processes. Adv Food Nutr Res 78:153–192. https://doi.org/10.1016/bs.afnr.2016.06.005
Loi M, Fanelli F, Zucca P, Liuzzi VC, Quintieri L, Cimmarusti MT, Monaci L, Haidukowski M, Logrieco AF, Sanjust E, Mule G (2016) Aflatoxin B(1) and M(1) degradation by Lac2 from Pleurotus pulmonarius and Redox Mediators. Toxins 8. https://doi.org/10.3390/toxins8090245
Lucero Camacho-Morales R, Garcia-Fontana C, Fernandez-Irigoyen J, Santamaria E, Gonzalez-Lopez J, Manzanera M, Aranda E (2018) Anthracene drives sub-cellular proteome-wide alterations in the degradative system of Penicillium oxalicum. Ecotoxicol Environ Saf 159:127–135. https://doi.org/10.1016/j.ecoenv.2018.04.051
Ma X, Zhang L, Xia M, Li S, Zhang X, Zhang Y (2017) Mimicking the active sites of organophosphorus hydrolase on the backbone of graphene oxide to destroy nerve agent simulants. ACS Appl Mater Interfaces 9:21089–21093. https://doi.org/10.1021/acsami.7b07770
Macellaro G, Pezzella C (2014) Fungal laccases degradation of endocrine disrupting compounds. Biomed Res Int 2014:614038. https://doi.org/10.1155/2014/614038
Margot J, Bennati-Granier C, Maillard J, Blanquez P, Barry DA, Holliger C (2013) Bacterial versus fungal laccase: potential for micropollutant degradation. AMB Express 3:63. https://doi.org/10.1186/2191-0855-3-63
Mate DM, Alcalde M (2015) Laccase engineering: from rational design to directed evolution. Biotechnol Adv 33:25–40. https://doi.org/10.1016/j.biotechadv.2014.12.007
Miele A, Giardina P, Sannia G, Faraco V (2010) Random mutants of a Pleurotus ostreatus laccase as new biocatalysts for industrial effluents bioremediation. J Appl Microbiol 108:998–1006. https://doi.org/10.1111/j.1365-2672.2009.04505.x
Minussi RC, Pastore GM, Duran N (2007) Laccase induction in fungi and laccase/N-OH mediator systems applied in paper mill effluent. Bioresour Technol 98:158–164. https://doi.org/10.1016/j.biortech.2005.11.008
Morel M, Meux E, Mathieu Y, Thuillier A, Chibani K, Harvengt L, Jacquot JP, Gelhaye E (2013) Xenomic networks variability and adaptation traits in wood decaying fungi. Microb Biotechnol 6:248–263. https://doi.org/10.1111/1751-7915.12015
Mougin C, Boukcim H, Jolivalt C (2009) Soil bioremediation strategies based on the use of fungal enzymes. In: Singh A, Kuhad RC, Ward OP (eds) Advances in applied bioremediation. Springer, Berlin/Heidelberg, pp 123–149. https://doi.org/10.1007/978-3-540-89621-0_7
Naranjo-Briceno L, Pernia B, Guerra M, Demey JR, De Sisto A, Inojosa Y, Gonzalez M, Fusella E, Freites M, Yegres F (2013) Potential role of oxidative exoenzymes of the extremophilic fungus Pestalotiopsis palmarum BM-04 in biotransformation of extra-heavy crude oil. Microb Biotechnol 6:720–730. https://doi.org/10.1111/1751-7915.12067
Nicell JA, Wright H (1997) A model of peroxidase activity with inhibition by hydrogen peroxide. Enzym Microb Technol 21:302–310. https://doi.org/10.1016/S0141-0229(97)00001-X
Olicon-Hernandez DR, Gonzalez-Lopez J, Aranda E (2017) Overview on the biochemical potential of filamentous fungi to degrade pharmaceutical compounds. Front Microbiol 8:1792. https://doi.org/10.3389/fmicb.2017.01792
Piontek K, Smith AT, Blodig W (2001) Lignin peroxidase structure and function. Biochem Soc Trans 29:111–116. https://doi.org/10.1042/bst0290111
Pointing SB (2001) Feasibility of bioremediation by white-rot fungi. Appl Microbiol Biotechnol 57:20–33. https://doi.org/10.1007/s002530100745
Pozdnyakova NN (2012) Involvement of the ligninolytic system of white-rot and litter-decomposing fungi in the degradation of polycyclic aromatic hydrocarbons. Biotechnol Res Int 2012:243217. https://doi.org/10.1155/2012/243217
Pozdnyakova N, Dubrovskaya E, Chernyshova M, Makarov O, Golubev S, Balandina S, Turkovskaya O (2018a) The degradation of three-ringed polycyclic aromatic hydrocarbons by wood-inhabiting fungus Pleurotus ostreatus and soil-inhabiting fungus Agaricus bisporus. Fungal Biol 122:363–372. https://doi.org/10.1016/j.funbio.2018.02.007
Pozdnyakova NN, Balandina SA, Dubrovskaya EV, Golubev CN, Turkovskaya OV (2018b) Ligninolytic basidiomycetes as promising organisms for the mycoremediation of PAH-contaminated environments. IOP Conf Ser Earth Environ Sci 107:012071. https://doi.org/10.1088/1755-1315/107/1/012071
Qin X, Zhang J, Zhang X, Yang Y (2014) Induction, purification and characterization of a novel manganese peroxidase from Irpex lacteus CD2 and its application in the decolorization of different types of dye. PLoS One 9:e113282. https://doi.org/10.1371/journal.pone.0113282
Rana KL, Kour D, Sheikh I, Dhiman A, Yadav N, Yadav AN, Rastegari AA, Singh K, Saxena AK (2019a) Endophytic fungi: biodiversity, ecological significance, and potential industrial applications. In: Yadav AN, Mishra S, Singh S, Gupta A (eds) Recent advancement in white biotechnology through fungi: Volume 1: diversity and enzymes perspectives. Springer International Publishing, Cham, pp 1–62. https://doi.org/10.1007/978-3-030-10480-1_1
Rana KL, Kour D, Sheikh I, Yadav N, Yadav AN, Kumar V, Singh BP, Dhaliwal HS, Saxena AK (2019b) Biodiversity of endophytic fungi from diverse niches and their biotechnological applications. In: Singh BP (ed) Advances in endophytic fungal research: present status and future challenges. Springer International Publishing, Cham, pp 105–144. https://doi.org/10.1007/978-3-030-03589-1_6
Rao MA, Scelza R, Scotti R, Gianfreda L (2010) Role of enzymes in the remediation of polluted environments. J Soil Sci Plant Nutr 10:333–353. https://doi.org/10.4067/S0718-95162010000100008
Rastegari AA, Yadav AN, Gupta A (2019) Prospects of renewable bioprocessing in future energy systems. Springer International Publishing, Cham
Rayu S, Karpouzas DG, Singh BK (2012) Emerging technologies in bioremediation: constraints and opportunities. Biodegradation 23:917–926. https://doi.org/10.1007/s10532-012-9576-3
Salony, Mishra S, Bisaria VS (2006) Production and characterization of laccase from Cyathus bulleri and its use in decolourization of recalcitrant textile dyes. Appl Microbiol Biotechnol 71:646–653. https://doi.org/10.1007/s00253-005-0206-4
Schlosser D, Hofer C (2002) Laccase-catalyzed oxidation of Mn(2+) in the presence of natural Mn(3+) chelators as a novel source of extracellular H(2)O(2) production and its impact on manganese peroxidase. Appl Environ Microbiol 68:3514–3521. https://doi.org/10.1128/AEM.68.7.3514-3521.2002
Sharma B, Dangi AK, Shukla P (2018) Contemporary enzyme based technologies for bioremediation: a review. J Environ Manag 210:10–22. https://doi.org/10.1016/j.jenvman.2017.12.075
Shedbalkar U, Dhanve R, Jadhav J (2008) Biodegradation of triphenylmethane dye cotton blue by Penicillium ochrochloron MTCC 517. J Hazard Mater 157:472–479. https://doi.org/10.1016/j.jhazmat.2008.01.023
Shrivastava R, Christian V, Vyas BRM (2005) Enzymatic decolorization of sulfonphthalein dyes. Enzym Microb Technol 36:333–337. https://doi.org/10.1016/j.enzmictec.2004.09.004
Sirisha VL, Jain A, Jain A (2016) Chapter nine - Enzyme immobilization: an overview on methods, support material, and applications of immobilized enzymes. In: Kim S-K, Toldrá F (eds) Advances in food and nutrition research, vol 79. Academic Press, pp 179–211. https://doi.org/10.1016/bs.afnr.2016.07.004
Su J, Fu JJ, Wang Q, Silva C, Cavaco-Paulo A (2018) Laccase: a green catalyst for the biosynthesis of poly-phenols. Crit Rev Biotechnol 38:294–307. https://doi.org/10.1080/07388551.2017.1354353
Suresh PS, Kumar A, Kumar R, Singh VP (2008) An in silico [correction of insilico] approach to bioremediation: laccase as a case study. J Mol Graph Model 26:845–849. https://doi.org/10.1016/j.jmgm.2007.05.005
Syed K, Yadav JS (2012) P450 monooxygenases (P450ome) of the model white rot fungus Phanerochaete chrysosporium. Crit Rev Microbiol 38:339–363. https://doi.org/10.3109/1040841X.2012.682050
Syed K, Porollo A, Lam YW, Yadav JS (2011) A fungal P450 (CYP5136A3) capable of oxidizing polycyclic aromatic hydrocarbons and endocrine disrupting alkylphenols: role of Trp(129) and Leu(324). PLoS One 6:e28286. https://doi.org/10.1371/journal.pone.0028286
Theerachat M, Emond S, Cambon E, Bordes F, Marty A, Nicaud JM, Chulalaksananukul W, Guieysse D, Remaud-Simeon M, Morel S (2012) Engineering and production of laccase from Trametes versicolor in the yeast Yarrowia lipolytica. Bioresour Technol 125:267–274. https://doi.org/10.1016/j.biortech.2012.07.117
Tongpim S, Pickard MA (1999) Cometabolic oxidation of phenanthrene to phenanthrene trans-9,10-dihydrodiol by Mycobacterium strain S1 growing on anthracene in the presence of phenanthrene. Can J Microbiol 45:369–376. https://doi.org/10.1139/cjm-45-5-369
Torres-Duarte C, Roman R, Tinoco R, Vazquez-Duhalt R (2009) Halogenated pesticide transformation by a laccase–mediator system. Chemosphere 77:687–692. https://doi.org/10.1016/j.chemosphere.2009.07.039
Torres-Farrada G, Manzano Leon AM, Rineau F, Ledo Alonso LL, Sanchez-Lopez MI, Thijs S, Colpaert J, Ramos-Leal M, Guerra G, Vangronsveld J (2017) Diversity of ligninolytic enzymes and their genes in strains of the Genus Ganoderma: applicable for biodegradation of xenobiotic compounds? Front Microbiol 8:898. https://doi.org/10.3389/fmicb.2017.00898
Tsutsumi Y, Haneda T, Nishida T (2001) Removal of estrogenic activities of bisphenol A and nonylphenol by oxidative enzymes from lignin-degrading basidiomycetes. Chemosphere 42:271–276. https://doi.org/10.1016/S0045-6535(00)00081-3
Valderrama B, Ayala M, Vazquez-Duhalt R (2002) Suicide inactivation of peroxidases and the challenge of engineering more robust enzymes. Chem Biol 9:555–565 . Pii S1074-5521(02)00149-7. https://doi.org/10.1016/S1074-5521(02)00149-7
Van den Brink HM, van Gorcom RF, van den Hondel CA, Punt PJ (1998) Cytochrome P450 enzyme systems in fungi. Fungal Genet Biol 23:1–17. https://doi.org/10.1006/fgbi.1997.1021
Verma P, Madamwar D (2002) Production of ligninolytic enzymes for dye decolorization by cocultivation of white-rot fungi Pleurotus ostreatus and phanerochaete chrysosporium under solid-state fermentation. Appl Biochem Biotechnol 102-103:109–118. https://doi.org/10.1385/ABAB:102-103:1-6:109
Viswanath B, Rajesh B, Janardhan A, Kumar AP, Narasimha G (2014) Fungal laccases and their applications in bioremediation. Enzyme Res 2014:163242. https://doi.org/10.1155/2014/163242
Wang M, Abad D, Kickhoefer VA, Rome LH, Mahendra S (2015) Vault nanoparticles packaged with enzymes as an efficient pollutant biodegradation technology. ACS Nano 9:10931–10940. https://doi.org/10.1021/acsnano.5b04073
Watanabe K (2001) Microorganisms relevant to bioremediation. Curr Opin Biotechnol 12:237–241. https://doi.org/10.1016/S0958-1669(00)00205-6
Wong KS, Cheung MK, Au CH, Kwan HS (2013a) A novel Lentinula edodes laccase and its comparative enzymology suggests guaiacol-based laccase engineering for bioremediation. PLoS One 8:1–12. https://doi.org/10.1371/journal.pone.0066426.t001
Wong KS, Cheung MK, Au CH, Kwan HS (2013b) A novel Lentinula edodes laccase and its comparative enzymology suggest guaiacol-based laccase engineering for bioremediation. PLoS One 8:e66426. https://doi.org/10.1371/journal.pone.0066426
Wu Y, Teng Y, Li Z, Liao X, Luo Y (2008) Potential role of polycyclic aromatic hydrocarbons (PAHs) oxidation by fungal laccase in the remediation of an aged contaminated soil. Soil Biol Biochem 40:789–796. https://doi.org/10.1016/j.soilbio.2007.10.013
Wu J, Xiao D, Zhao H, He H, Peng J, Wang C, Zhang C, He J (2015) A nanocomposite consisting of graphene oxide and Fe3O4 magnetic nanoparticles for the extraction of flavonoids from tea, wine and urine samples. Mikrochim Acta 182:2299–2306. https://doi.org/10.1007/s00604-015-1575-8
Xu F (1996) Oxidation of phenols, anilines, and benzenethiols by fungal laccases: correlation between activity and redox potentials as well as halide inhibition. Biochemistry 35:7608–7614. https://doi.org/10.1021/bi952971a
Yadav AN, Sachan SG, Verma P, Kaushik R, Saxena AK (2016) Cold active hydrolytic enzymes production by psychrotrophic Bacilli isolated from three sub-glacial lakes of NW Indian Himalayas. J Basic Microbiol 56:294–307
Yadav A, Verma P, Kumar R, Kumar V, Kumar K (2017) Current applications and future prospects of eco-friendly microbes. EU Voice 3:21–22
Yadav AN, Verma P, Kumar V, Sangwan P, Mishra S, Panjiar N, Gupta VK, Saxena AK (2018) Biodiversity of the genus Penicillium in different habitats. In: Gupta VK, Rodriguez-Couto S (eds) New and future developments in microbial biotechnology and bioengineering, Penicillium system properties and applications. Elsevier, Amsterdam, pp 3–18. https://doi.org/10.1016/B978-0-444-63501-3.00001-6
Yadav AN, Mishra S, Singh S, Gupta A (2019a) Recent advancement in white biotechnology through fungi Volume 1: diversity and enzymes perspectives. Springer International Publishing, Cham
Yadav AN, Mishra S, Singh S, Gupta A (2019b) Recent advancement in white biotechnology through fungi. Volume 2: perspective for value-added products and environments. Springer International Publishing, Cham
Yang J, Li W, Ng TB, Deng X, Lin J, Ye X (2017) Laccases: production, expression regulation, and applications in pharmaceutical biodegradation. Front Microbiol 6. https://doi.org/10.3389/fmicb.2017.00832
Zhang C, Li M, Chen X, Li M (2015) Edible fungus degrade bisphenol A with no harmful effect on its fatty acid composition. Ecotoxicol Environ Saf 118:126–132. https://doi.org/10.1016/j.ecoenv.2015.04.020
Zhang H, Zhang S, He F, Qin X, Zhang X, Yang Y (2016) Characterization of a manganese peroxidase from white-rot fungus Trametes sp.48424 with strong ability of degrading different types of dyes and polycyclic aromatic hydrocarbons. J Hazard Mater 320:265–277. https://doi.org/10.1016/j.jhazmat.2016.07.065
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Baker, P., Tiroumalechetty, A., Mohan, R. (2019). Fungal Enzymes for Bioremediation of Xenobiotic Compounds. In: Yadav, A., Singh, S., Mishra, S., Gupta, A. (eds) Recent Advancement in White Biotechnology Through Fungi. Fungal Biology. Springer, Cham. https://doi.org/10.1007/978-3-030-25506-0_19
Download citation
DOI: https://doi.org/10.1007/978-3-030-25506-0_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-25505-3
Online ISBN: 978-3-030-25506-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)